Abstract
Jaspamide, a naturally occurring cyclic peptide isolated from the marine sponge Hemiastrella minor, has fungicidal and growth-inhibiting activities. Exposure of promyelocytic HL-60 cells and human monocytes to jaspamide induces a dramatic reorganization of actin from a typical fibrous network to focal aggregates. HL-60 cells exposed to 5 × 10−8 mol/L or 10−7 mol/L jaspamide exhibited a reduced proliferation rate. In addition, 10−7mol/L jaspamide induced maturation of HL-60 cells as indicated by the appearance of a lobulated nucleus in 55% ± 5% of the cells and immunophenotypic maturation of the leukemia cells (upregulation of CD16 and CD14 B antigens). Further characterization has shown that F-actin is aggregated both in HL-60 cells and in human monocytes exposed to 10−7 mol/L jaspamide. Well-spread cultured human monocytes contracted and adopted round shapes after treatment with jaspamide. Moreover, a dose-dependent increase in both total actin and de novo synthesized portions of the soluble actin was observed in jaspamide-treated HL-60 cells. Jaspamide treatment inhibits ruffling and intracellular movement in HL-60 cells and monocytes, but does not affect phagocytic activity or respiratory burst activity. The consequential effects of jaspamide-induced actin reorganization on ruffling, versus its negligible effect on phagocytosis and oxidative burst, may shed light on molecular mechanisms of actin involvement in these processes. Jaspamide disrupts the actin cytoskeleton of normal and malignant mammalian cells with no significant effect on phagocytic activity and may, therefore, be considered as a novel therapeutic agent.
ACTIN IS A ubiquitous eukaryotic cytoskeletal protein, critical for many aspects of cell activity. In addition to maintaining cell morphology, it is required for cell motility, cell division, and intracellular transport.1Actin reorganization is rapidly induced by many extracellular factors and/or by adhesion to the extracellular matrix.1-3 Cellular actin rapidly alternates between two forms: monomeric G-actin (globular) and polymeric F-actin (fibrous). The dynamics of G-: F-actin transition may be critical to many of its specialized cellular functions including regulation of cell shape, motility, secretion, intracellular transport, endocytosis, exocytosis, and cell division.2 3
HL-60 human promyelocytic leukemia cells are a well-established model for studying the effect of different physiologic and pharmacologic agents on cell growth and maturation.4-6 Changes in F-actin organization and actin-binding proteins have been observed during the differentiation of HL-60 cells along the granulocytic pathway.7 An increase in levels of actin and actin binding proteins has also been observed in both murine8 and human9 differentiated myeloid leukemia cells.
In a recent study we have shown that jaspamide, a peptide isolated from the marine sponge Hemiastrella minor, induces growth modulation and differentiation in acute myeloid leukemia (AML) cells.10 We found jaspamide, similarly to cytosine arabinoside, to be effective in suppressing leukemic blast colony formation and inducing immunophenotypic maturation of three leukemic cell lines (including HL-60 cells) and blasts of AML patients.10
In the present study we examine the effects of jaspamide on cellular growth and functional activity of HL-60 cells and human monocytes. We show that jaspamide induces cellular maturation accompanied by a dramatic reorganization of actin in both HL-60 cells and human monocytes. Jaspamide-induced actin reorganization (aggregation) inhibits intrinsic cell movement and ruffling but does not affect phagocytosis or oxidative burst, biological activities that are considered actin dependent.
MATERIALS AND METHODS
Cell Culture, Antibodies, and Treatments
HL-60 human promyelocytic leukemia cells were grown in 10% fetal bovine serum (FBS) (Hyclone Laboratories, Logan, UT) with Iscove’s modified Dulbecco’s medium (IMDM) (GIBCO, Grand Island, NY).10 Stock cell cultures were passaged twice weekly and maintained at 37°C in a humidified atmosphere containing 5% CO2. Cells (5 × 105/mL) were incubated with different concentrations (10−8 to 10−6 mol/L) of jaspamide (Molecular Probes, Eugene, OR) for 1 to 48 hours. A stock solution of 10−3 mol/L jaspamide was prepared in dimethyl sulfoxide (DMSO) and stored at −20°C. The drug was diluted before use with IMDM to the appropriate concentrations. Experiments of cytochalasin treatment were performed with cytochalasin D (3.94 μmol/L) (Sigma Chemical Co, St Louis, MO) added to the cell cultures for 24 hours. The following antibodies were used: CD16 (code no. 306) and CD14 B (code no. 253) (a generous gift from Dr E. Gazit, Tel Aviv, Israel). These monoclonal antibodies (MoAbs) were studied at the International Workshop on Human Leukocyte Differentiation Antigens (Leukocyte typing III) in Oxford, UK, September 21-26, 1986. Mouse anti-actin MoAb was purchased from Boehringer Mannheim (Mannheim, Germany). Rabbit anti-actin Ab was obtained from Sigma. Rhodamine-conjugated donkey anti-mouse Ab and fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse IgG were purchased from Jackson Immunoresearch Laboratories (West Grove, PA). Anti-mouse IgG-HRP was purchased from Amersham (Buckinghamshire, UK).
Human Blood Monocytes
Heparinized blood, obtained with informed consent from healthy donors, was layered on Ficoll-Hypaque (Pharmacia Fine Chemicals, Uppsala, Sweden), centrifuged at 400g for 30 minutes at room temperature; subsequently, the mononuclear cell layer was collected. The cells were resuspended in IMDM containing 10% FBS and plated at a concentration of 5 × 106/mL in 75-cm2flasks. After a 24-hour incubation at 37°C, the cells were washed with phosphate-buffered saline (PBS) to remove nonadherent cells and the adherent cells were detached by vigorous washing with cold Ca2+Mg2+-free PBS, and resuspended in IMDM containing 10% FBS as previously described.11 Monocyte purity was greater than 85% as determined by morphology (Wright-Giemsa staining) of cytocentrifugated specimens. The cells were plated overnight at 106 cells/mL in flat-based tissue culture tubes (Nunc, Roskilde, Denmark) containing coverslips, for F-actin staining and phagocytosis studies. After incubation, 10−7 mol/L jaspamide or 0.01% DMSO was added to the monocyte cultures, and treated cultures were incubated for an additional 24 hours.
For NADPH-dependent O2−production studies, monocytes were plated in 96 flat-bottom well tissue-culture plates at a concentration of 2.5 × 105cells/100 μL/well and incubated at 37°C for 3 hours. After incubation, 10−7 mol/L jaspamide, 0.01% DMSO, or medium was added to appropriate wells and plates were incubated for an additional 24 hours.
Cell Growth and Viability
104 HL-60 cells were aliquoted to 96-well plates in medium with 10−8 to 10−7 mol/L jaspamide at a final volume of 200 μL per well. Cells grown in the presence of 0.01% DMSO or medium were used as controls. After 24 or 48 hours, cultured cells were stained by trypan blue and viable cells were quantified by phase-contrast microscopy.
3H-thymidine incorporation experiments were carried out as previously described.6 In brief, 104 HL-60 cells were cultured in 96-well plates in medium with 10−8 to 10−7 mol/L jaspamide at a final volume of 200 μL per well. Cells grown in the presence of 0.01% DMSO or medium were used as controls. Cultures were pulsed on days 1 and 2 with 1 μCi/mL 3H thymidine (1 mCi/mol/L; Nuclear Research Center, Negev, Israel) and harvested 6 hours later onto glass filters with an automated cell harvester under hypotonic conditions. Filter discs were dried and counted in a Packard Tri-Carb liquid scintillation counter (Downers Grove, IL).
Cell Differentiation
HL-60 cells were grown in liquid culture for 48 hours in the presence of 10−7 mol/L jaspamide or 0.01% DMSO, and cell differentiation was evaluated using cytospin (Shandon, Runcorn, UK) slide preparations stained with Wright-Giemsa. Differential counts were performed on at least 200 cells using a light microscope at 1,000× magnification.
Fluorescence-Activated Cell Sorter (FACS) Analysis of Cell-Surface Antigens
Expression of the myeloid cell-surface antigens CD16 and CD14 B was determined by FACS analysis of indirect immunofluorescence using MoAbs. HL-60 cells grown in the presence of 10−7 mol/L jaspamide or 0.01% DMSO for 48 hours were incubated with the CD16 or CD14 B MoAb, and subsequently labeled with FITC-conjugated goat anti-mouse IgG (Bio-Yead, Rehovot, Israel) as previously described.10 Fluorescent cells were detected by flow cytometer (Becton Dickinson, San Jose, CA).
Actin Staining and Confocal Laser Scanning Microscopy Analyses
Total actin staining was performed on cytospin preparations of cells fixed with ice-cold 100% acetone and 100% methanol (10 minutes each). After washing, cells were blocked (1% normal donkey serum and 0.1% bovine serum albumin [BSA] in PBS) for 1 hour; incubated with mouse anti-actin MoAb (10 μg/mL) for 1 hour and subsequently labeled with rhodamine-conjugated donkey anti-mouse-antibodies (diluted 1:100) for 1 hour. All steps were performed at room temperature. F-actin was detected using rhodamine phalloidin (Rh-phalloidin) (Molecular Probes) as previously described12 with slight modifications. After incubation with the drug, cytospin preparations of HL-60 cells and monocytes grown on coverslips in flat-based tissue-culture tubes were fixed, permeabilized, and stained in a one-step procedure, as suggested by the manufacturer (Molecular Probes; simultaneous fixation protocol). In brief, treated and untreated cells were incubated for 20 minutes with 3.7% formaldehyde containing 100 μg/mL lysophosphatidyl choline (Sigma) and 0.3 μmol/L Rh-phalloidin and then washed. Slides and coverslips were mounted using Gel Mount (Biomeda, Foster City, CA). Staining analyses was performed using a Zeiss (Oberkochen, Germany) confocal laser scanning microscope (CLSM). The Zeiss LSM 410 is equipped with a 25-mW Krypton-Argon laser and a 10-mW HeNe laser (488, 543, and 633 maximum lines). Images were stored on an optical disk drive and printed using a Codonics NP1600 printer (Codonics, Middleburg Heights, OH). Competition experiments of jaspamide and phalloidin were performed with HL-60 cells by exposure of untreated cells to Rh-phalloidin in the presence of 10−7 mol/L or 10−8 mol/L jaspamide for 30 minutes. In other experiments, cells were incubated for 24 hours with 10−7 mol/L jaspamide, subsequently washed, and double staining for F-actin and total actin was performed using Rh-phalloidin (Molecular Probes; simultaneous fixation protocol) and anti-actin MoAb followed by FITC-conjugated donkey anti-mouse antibodies (diluted 1:50). CLSM analysis was performed as described above.
Analysis of Actin
Sample preparation for total actin analysis.
HL-60 cells were exposed to 10−7 mol/L jaspamide, 0.01% DMSO, or medium for 24 hours. Cells were then washed twice with PBS and lysed (20 mmol/L Tris-HCl, pH 7.8, 100 mmol/L NaCl, 50 mmol/L NaF, 1% NP40, 0.1% sodium dodecyl sulfate (SDS), 2 mmol/L EDTA, 10% glycerol) with protease inhibitor cocktail (Boehringer Mannheim). Protein concentrations were determined using Pierce BCA assay (Pierce Chemical Co, Rockford, IL). Cell lysates were solubilized in X2 loading buffer (20% glycerol, 0.2% bromophenol blue, 4% SDS, 100 mmol/L Tris-Cl and 2% β-mercaptoethanol) and boiled at 100°C for 5 minutes.
Sample preparation for soluble, insoluble, and total actin analysis.
Cellular soluble, insoluble, and total fractions of actin were prepared as described by Hallows et al with slight modifications.13In short, an equal number of HL-60 cells were placed in three tubes (A, B, and C); 100 μL of lysis buffer was added to each tube. The lysed cells were centrifuged 15,000g for 5 minutes. To obtain soluble actin, 100 μL of the supernatant from tube A was transferred to a new tube. The supernatant from tube B was discarded, the precipitate saved, and 100 μL of fresh lysis buffer was added to tube B. Tube C contained total actin, where both supernatant and precipitate were saved. Boiling X2 loading buffer was subsequently added to the three tubes.
Coomassie staining of SDS-polyacrylamide gel electrophoresis (PAGE) gels.
Cell lysates in X2 loading buffer were boiled for 15 minutes, then loaded and electrophoresed on 12% polyacrylamide gel. Gels were stained for 2 to 6 hours with Coomassie brilliant blue R250 stain (Bio-Rad, Hercules, CA; 0.1% dissolved in 50% methanol and 10% acetic acid) and then destained in a 10% methanol 7% acetic acid solution. The gel was subsequently scanned using a Microtek scanner.
Western blot analysis of actin.
Cell lysates were loaded onto a 10% SDS-PAGE, electrophoresed, and transferred onto nitrocellulose. The nitrocellulose membrane blots were blocked (2% BSA in PBS, pH 7.4), rinsed in PBS-T (0.2% Tween-20 in PBS), and incubated with mouse anti-actin MoAb at 1:1,500 dilution in PBS-BT (1% BSA, 0.1% Tween 20 in PBS) for 1 hour at room temperature. The blots were washed three times with PBS-T and incubated with anti-mouse IgG-HRP diluted 1:5,000 in PBS-BT for 1 hour at room temperature. Blots were washed, incubated with enhanced chemiluminescence peroxidase substrates (Amersham, Arlington Heights, IL), and exposed to Fuji X-ray film RX (Fuji, Tokyo, Japan) for varying exposure times.
Actin Immunoprecipitation of [35S]-Cysteine-Methionine–Labeled HL-60 Cells
HL-60 cells were metabolically labeled with 0.3 μCi/mL [35S]-cysteine-methionine (Amersham, Buckinghamshire, UK) in supplemented cysteine-methionine-free DMEM (GIBCO) and exposed to 10−7 mol/L jaspamide or 0.01% DMSO for 1 hours. Labeled cells were extracted in 1 mL lysis buffer (as described above). Clarified cell lysates (2 × 107 cpm) were immunoprecipitated with rabbit anti-actin Ab overnight at 4°C. Immunocomplexes were collected on protein A-sepharose (Pharmacia, Uppsala, Sweden). Each sample was microfuged (10,000 rpm, 5 minutes), the supernatants were decanted, and the pellets were washed three times, resuspended in loading buffer and boiled at 100°C for 10 minutes. Supernatants were collected from microfuged samples, analyzed on 10% SDS-PAGE, and quantified using phosphor-imager (Molecular Dynamics, Sunnyvale, CA) or exposed to Fuji X-ray film RX for varying exposure times.
Phagocytosis of Candida albicans
C albicans was grown in Sabouraud’s dextrose broth for 5 days, washed twice in saline, and resuspended in Hanks’ Balanced Salt Solution (HBSS) to a concentration of 2 × 107 yeast particles/mL as previously described.6 Two types of experiments were performed:
(1) To determine the percentage of phagocytosing cells and the phagocytosis index, HL-60 cells or monocytes were treated for 24 hours with 10−7 mol/L jaspamide, DMSO or medium, washed, and resuspended in HBSS. One hundred microliters of HL-60 cells (containing 106 cells) were placed in capped plastic tubes (12 × 75 mm) with 100 μL AB serum, 200 μL yeast suspension, and HBSS (final volume, 500 μL). Tubes were incubated in a shaking water bath at 37°C for 60 minutes. Cells were then diluted 1:5 with cold HBSS, and cytocentrifuge preparations were collected and stained with Giemsa. At least 200 phagocytosing cells were surveyed to calculate the mean phagocytosing yeast/HL-60 cell (phagocytosing index). We also determined the percentage of cells phagocytosing the C albicans.6
Monocytes were grown on coverslips; 600 μL HBSS, 200 μL AB serum, and 200 μL yeast suspension were added (final volume, 1 mL). The tubes were incubated for 60 minutes. After incubation, cells were washed, stained with Giemsa, and examined as described above.
(2) Fluorescence labeling of F-actin and phagocytosed C albicans in experiments requiring simultaneous visualization were performed in two stages: C albicans (2 × 107) were centrifuged at 2,000 rpm for 10 minutes and the yeast pellet was stained with PKH2 green fluorescent general cell linker kit (Sigma) according to the manufacturer’s instructions. HL-60 cells or monocytes preincubated with 10−7 mol/L jaspamide were incubated with the stained C albicans as described above for 30 minutes. After incubation, the cells were washed with HBSS and stained with Rh-phalloidin as described above.
Electron Microscopy
Monocytes preincubated with 10−7 mol/L jaspamide were incubated with C albicans for 30 minutes as described above. After incubation, the cells were rinsed three times with PBS and fixed in 2% glutaraldehyde for 2 hours at 4°C. The samples were rapidly rinsed three times in 0.1 mol/L cacodylate buffer, pH 7.4, postfixed in osmium tetroxide for 2 hours, and then washed extensively in cacodylate buffer. Samples were dehydrated through a graded series of ethanols, then treated with propylene oxide for 1 hour and embedded in araldite. Thin sections were cut with a diamond knife, stained with uranyl acetate and lead citrate, and examined in a JEOL 100B electron microscope (JEOL, Tokyo, Japan).
Assay of O2− Production by HL-60 Cells and Monocytes
HL-60 cells.
Generation of O2− was measured by determining the rate of superoxide dismutase-inhibitable ferricytochrome C reduction as previously described.6 In brief, HL-60 cells (5 × 105/mL) were grown for 24 hours in the presence of 10−7 mol/L jaspamide, 0.01% DMSO, or medium. After incubation, the cells were washed twice in HBSS containing 5% FBS, and suspended in a 160 μmol/L solution of ferricytochrome C (Sigma) in HBSS. The cells (2.5 × 106 cells/mL) were plated in 96-well flat-bottom tissue-culture plates, 100 μL per well. Phorbol myristate acetate (PMA) (Consolidated Midland Corp, Brewster, NY) (50 mmol/L) was added and the cells were incubated for various periods of time (30 to 90 minutes) at 37°C. Subsequently, the plates were transferred to a Multiskan apparatus (Flow Laboratories, McLean, VA) and the absorbence read at 500 nm against a blank of cytochrome C incubated for the indicated time at 37°C in the absence of cells. The amount of O2− produced was calculated using the equation:
Results were expressed as nanomoles of O2− per 106 cells per minute. Specificity of cytochrome C reduction was verified by its elimination in the presence of 300 U/mL of superoxide dismutase (Sigma).
Monocytes.
Monocytes (2.5 × 106 cells/mL) were plated in 96-well flat-bottom tissue-culture plates, 100 μL per well, and exposed to 10−7 mol/L jaspamide, DMSO, or medium for 24 hours. After incubation, the cells were washed twice with HBSS containing 5% FBS, and O2− production was determined as described for HL-60 cells.
Statistical Analyses
Statistical analyses were performed using Student’s t-test.
RESULTS
Effect of Jaspamide on Cell Proliferation, Differentiation, and Cell-Surface Antigen Expression
We have previously shown that jaspamide suppressed primary colony formation in agar and the recovery of clonogenic cells from suspension cultures of leukemic cell lines in a dose-dependent manner.10 In this study we show that incubation of HL-60 cells with 5 × 10−8 mol/L jaspamide for 24 or 48 hours resulted in 30% and 38% proliferation inhibition, respectively (P < .05) as determined by3H-thymidine incorporation (Table 1). Incubation with 10−7 mol/L jaspamide for 24 or 48 hours resulted in 46% and 77% inhibition, respectively (P < .05) (Table 1). The inhibitory effect of 10−8 mol/L jaspamide was not statistically significant. These results indicate that jaspamide inhibits HL-60 cell proliferation in a dose- and time-dependent manner.
Effect of Jaspamide on the Proliferation of HL-60 Cells
| Treatment . | Incubation Time . | |||
|---|---|---|---|---|
| 24 h . | 48 h . | |||
| Cell No./Well . | cpm . | Cell No./Well . | cpm . | |
| Nil | 1.5 ± 0.1 | 5,153 ± 129 | 2.6 ± 0.2 | 7,078 ± 846 |
| DMSO 0.01% | 1.4 ± 0.01 | 4,984 ± 413 | 2.0 ± 0.1 | 6,452 ± 542 |
| Jaspamide 10−8 mol/L | 1.2 ± 0.1 | 4,729 ± 318 | 2.2 ± 0.1 | 7,078 ± 846 |
| Jaspamide 5 × 10−8 mol/L | 1.1 ± 0.06 | 3,629 ± 1,125* | 1.2 ± 0.1* | 4,386 ± 348* |
| Jaspamide 10−7 mol/L | 1.0 ± 0.14* | 2,783 ± 181* | 0.36 ± 0.06* | 1,625 ± 576* |
| Treatment . | Incubation Time . | |||
|---|---|---|---|---|
| 24 h . | 48 h . | |||
| Cell No./Well . | cpm . | Cell No./Well . | cpm . | |
| Nil | 1.5 ± 0.1 | 5,153 ± 129 | 2.6 ± 0.2 | 7,078 ± 846 |
| DMSO 0.01% | 1.4 ± 0.01 | 4,984 ± 413 | 2.0 ± 0.1 | 6,452 ± 542 |
| Jaspamide 10−8 mol/L | 1.2 ± 0.1 | 4,729 ± 318 | 2.2 ± 0.1 | 7,078 ± 846 |
| Jaspamide 5 × 10−8 mol/L | 1.1 ± 0.06 | 3,629 ± 1,125* | 1.2 ± 0.1* | 4,386 ± 348* |
| Jaspamide 10−7 mol/L | 1.0 ± 0.14* | 2,783 ± 181* | 0.36 ± 0.06* | 1,625 ± 576* |
Cells were incubated with jaspamide for 24 and 48 hours in liquid cultures at an initial concentration of 104 HL-60 cells/200 μL. The cells were counted by trypan blue exclusion.3H-thymidine incorporation studies were performed as described in Materials and Methods. Results are the mean ± SE of three experiments performed in triplicate.
Indicated P < .05.
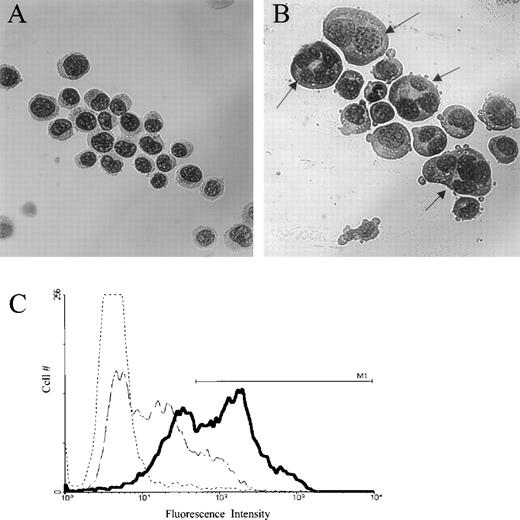
To study the effect of jaspamide on cell differentiation, HL-60 cells were treated with 10−7 mol/L jaspamide for 48 hours and examined for changes in cell morphology and surface differentiation markers. DMSO-treated cells served as control. After jaspamide treatment, 55% ± 5% (n = 3) of the cells exhibited myeloid differentiation as indicated by a lobulated nucleus (Fig 1B). This lobulation was not apparent in the DMSO-treated cells (Fig 1A). FACS analysis of surface antigen CD16 shows 53% upregulation of this differentiation marker in jaspamide-treated cells (Table 2 and Fig1C). Treatment with jaspamide induced 26.6% upregulation in CD14B expression (Table 2). These results indicate that jaspamide induced maturation of HL-60 cells to the granulocyte monocyte lineage.
Jaspamide induces differentiation in HL-60 cells. HL-60 cells were grown for 48 hours with (A) 0.01% DMSO, or (B) 10−7 mol/L jaspamide. Cell differentiation was determined on Wright-Giemsa–stained cytocentrifuge preparations. (C) FACS analysis and overlay histograms of fluorescence intensity showing the upregulation of the differentiation antigen CD16 in HL-60 cells after exposure of the cells for 48 hours to 10−7 mol/L jaspamide (thick line) or 0.01% DMSO (broken thin line). An irrelevant antibody was used as background control (dotted line). M1line defines the positive intensities range (A, B: original magnification × 475).
Jaspamide induces differentiation in HL-60 cells. HL-60 cells were grown for 48 hours with (A) 0.01% DMSO, or (B) 10−7 mol/L jaspamide. Cell differentiation was determined on Wright-Giemsa–stained cytocentrifuge preparations. (C) FACS analysis and overlay histograms of fluorescence intensity showing the upregulation of the differentiation antigen CD16 in HL-60 cells after exposure of the cells for 48 hours to 10−7 mol/L jaspamide (thick line) or 0.01% DMSO (broken thin line). An irrelevant antibody was used as background control (dotted line). M1line defines the positive intensities range (A, B: original magnification × 475).
Effect of Jaspamide on the Expression of CD16 and CD14B Antigens by HL-60 Cells
| Antigen . | Positive Cells (%) . | Delta % . | Reactivity . | ||
|---|---|---|---|---|---|
| DMSO Treated . | Jaspamide Treated . | Monocytes . | Granulocytes . | ||
| CD16 (306) | 28.5 | 81.5 | 53.0 | + | +++ |
| CD14B (253) | 20.7 | 47.3 | 26.6 | +++ | − |
| Antigen . | Positive Cells (%) . | Delta % . | Reactivity . | ||
|---|---|---|---|---|---|
| DMSO Treated . | Jaspamide Treated . | Monocytes . | Granulocytes . | ||
| CD16 (306) | 28.5 | 81.5 | 53.0 | + | +++ |
| CD14B (253) | 20.7 | 47.3 | 26.6 | +++ | − |
Cells were incubated for 48 hours with 10−7 mol/L jaspamide or 0.01% DMSO. FACS analysis was performed as described in Materials and Methods. Similar results were observed in three additional experiments.
Effects of Jaspamide on Actin Organization
The jaspamide-induced reorganization of actin in HL-60 cells was examined using anti-actin MoAb staining and CLSM analysis. Exposure of HL-60 cells to jaspamide for 24 hours resulted in a dramatic reorganization of actin from the short filamentous actin network ordinarily found in HL-60 cells (Fig2A) to focal aggregates (Fig 2B, C, C’). This effect was dose dependent: detectable at 10−8 mol/L (Fig 2B) and observed to be most prominent at 10−7 mol/L jaspamide (Fig 2C and C’). Jaspamide was toxic at concentrations of 10−6 mol/L (data not shown).
Jaspamide induces actin aggregation in HL-60 cells in a dose- and time-dependent manner. HL-60 cells were treated with jaspamide and stained for actin (with mouse anti-actin MoAb labeled with rhodamine-conjugated donkey anti-mouse antibody) and analyzed by CLSM. (A) Untreated cells. (B) Cells grown for 24 hours with 10−8 mol/L jaspamide. (C, C’) Cells grown for 24 hours with 10−7 mol/L jaspamide; in C’ the actin staining (red) is overlaid on the Nomarski image (gray). (D) Cells grown for 48 hours in the presence of 10−8 mol/L jaspamide. (E) Cells grown for 48 hours in the presence of 10−7 mol/L jaspamide (A through E: original magnification × 250) (C’: original magnification × 2,100).
Jaspamide induces actin aggregation in HL-60 cells in a dose- and time-dependent manner. HL-60 cells were treated with jaspamide and stained for actin (with mouse anti-actin MoAb labeled with rhodamine-conjugated donkey anti-mouse antibody) and analyzed by CLSM. (A) Untreated cells. (B) Cells grown for 24 hours with 10−8 mol/L jaspamide. (C, C’) Cells grown for 24 hours with 10−7 mol/L jaspamide; in C’ the actin staining (red) is overlaid on the Nomarski image (gray). (D) Cells grown for 48 hours in the presence of 10−8 mol/L jaspamide. (E) Cells grown for 48 hours in the presence of 10−7 mol/L jaspamide (A through E: original magnification × 250) (C’: original magnification × 2,100).
To evaluate the time-course of jaspamide-induced actin reorganization, HL-60 cells were incubated with 10−7 mol/L jaspamide for 1, 4, 24, and 48 hours. At 1 and 4 hours, approximately 10% and 50%, respectively, of the HL-60 cells exhibited actin aggregation (data not shown). The percentage of cells showing actin aggregation increased over time, with greater than 80% of the cells exhibiting large focal aggregates of actin at 24 hours (Fig 2C). Similar large focal aggregates of actin were observed upon incubation of HL-60 cells for 48 hours (Fig 2E). HL-60 cells exposed to 10−7mol/L jaspamide for 24 hours, followed by extensive washing and subsequent examination after an additional 48 hours in media without jaspamide, exhibited no reversal of the jaspamide-induced actin aggregation (data not shown).
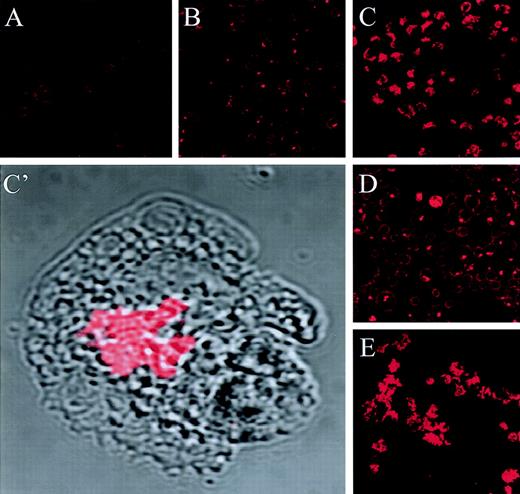
Previous studies have shown that jaspamide inhibits binding of phalloidin to purified F-actin.14 Jaspamide is capable of entering cells unfacilitated.15,16 It was previously shown that phalloidin does not penetrate living cells without cell permeabilization.17,18 However, it was recently shown that phalloidin can penetrate living cells.19 We did not observe significant penetration of phalloidin in nonpereablized cells (data not shown). We proceeded to investigate whether jaspamide also inhibits phalloidin binding to actin in permeabilized cells. Phalloidin staining of HL-60 cells was performed in the presence of 10−8mol/L (Fig 3B) or 10−7mol/L jaspamide (Fig 3C) and compared with phalloidin staining in the absence of jaspamide (Fig 3A). These experiments show that 10−8 mol/L jaspamide partially reduced Rh-phalloidin staining whereas 10−7 mol/L jaspamide further reduced Rh-phalloidin labeling. Conversely, HL-60 cells, treated with 10−7 mol/L jaspamide for 24 hours followed by their extensive washing and then Rh-phalloidin staining, did not exhibit reduced actin labeling by Rh-phalloidin, compared with untreated cells. Moreover, these cells exhibited aggregated staining of actin similar to that observed in treated cells labeled with anti-actin MoAb (Fig 3E). From these results we conclude that jaspamide competes with phalloidin binding to actin in cells only if present in the labeling reaction. To show that the aggregated phalloidin staining depicts actin rearrangement, we performed double staining with Rh-phalloidin and anti-actin MoAb (Fig 3G through I). CLSM analysis of double-labeled HL-60 cells (treated with 10−7 mol/L jaspamide for 24 hours) is shown in Fig 3G (green, MoAb staining) and Fig 3H (red, Rh-phalloidin staining). The overlay of red and green images shows that most of the anti-actin MoAb and Rh-phalloidin colocalize (yellow) (Fig3I). These results demonstrate that jaspamide induces cellular F-actin rearrangement in HL-60 cells.
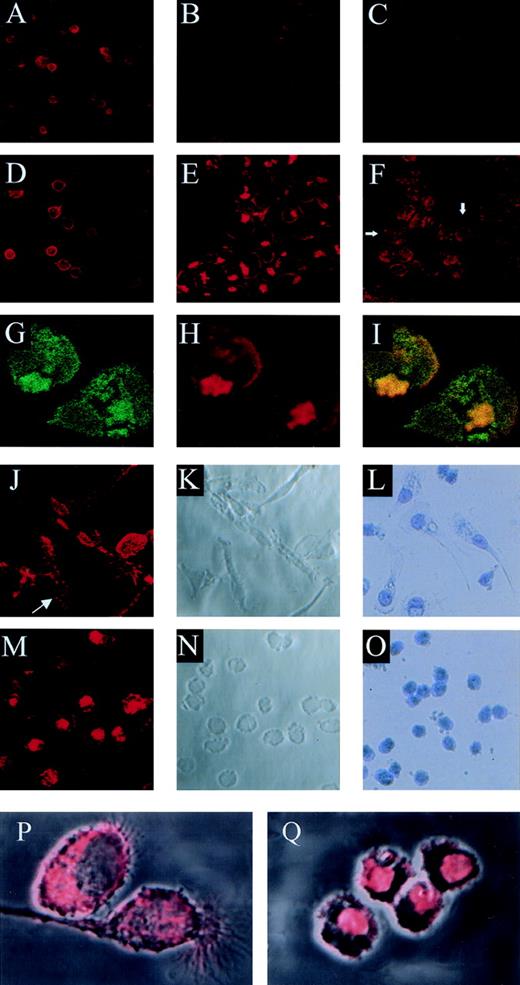
Competitive binding studies between jaspamide and Rh-phalloidin. Jaspamide induces F-actin aggregation in HL-60 cells and in human monocytes. Competitive studies (A through C). HL-60 cells were stained with Rh-phalloidin in the presence of (A) medium (control). (B) 10−8 mol/L jaspamide and (C) 10−7 mol/L jaspamide; analyzed by CLSM. No actin aggregation is seen. Aggregation of F-actin in HL-60 cells (D through F). HL-60 cells were incubated for 24 hours with (D) medium. (E) 10−7 mol/L jaspamide subsequently washed and stained with Rh-phalloidin (no jaspamide added during staining with Rh-phalloidin). Aggregation of F-actin is seen in (E). (F) HL-60 cells grown for 24 hours in the presence of 3.94 μmol/L CD were stained with Rh-phalloidin (arrows indicate cytochalasin-induced small focal F-actin clusters). Costaining of HL-60 cells with Rh-phalloidin and anti-actin MoAb (G through I). HL-60 cells grown for 24 hours in the presence of 10−7 mol/L jaspamide were costained for F-actin (Rh-phalloidin) and total actin (anti-actin MoAb labeled with anti-mouse–FITC). Colocalization analysis was performed: (G) Actin labeled with anti-actin MoAb (green), (H) F-actin labeled with Rh-phalloidin (red), (I) colocalization of the total actin and F-actin staining is observed (yellow). Effect of jaspamide on monocytes (J through Q). Freshly isolated monocytes were grown for 24 hours in IMDM containing 10% FBS. The cells were incubated for additional 24 hours with the following: (J through L, P) medium; (M through O, Q) 10−7 mol/L jaspamide. (J, M, P, and Q) Monocytes stained with Rh-phalloidin to show the distribution of F-actin. In (P) and (Q) the actin staining (red) is overlaid on the Nomarski image (gray) phase microscopy of the same cells. (L and O) Monocytes stained with Giemsa to show the changes in cell morphology. (A through F: original magnification × 250; G through H: original magnification × 750; J through O: original magnification × 250; P and Q: original magnification × 2,000).
Competitive binding studies between jaspamide and Rh-phalloidin. Jaspamide induces F-actin aggregation in HL-60 cells and in human monocytes. Competitive studies (A through C). HL-60 cells were stained with Rh-phalloidin in the presence of (A) medium (control). (B) 10−8 mol/L jaspamide and (C) 10−7 mol/L jaspamide; analyzed by CLSM. No actin aggregation is seen. Aggregation of F-actin in HL-60 cells (D through F). HL-60 cells were incubated for 24 hours with (D) medium. (E) 10−7 mol/L jaspamide subsequently washed and stained with Rh-phalloidin (no jaspamide added during staining with Rh-phalloidin). Aggregation of F-actin is seen in (E). (F) HL-60 cells grown for 24 hours in the presence of 3.94 μmol/L CD were stained with Rh-phalloidin (arrows indicate cytochalasin-induced small focal F-actin clusters). Costaining of HL-60 cells with Rh-phalloidin and anti-actin MoAb (G through I). HL-60 cells grown for 24 hours in the presence of 10−7 mol/L jaspamide were costained for F-actin (Rh-phalloidin) and total actin (anti-actin MoAb labeled with anti-mouse–FITC). Colocalization analysis was performed: (G) Actin labeled with anti-actin MoAb (green), (H) F-actin labeled with Rh-phalloidin (red), (I) colocalization of the total actin and F-actin staining is observed (yellow). Effect of jaspamide on monocytes (J through Q). Freshly isolated monocytes were grown for 24 hours in IMDM containing 10% FBS. The cells were incubated for additional 24 hours with the following: (J through L, P) medium; (M through O, Q) 10−7 mol/L jaspamide. (J, M, P, and Q) Monocytes stained with Rh-phalloidin to show the distribution of F-actin. In (P) and (Q) the actin staining (red) is overlaid on the Nomarski image (gray) phase microscopy of the same cells. (L and O) Monocytes stained with Giemsa to show the changes in cell morphology. (A through F: original magnification × 250; G through H: original magnification × 750; J through O: original magnification × 250; P and Q: original magnification × 2,000).
Cytochalasin D (CD), a fungal toxin, inhibits microfilament function and polymerization by blocking monomer addition at the rapidly growing ends of F-actin filaments. Incubation of HL-60 cells with 3.94 μmol/L CD for 24 hours resulted in the appearance of small actin clusters distributed throughout the cytoplasm (Fig 3F), compared with the large bulky aggregation induced by jaspamide (Fig 3E and H).
Jaspamide also induced dramatic changes in the cellular morphology of human monocytes. Well-spread cultured monocytes (Fig 3J through L) treated with 10−7 mol/L jaspamide for 24 hours drastically contracted and adopted round shapes (Fig 3M through O). To study the effect of jaspamide on F-actin organization in these cells, jaspamide-treated monocytes and untreated control cells were stained with Rh-phalloidin. In untreated monocytes F-actin is organized in a network and is condensed in focal adhesion regions (Fig 3J, see arrow and P). In jaspamide-treated cells, F-actin was dramatically rearranged to form aggregated clumps (Fig 3M and Q). The clustering of F-actin in jaspamide-treated human monocytes was similar to the actin reorganization observed in jaspamide-treated HL-60 cells (Fig 3E and H).
Effect of Jaspamide on Total Actin Content and Synthesis in HL-60 Cells
The effects of jaspamide treatment on total cellular actin content and de novo synthesis of soluble actin in HL-60 cells were determined using SDS-PAGE, Western blot, and immunoprecipitation studies. Exposure of HL-60 cells to 10−7 mol/L jaspamide for 48 hours induces a dramatic increase in a 42-kD band corresponding with actin, compared with untreated cells as determined by SDS-PAGE (Fig 4Aa, lanes 2 and 1, respectively). Western blot analysis with anti-actin MoAb verified a specific dose-dependent increase in total actin content after incubation of the cells with 10−8 to 10−7 mol/L jaspamide for 24 hours, as compared with DMSO (Fig 4Ab, lanes 4, 3, and 2, respectively). To study the time course of jaspamide-induced total actin accumulation, we measured total actin in HL-60 cells after their exposure to 10−7 mol/L jaspamide for various periods of time (1 to 24 hours) (Fig 4B). A slight increase in total actin was observed after 4 hours of incubation with jaspamide compared to DMSO (Fig 4B, lanes 5 and 4, respectively). A further increase was observed after 24 hours of exposure to jaspamide compared with DMSO (Fig 4B, lanes 7 and 6, respectively). No significant increase in total actin was observed after a 1-hour incubation of the cells with the drug compared with DMSO (Fig 4B, lanes 3 and 2, respectively).
Jaspamide induces increase in total cellular actin and de novo actin synthesis in HL-60 cells. (Aa) Coomassie-stained 12% SDS-PAGE gel showing total cellular protein of HL-60 cells exposed for 48 hours to DMSO 0.01% (lane 1) or jaspamide 10−7 mol/L (lane 2). (Ab) Dose-dependent actin accumulation: HL-60 cell extracts obtained from cells exposed for 24 hours to medium (lane 1), DMSO 0.01% (lane 2), jaspamide 10−7 mol/L (lane 3), jaspamide 10−8 mol/L (lane 4) were resolved on SDS-PAGE, and analyzed by Western blot using anti-actin MoAb. (B) Time course of actin accumulation. HL-60 cell extracts obtained from cells exposed to medium (lane 1), DMSO 0.01% (lanes 2, 4, and 6), or jaspamide 10−7 mol/L (lanes 3, 5, and 7) for 1 hour (lanes 2 and 3), 4 hours (lanes 4 and 5), or 24 hours (lanes 6 and 7) were resolved on SDS-PAGE, and analyzed by Western blot using anti-actin MoAb. (C) Effect of jaspamide on soluble and insoluble actin: Soluble, insoluble, and total actin fractions were prepared as described in Materials and Methods. (a) Coomassie blue and (b) immunoblot with anti-actin MoAb studies were performed (the lanes indicated are for both methods; ie, Fig 4Ca and Fig 4Cb). HL-60 cells treated with DMSO (lanes 1 through 3) or jaspamide 10−7mol/L for 24 hours (lanes 4 through 6) were examined for total (lanes 1 and 4) soluble (lanes 2 and 5) insoluble (lanes 3 and 6) actin. (D) De novo actin synthesis. (a) Effect of jaspamide on de novo actin synthesis. [S35]-cystein-methionine-labeled HL-60 cells were exposed to medium (lane 1), DMSO (lane 2), or to jaspamide 10−7 mol/L (lane 3) for 1 hour, immunoprecipitated with rabbit anti-actin Ab, and the immunoprecipitates were resolved on SDS-PAGE and exposed to Fuji X-ray film. (b) Coomassie-stained SDS-PAGE of HL-60 cells treated for 1 hour with DMSO 0.01% (lanes 1 through 3) or jaspamide 10−7 mol/L (lanes 4 through 6) showing total (lanes 1 and 4) soluble (lanes 2 and 5) and insoluble (lanes 3 and 6) actin.
Jaspamide induces increase in total cellular actin and de novo actin synthesis in HL-60 cells. (Aa) Coomassie-stained 12% SDS-PAGE gel showing total cellular protein of HL-60 cells exposed for 48 hours to DMSO 0.01% (lane 1) or jaspamide 10−7 mol/L (lane 2). (Ab) Dose-dependent actin accumulation: HL-60 cell extracts obtained from cells exposed for 24 hours to medium (lane 1), DMSO 0.01% (lane 2), jaspamide 10−7 mol/L (lane 3), jaspamide 10−8 mol/L (lane 4) were resolved on SDS-PAGE, and analyzed by Western blot using anti-actin MoAb. (B) Time course of actin accumulation. HL-60 cell extracts obtained from cells exposed to medium (lane 1), DMSO 0.01% (lanes 2, 4, and 6), or jaspamide 10−7 mol/L (lanes 3, 5, and 7) for 1 hour (lanes 2 and 3), 4 hours (lanes 4 and 5), or 24 hours (lanes 6 and 7) were resolved on SDS-PAGE, and analyzed by Western blot using anti-actin MoAb. (C) Effect of jaspamide on soluble and insoluble actin: Soluble, insoluble, and total actin fractions were prepared as described in Materials and Methods. (a) Coomassie blue and (b) immunoblot with anti-actin MoAb studies were performed (the lanes indicated are for both methods; ie, Fig 4Ca and Fig 4Cb). HL-60 cells treated with DMSO (lanes 1 through 3) or jaspamide 10−7mol/L for 24 hours (lanes 4 through 6) were examined for total (lanes 1 and 4) soluble (lanes 2 and 5) insoluble (lanes 3 and 6) actin. (D) De novo actin synthesis. (a) Effect of jaspamide on de novo actin synthesis. [S35]-cystein-methionine-labeled HL-60 cells were exposed to medium (lane 1), DMSO (lane 2), or to jaspamide 10−7 mol/L (lane 3) for 1 hour, immunoprecipitated with rabbit anti-actin Ab, and the immunoprecipitates were resolved on SDS-PAGE and exposed to Fuji X-ray film. (b) Coomassie-stained SDS-PAGE of HL-60 cells treated for 1 hour with DMSO 0.01% (lanes 1 through 3) or jaspamide 10−7 mol/L (lanes 4 through 6) showing total (lanes 1 and 4) soluble (lanes 2 and 5) and insoluble (lanes 3 and 6) actin.
HL-60 cell actin is considered primarily (≈75%) to be composed of “Triton-soluble” fraction.13 We proceeded to determine the effect of jaspamide on the soluble and insoluble actin fractions. Coomassie staining of SDS-PAGE (Fig 4Ca) and Western blot analysis (Fig 4Cb) both show a significant shift of actin from the soluble to the insoluble fraction in jaspamide-treated cells (Fig 4Cb, lanes 5 and 6, respectively) compared with DMSO-treated cells (lanes 2 and 3, respectively).
The identification of newly synthesized actin using immunoprecipitation is a complicated experiment due to the fact that portions of actin are insoluble. However, we proceeded to analyze de novo actin synthesis by a standard immunoprecipitation procedure using short-term (1 hour) labeling to limit the effect of an increase in the insoluble fraction in treated cells. Results show a fivefold increase in short-term35S-labeled immunoprecipitation actin. We verified the limited increase in insoluble actin during the 1-hour treatment by SDS-PAGE (Fig 4Da and b). These results indicate that a significant portion of the increase in total actin levels is caused by de novo synthesis.
Effect of Jaspamide on Cellular Morphology, Intracellular Movement, and Ruffling
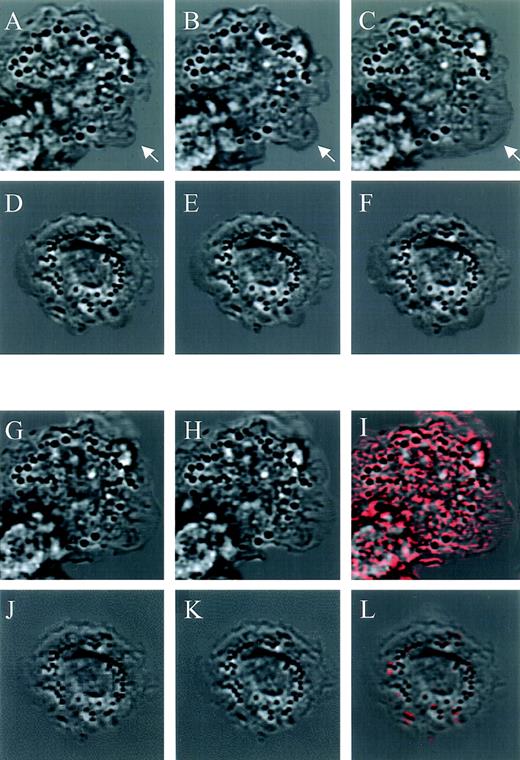
To study jaspamide’s effect on cellular movement, we compared CLSM time-lapse photography (1-second interval) of monocytes treated for 1 hour with 10−7 mol/L jaspamide and untreated cells. The untreated monocytes moved extensively and spread on the substrate (Fig 5A through C, see arrow) while the jaspamide-treated cells moved significantly less and pseudopodia formation was rarely detected (Fig 5D through F).
Effect of jaspamide on intracellular movement and ruffling in monocytes. Nomarski time-lapse photography of monocytes treated for 1 hour with (D through F and J through L) 10−7 mol/L jaspamide compared with (A through C, G through I) untreated cells. Cellular movement (A through F). Nomarski images of a 1-second interval of the (A through C) untreated cells exhibiting movement on the substrate (arrow indicating spreading region); (D through F) jaspamide-treated cells that exhibited no significant movement. Ruffling and intracellular movement: (G through L) 0.05-second interval of treated and untreated cells, after (G and J) 10 minutes and (H and K) 10 minutes + 0.05 seconds were analyzed. Using the CLSM program, we determined the cellular movement that occurred in 0.05 seconds by subtracting the 10-minute images from the images of 10 minute + 0.05 seconds (images 5H minus 5G and images 5K minus 5J) for untreated and treated cells, respectively. The resulting calculation of movement is shown as red areas superimposed on 10-minute + 0.05-second untreated and treated images (I and L, respectively) (original magnification × 1,400).
Effect of jaspamide on intracellular movement and ruffling in monocytes. Nomarski time-lapse photography of monocytes treated for 1 hour with (D through F and J through L) 10−7 mol/L jaspamide compared with (A through C, G through I) untreated cells. Cellular movement (A through F). Nomarski images of a 1-second interval of the (A through C) untreated cells exhibiting movement on the substrate (arrow indicating spreading region); (D through F) jaspamide-treated cells that exhibited no significant movement. Ruffling and intracellular movement: (G through L) 0.05-second interval of treated and untreated cells, after (G and J) 10 minutes and (H and K) 10 minutes + 0.05 seconds were analyzed. Using the CLSM program, we determined the cellular movement that occurred in 0.05 seconds by subtracting the 10-minute images from the images of 10 minute + 0.05 seconds (images 5H minus 5G and images 5K minus 5J) for untreated and treated cells, respectively. The resulting calculation of movement is shown as red areas superimposed on 10-minute + 0.05-second untreated and treated images (I and L, respectively) (original magnification × 1,400).
To analyze the effect of jaspamide on membrane ruffling, we compared rapid CLSM time-lapse photography (0.05-second interval) of untreated and treated monocytes. CLSM images of 10−7 mol/L jaspamide-treated and untreated cells after 10 minutes (Fig 5G and J, respectively) and 10 minutes + 0.05 seconds were acquired (Fig 5H and K, respectively). Using the CLSM program we subtracted the 10-minute images from the 10-minute + 0.05-second images (Fig 5H minus 5G and 5K minus 5J). The differences between images were calculated, and then areas where changes occurred were superimposed on the corresponding 10-minute time image. These areas (shown in red Fig 5I and Fig 5L) represent positional changes of cellular organelle and membrane, thus indicating that movement took place. The movement and ruffling in untreated cells were extensive as shown by the significant amount of red in Fig 5I. Furthermore, untreated cells show extensive formation of motile cell-surface protrusions-ruffling. No significant ruffling or intracellular movement was observed in the jaspamide-treated cells; therefore, the image scarcely features any red (Fig 5L). Moreover, the internal movement of organelles is dramatically inhibited (Fig 5L). We performed 10 experiments to determine number of cells exhibiting membrane ruffling for at least 50 cells per experiment. Ninety-five percent of the untreated monocytes exhibit membrane ruffling while only 4% of the jaspamide-treated cells exhibited ruffling. None of the cells with significant actin aggregation was observed to exhibit ruffling. These results suggest that proper F-actin organization is essential for overall motility of the cell.
Effects of Jaspamide on Phagocytosis and O2− Production
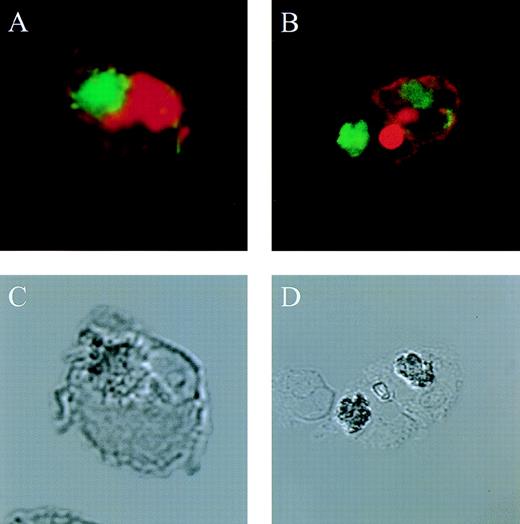
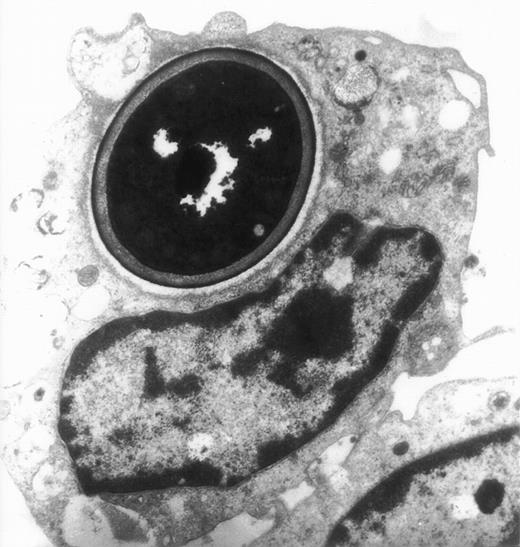
The actin network plays a major role in chemotaxis and phagocytosis. To study the effects of jaspamide on phagocytosis, HL-60 cells and monocytes were exposed for 24 hours to 10−7 mol/L jaspamide, incubated with C albicans for 60 minutes, and examined for phagocytosis. Jaspamide did not affect the percentage of phagocytosing cells or the number of yeast particles phagocytosed by each cell (Table 3). To ascertain that aggregated actin clusters did not affect phagocytosis, fluorescein-labeled C albicans was incubated with jaspamide-treated HL-60 cells and monocytes. The cells were fixed, Rh-phalloidin stained, and analyzed by CLSM. HL-60 cells (Fig 6A and C [see page 3998]) and human monocytes (Fig 6B and D [see page 3998]) that show jaspamide-induced actin aggregates phagocytose C albicans. Electron microscopy analysis of jaspamide-treated monocytes incubated with C albicans shows that the C albicans was phagocytosed and is located inside the cell (Fig 7).
Effect of Jaspamide on Phagocytosis of C albicans by HL-60 Cells and Monocytes
| Treatment . | HL-60 Cells . | Monocytes . | ||
|---|---|---|---|---|
| (%) Cells . | Phagocytosis Index . | (%) Cells . | Phagocytosis Index . | |
| Nil | 6.3 ± 2.2 | 0.17 ± 0.02 | 86.3 ± 4.7 | 3.0 ± 0.2 |
| DMSO (0.01%) | 6.6 ± 0.6 | 0.17 ± 0.02 | 80.2 ± 7.0 | 2.5 ± 0.55 |
| Jaspamide 10−7 mol/L | 9.3 ± 1.3 | 0.22 ± 0.02 | 87.5 ± 4.0 | 2.4 ± 0.55 |
| Treatment . | HL-60 Cells . | Monocytes . | ||
|---|---|---|---|---|
| (%) Cells . | Phagocytosis Index . | (%) Cells . | Phagocytosis Index . | |
| Nil | 6.3 ± 2.2 | 0.17 ± 0.02 | 86.3 ± 4.7 | 3.0 ± 0.2 |
| DMSO (0.01%) | 6.6 ± 0.6 | 0.17 ± 0.02 | 80.2 ± 7.0 | 2.5 ± 0.55 |
| Jaspamide 10−7 mol/L | 9.3 ± 1.3 | 0.22 ± 0.02 | 87.5 ± 4.0 | 2.4 ± 0.55 |
HL-60 cells and monocytes were incubated with C albicansfor 60 minutes and tested for percentage of phagocytosing cells and phagocytosis index. Values represent mean ± SE of 3 experiments.
Effect of jaspamide on phagocytosis of C albicansby HL-60 cells and monocytes. (A and C) HL-60 cells or (B and D) monocytes were grown for 24 hours in the presence of jaspamide 10−7 mol/L and exposed to fluorescence-stained C albicans for 30 minutes. Cells were stained with Rh-phalloidin and analyzed by CLSM for (A and B) fluorescence or (C and D) Nomarski (original magnification × 1,700).
Effect of jaspamide on phagocytosis of C albicansby HL-60 cells and monocytes. (A and C) HL-60 cells or (B and D) monocytes were grown for 24 hours in the presence of jaspamide 10−7 mol/L and exposed to fluorescence-stained C albicans for 30 minutes. Cells were stained with Rh-phalloidin and analyzed by CLSM for (A and B) fluorescence or (C and D) Nomarski (original magnification × 1,700).
Electron migrograph of a monocyte that phagocytosed C albicans. A thin section through monocytes following their growth for 24 hours in the presence of 10−7 mol/L jaspamide and their exposure for 30 minutes to C albicans (original magnification × 9,250).
Electron migrograph of a monocyte that phagocytosed C albicans. A thin section through monocytes following their growth for 24 hours in the presence of 10−7 mol/L jaspamide and their exposure for 30 minutes to C albicans (original magnification × 9,250).
Superoxide anion production during the respiratory burst in monocytes is presumed to be actin-dependent.20 We examined O2− production by HL-60 cells and by monocytes following their exposure to 10−7 mol/L jaspamide for 24 hours. Jaspamide did not affect release of superoxide anion upon PMA stimulation as indicated by the SOD-inhibitable cytochrome C reduction assay (Table 4). Overall production of O2− by HL-60 cells was very low, compared with monocytes.
Effect of Jaspamide on Production of Superoxide by Monocytes and HL-60 Cells Incubated in the Absence or Presence of PMA
| Treatment . | production (nmol) per 106 Cells During 60 min . | |||
|---|---|---|---|---|
| Monocytes . | HL-60 Cells . | |||
| −PMA . | +PMA . | −PMA . | +PMA . | |
| Nil | 3.8 ± 0.3 | 38.3 ± 7.8 | 0 | 1.7 ± 0.35 |
| DMSO 0.01% | 5.0 ± 2.0 | 42.3 ± 3.7 | 0 | 1.8 ± 0.05 |
| Jaspamide 10−7 mol/L | 3.8 ± 0.1 | 43.1 ± 5.2 | 0 | 2.35 ± 0.4 |
| Treatment . | production (nmol) per 106 Cells During 60 min . | |||
|---|---|---|---|---|
| Monocytes . | HL-60 Cells . | |||
| −PMA . | +PMA . | −PMA . | +PMA . | |
| Nil | 3.8 ± 0.3 | 38.3 ± 7.8 | 0 | 1.7 ± 0.35 |
| DMSO 0.01% | 5.0 ± 2.0 | 42.3 ± 3.7 | 0 | 1.8 ± 0.05 |
| Jaspamide 10−7 mol/L | 3.8 ± 0.1 | 43.1 ± 5.2 | 0 | 2.35 ± 0.4 |
production (nmol) per 106 cells during 60 minutes. Results expressed as mean ± SE of 3 experiments performed in quadruplicate.
DISCUSSION
HL-60 cells have been used as a model to study morphological, functional, and biochemical changes after differentiation induced by various agents.4,5,21 Previous studies have shown that actin synthesis undergoes changes during induced myeloid maturation,9 and that actin polymerization affects a variety of cell functions including locomotion, phagocytosis, and maturation in a wide range of cells including HL-60 cells.22
In recent years, a number of molecules have been shown to affect actin organization and cell functions.23,24 One such molecule, jaspamide, has been shown to directly interact with purified actin to induce actin polymerization.14 In a previous study, we have shown that jaspamide induces growth modulation and differentiation of AML cells.10 The present study was undertaken to investigate the effects of jaspamide on cellular actin levels and organization along with its effect on membrane ruffling, oxidative burst, and phagocytic activity in both HL-60 cells and monocytes.
It has been previously shown that jaspamide has antiproliferative activity and a cytotoxic effect on a number of tumor cell lines, including breast and prostatic cancer cells.15,16 Here we show that jaspamide inhibits the growth of human promyelocytic leukemic HL-60 cells, induces differentiation, and acts on the actin-based cytoskeleton. It is not clear whether growth inhibition by jaspamide is directly induced by actin aggregation or indirectly by affecting other proteins that induce growth inhibition. A recent report showing that jasplakinolide inhibited bombesin-stimulated phosphorylation of FAK and inhibited PC-3 cell growth25 offers support to the indirect inhibition hypothesis. Moreover, these investigators suggest that additional actin-disrupting agents can block FAK signal transduction, which may be critical to their antitumor activity in prostate carcinoma.25
Jaspamide has been recently shown to also markedly influence the morphogenetic process in the green alga Micrasterias when used in concentrations higher than 3 μmol/L. Development ofMicrasterias was found to be inhibited or strongly retarded, and malformations occurred and large vacuoles were formed. At the ultrastructural level, dense abnormal accumulations of filamentous structures have been found, indicating actin filament polymerizing activities of the drug in situ.26
The present data show that jaspamide induces actin accumulation in HL-60 cells in a dose- and time-dependent manner. This increase in total actin is accompanied by a shift of the major portion of actin from the soluble to the insoluble form in jaspamide-treated HL-60 cells. We proceeded to measure de novo actin synthesis using35S-labeling and immunoprecipitation. Although this method does not take into account newly synthesized actin that was immediately polymerized to insoluble actin, we suggest that these results are a good indication that jaspamide treatment increases actin synthesis in HL-60 cells. This is based on the comparatively short labeling time experiments and the assumption that the rate of actin polymerization to insoluble form is greater in the jaspamide-treated cells as suggested by the results of SDS-PAGE and Western blot of different actin fractions (Fig 4C).
The increase in actin synthesis can be attributed to actin clustering that may signal for jaspamide-induced reduction of the cellular actin pool. Our results are in line with previous studies showing that HL-60 cells induced toward myeloid maturation by a 5-day exposure to dimethylformamide contain about twice as much actin per milligram protein as do uninduced HL-60 cells.9 An increase in the content of total cellular actin has been reported in differentiated myeloid leukemia cells compared with their nondifferentiated counterparts.8 27-29
We show that jaspamide not only induces actin accumulation but also induces the clustering of actin in HL-60 cells in a dose-and time-dependent manner. Our results also demonstrate that jaspamide inhibits phalloidin binding to F-actin in cells if present in the labeling mix. However, after a 24-hour incubation and extensive washing, jaspamide did not inhibit phalloidin biding to F-actin. This loss of inhibition was probably caused by reduced jaspamide concentration resulting from the washing or its instability. Other drugs known to affect actin organization, as well as cell function, are the cytochalasins.30 Cytochalasins disrupt actin organization, inhibit various cell movements,31 bind to the growing end of F-actin filaments, and block all association and dissociation reactions at those ends.32 We compared the effects of jaspamide in HL-60 cells to those of cytochalasin D and have shown that while jaspamide induced large F-actin aggregates, cytochalasin D induced the formation of only small F-actin clusters distributed throughout the HL-60 cytoplasm. The different effect of these compounds on actin organization indicates that the mode of action of the two classes of drugs is different. Our study also shows that jaspamide has similar effects on the F-actin organization in human monocytes.
In membrane ruffling areas, the actin filaments are tightly associated with the plasma membrane and a number of proteins have been shown to be associated with these sites.33 The present results show that aggregation of actin inhibits cell ruffling. A similar inhibition is induced by other factors such as cytochalasin D.34 These results indicate that ruffling, which is a well-coordinated membrane cytoskeleton procedure, requires the presence of an organized membrane-bound actin cytoskeleton. Jasplakinolide was recently shown to cause actin polymerization within neutrophils and an increase in association of actin with the Triton-insoluble cytoskeleton. Actin polymerization in neutrophils induce marked increase in rigidity and affect adhesive and mechanical properties of the cells.35 36
In the present study we have shown that jaspamide does not affect phagocytosis of C albicans by HL-60 cells or by monocytes. The functions of mature neutrophils, such as phagocytosis,37and expression of the oxidase involved in respiratory burst38 have all been shown to be associated with F-actin and the actin-binding protein network. Recent studies have shown that although fibronectin enhances actin polymerization in neutrophils, monocytes, and macrophages,39 it promotes phagocytosis of unopsonized Staphylococci, IgG-opsonized bacteria,40 IgA-opsonized bacteria,41 and complement receptor-mediated phagocytosis42 by phagocytes.
The role of actin in microorganism phagocytosis is still not fully understood. Several works have shown that there is a dramatic change of actin organization upon phagocytosis, but it is not clear if the phagocytic process is dependent on actin polymerization. For example, double-immunofluorescence staining of gonococci and actin filaments in infected cells showed bacterium-associated accumulations of F-actin as an early signal of bacterial entry. The recruitment of F-actin was transient and disappeared once the bacteria were inside the cells. Cytochalasin D disrupted the actin cytoskeleton architecture but did not prevent the recruitment of F-actin by the bacteria.43 It was also shown that changes in the organization of actin occurred during the C albicansinternalization to fibroblastic cells.44 Other studies have shown that endothelial cell microfilaments polymerized around C albicans after phagocytosis of the organisms, suggesting that condensation of actin filaments around the organisms is required forC albicans phagocytosis.45 There is not a conclusive answer to the question if actin is directly involved and is necessary for phagocytosis. The results shown in this report suggest that normal filamentous arrangement of actin is not essential for phagocytosis.
Phagocytosis is usually accompanied by respiratory burst activity. Recent evidence suggests that the respiratory burst oxidase is associated with the cytoskeleton.38 Studies have shown that fibronectin has an adverse effect on superoxide production by neutrophils and macrophages compared with monocytes. Precoating microwells with fibronectin significantly suppressed neutrophil and macrophage release of O2− in response to fMLP while moderately enhancing O2−production by monocytes.39 Our study indicates that jaspamide did not affect O2− production by HL-60 cells or monocytes. Possible explanations for these observations are either that the residual actin filaments that are not clustered could participate in the oxidative burst process or, in contrast, that the clustered actin can provide the functional support. A remote possibility is that actin is not obligatory for the oxidative burst and other cytoskeletal components can replace its function.
In summary, the present study shows that jaspamide has a potent antiproliferative effect on HL-60 cells and that it induces phenotype differentiation. This antiproliferative activity correlates with reorganization of the actin cytoskeleton with no significant effect on phagocytic activity of HL-60 cells or monocytes. Taken together, our previous10 and current studies suggest that jaspamide may be considered as a novel therapeutic agent for the treatment of acute myeloid leukemia patients.
ACKNOWLEDGMENT
The authors thank Rina Socher for excellent technical assistance in conducting the electron microscopy studies and Michal Firon for reviewing the article.
Supported in part by the Tel Aviv University Basic Research Fund and the Israel Science Foundation founded by the Israel Academy of Sciences & Humanities.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Ina Fabian, PhD, Department of Cell Biology and Histology, Sackler School of Medicine, University of Tel Aviv, Ramat Aviv, Tel Aviv 69978, Israel.

![Fig. 4. Jaspamide induces increase in total cellular actin and de novo actin synthesis in HL-60 cells. (Aa) Coomassie-stained 12% SDS-PAGE gel showing total cellular protein of HL-60 cells exposed for 48 hours to DMSO 0.01% (lane 1) or jaspamide 10−7 mol/L (lane 2). (Ab) Dose-dependent actin accumulation: HL-60 cell extracts obtained from cells exposed for 24 hours to medium (lane 1), DMSO 0.01% (lane 2), jaspamide 10−7 mol/L (lane 3), jaspamide 10−8 mol/L (lane 4) were resolved on SDS-PAGE, and analyzed by Western blot using anti-actin MoAb. (B) Time course of actin accumulation. HL-60 cell extracts obtained from cells exposed to medium (lane 1), DMSO 0.01% (lanes 2, 4, and 6), or jaspamide 10−7 mol/L (lanes 3, 5, and 7) for 1 hour (lanes 2 and 3), 4 hours (lanes 4 and 5), or 24 hours (lanes 6 and 7) were resolved on SDS-PAGE, and analyzed by Western blot using anti-actin MoAb. (C) Effect of jaspamide on soluble and insoluble actin: Soluble, insoluble, and total actin fractions were prepared as described in Materials and Methods. (a) Coomassie blue and (b) immunoblot with anti-actin MoAb studies were performed (the lanes indicated are for both methods; ie, Fig 4Ca and Fig 4Cb). HL-60 cells treated with DMSO (lanes 1 through 3) or jaspamide 10−7mol/L for 24 hours (lanes 4 through 6) were examined for total (lanes 1 and 4) soluble (lanes 2 and 5) insoluble (lanes 3 and 6) actin. (D) De novo actin synthesis. (a) Effect of jaspamide on de novo actin synthesis. [S35]-cystein-methionine-labeled HL-60 cells were exposed to medium (lane 1), DMSO (lane 2), or to jaspamide 10−7 mol/L (lane 3) for 1 hour, immunoprecipitated with rabbit anti-actin Ab, and the immunoprecipitates were resolved on SDS-PAGE and exposed to Fuji X-ray film. (b) Coomassie-stained SDS-PAGE of HL-60 cells treated for 1 hour with DMSO 0.01% (lanes 1 through 3) or jaspamide 10−7 mol/L (lanes 4 through 6) showing total (lanes 1 and 4) soluble (lanes 2 and 5) and insoluble (lanes 3 and 6) actin.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/11/10.1182_blood.v93.11.3994/4/m_blod41139004w.jpeg?Expires=1769094234&Signature=3dxt2KbjmmP6AyWND~TVXpNaPsUiVdWZs5XBmpdIQ18-gXS9vWDWcoBGNOEUrW~PsfypN-VMl5q6prGGzbko5nwSnTax~BehFVfhYQmGTDyN2gVKp-h83Wbkk0av6PCVvZFU3IsP6akMIzjSwR2pk-pSb3ML~bCPPGueHWZsBoO~rezQgRKFSPn5u~yeR6--18IfEu-c1YoOTZqXeyZNYVim1SEqjf-y1FX8QvfDbR755QuxMEHOVMz4OhTM8kJnmJqb9oBe4AngkC1H6W3psMQpW0ps4X~4P3nQ6qlQEjvwIbfU873oPvLxUNSeYi14p13-brnDkAEXMwNuj3oywA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal