Abstract
The transcription factors of the Myc/Max/Mad network are important regulators of cell growth, differentiation, and apoptosis and are frequently involved in tumor development. Constitutive expression of v-Myc blocks phorbol ester (TPA)-induced differentiation of human U-937 monoblasts. However, costimulation with interferon-γ (IFN-γ) and TPA restores terminal differentiation and G1cell-cycle arrest despite continuous expression of v-Myc. The mechanism by which TPA + IFN-γ counteract v-Myc activity has not been unravelled. Our results show that TPA + IFN-γ treatment led to an inhibition of v-Myc– and c-Myc–dependent transcription, and a specific reduction of v-Myc:Max complexes and associated DNA-binding activity, whereas the steady state level of the v-Myc protein was only marginally affected. In contrast, TPA + IFN-γ costimulation neither increased the expression of Mad1 or other mad/mnt family genes nor altered heterodimerization or DNA-binding activity of Mad1. The reduced amount of v-Myc:Max heterodimers in response to treatment was accompanied by partial dephosphorylation of v-Myc and c-Myc. Phosphatase treatment of Myc:Max complexes lead to their dissociation, thus mimicking the effect of TPA + IFN-γ. In addition to modulation of the expression of Myc/Max/Mad network proteins, posttranslational negative regulation of Myc by external signals may, therefore, be an alternative biologically important level of control with potential therapeutic relevance for hematopoietic and other tumors with deregulated Myc expression.
THE MYC FAMILY of proto-oncogenes plays an important role in the regulation of cell proliferation, differentiation, and apoptosis. c-myc expression is normally tightly linked to the proliferative state of cells and its product is suggested to be one of the key nuclear transmitters of mitogenic signals. Constitutive c-Myc expression results in enforced cell-cycle progression in the absence of mitogens, blocks differentiation of various cell types, and may induce apoptosis if survival factors are lacking.1 Chromosomal aberrations involving mycfamily loci have been implicated in the generation of a variety of tumors in vertebrates, in particular within the hematopoietic system and are often strongly correlated to a poor prognosis.2
Myc proteins are transcription factors of the basic region (b)/helix-loop-helix (HLH)/leucine zipper (Zip)-family. The HLHZip motif of Myc mediates dimerization with the bHLHZip protein Max,3-5 an interaction that seems necessary for the biological activities of c-Myc.6-9 The basic regions enable Myc:Max heterodimers to bind specifically to a subclass of E-box DNA elements,10-12 whereas the N-terminal transactivation domain of c-Myc mediates stimulation of E box–driven promoters.6,7,13-15 Suggested target genes of c-Myc/Max complexes include α-prothymosine, ODC, cdc25A, eIF-2α, eIF-4E, CAD, and MrDb.16
In addition to the interaction with c-Myc, Max has recently been shown to form heterodimers with a number of other bHLHZip proteins, including Mad1, Mxi1, Mad3, Mad4, of the Mad family,17-19 and Mnt/Rox.20,21 These heterodimers bind to the same E-box sequence as Myc:Max complexes, and Mad/Mnt may, therefore, compete with Myc for binding to Max and/or to DNA. The Mad/Mnt proteins have been shown to inhibit cell growth22-24 and to repress transactivation and transformation by Myc.17,19,20,22,25-28These activities require an interaction with the repressor protein mSin3 and associated proteins including N-CoR/SMRT and the histone deacetylases HDAC1/HDAC2,29 suggesting that Mad/Mnt repress transcription at least, in part, through remodeling of chromatin.
The mad genes seem to be expressed primarily in differentiated, nonproliferative tissues.18,19,28,30-34 These observations have lead to the hypothesis that the Myc/Max/Mad network may constitute a molecular switch in which the prevalence of Myc-containing versus Mad-containing heterodimers determines whether cells enter a differentiation pathway or remain in a proliferative, undifferentiated state. This view has been further supported by recent observations that ectopic expression of Mad1 promotes differentiation of murine erythroleukemia cells,35 whereas targeted disruption ofmad1 inhibits cell-cycle exit and delays terminal differentiation of granulocytic precursor cells.36mxi1-deficient mice exhibit abnormalities in the homeostasis of several differentiated organs and increased cancer susceptibility.37
It is generally believed that the main regulation of the Myc/Max/Mad network occurs at the level of the expression of its components. Although posttranslational regulation in response to cellular signaling has been described for many other transcription factors,38this has not been clearly established for the Myc/Max/Mad proteins, and therefore remains an open question. With the U-937 monocytic differentiation model we have previously shown that interferon-γ (IFN-γ), if combined with classical inducers of differentiation such as the phorbol ester TPA, restores differentiation and G1cell-cycle arrest in v-Myc–transformed U-937 cells, despite a continuous expression and nuclear localization of v-Myc.39The mechanism by which TPA + IFN-γ costimulation counteracts the activity of Myc has not been unravelled. We hypothesized that this treatment could either increase the expression or activity of members of the mad-family and, thereby, restore the balance within the Myc/Max/Mad network or directly inhibit the activity or downregulate the steady state level of v-Myc.
Our results suggest that TPA + IFN-γ costimulation does not increase the expression of mad-family genes or the activity of Mad1, but interferes with Myc function as demonstrated by inhibition of the transactivating and DNA-binding ability of v-Myc and c-Myc. This likely occurs through the observed destabilization of Myc:Max heterodimers, a mechanism that is correlated with modification of Myc by dephosphorylation. Therefore, our findings provide evidence for an alternative signal-mediated pathway to interfere with Myc function in addition to the Mad/Mnt family of proteins, which potentially may have therapeutical relevance for hematopoietic and other tumors with deregulated c-Myc expression.
MATERIALS AND METHODS
Cell culture and differentiation assays.
U-937 human histiocytic lymphoma cells40 were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum (FCS) and antibiotics. BK3A chicken bursal lymphoma cells41 were grown in Dulbecco’s modified Eagle’s medium containing 5% FCS, 1% chicken serum, and 10% tryptase broth. The U-937 clones U-937-myc-6 (expressing the OK10 v-myc gene), U-937-neo-6 (lacking the v-myc gene), the parental clone U-937-GTB, and the U-937-1 clone have been described previously.31,42 Exponentially growing cells (105/mL) were induced to differentiate in medium containing 1.6 × 10−8 mol/L TPA (Sigma, St Louis, MO) and/or 100 U/mL IFN-γ (generously provided by Dr G.R. Adolf, Ernst-Boehringer Institute, Vienna, Austria). Differentiation was defined by the expression of mature monocytic antigens; the adhesion molecules (α-integrins) CD11a/LFA-1α, CD11b/Mac1, and CD11c/p150.95, and the receptor for monomeric IgG, FcγRI. Also CD4, which is expressed on immature U-937 cells and downregulated on differentiation, was analyzed. 3H-thymidine incorporation and the immunofluorescence studies were performed as described previously.39 For the analysis of CD4, CD11c, CD11a, CD11b, and FcγRI the monoclonal antibodies (MoAbs) OKT4 and LeuM5 (Becton Dickinson, Mountain View, CA), MHM24, 44, and 32.2 (from the IVth Leukocyte Typing Workshop collection43) were used respectively.
Immunoprecipitations and Western blot analysis.
For 35S and 32P in vivo cell labeling 5 × 106 cells were labeled for 40 minutes in 1 mL of methionine-free RPMI-1640 containing 0.15 mCi of35S-methionine or for 2 hours in 1 mL of phosphate-free RPMI-1640 containing 0.35 mCi of 32P-orthophosphate, respectively. For high and low stringency precipitations, cells were lysed in AB and Tris lysis buffer, respectively33 and immunoprecipitated with specific antibodies. An equal number of TCA-precipitable counts (for 35S-labeled proteins) or aliquots of lysates with equal amounts of protein (unlabeled proteins) were used for each sample. The washing procedure for high and low stringency immunoprecipitations has been described.33 The samples were analyzed on 10% to 15% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gels.
For phosphatase treatment, L-buffer lysates of 35S-labeled cells were immunoprecipitated with antibodies whereafter the immunocomplexes were washed in L-buffer and AP buffer (100 mmol/L Tris pH 8, 50 mmol/L MgCl2, 1% aprotinin) before treatment with 5 U alkaline phosphatase (Sigma) at 30°C for 20 minutes either in the presence or absence of 100 mmol/L β-glycerophosphate. The immunocomplexes were then extracted twice with 200 μL L-buffer and the proteins analyzed by SDS-PAGE.
The immunoblot and immunostaining procedure were performed as described previously.33 The blots were developed by enhanced chemiluminescence (Amersham Pharmacia Biotech, Uppsala, Sweden) using horseradish peroxidase (HRP) conjugated anti-rabbit Ig (Amersham) for rabbit antisera or streptavidin conjugated HRP (Amersham) for biotinylated antibodies.
For detection of Myc, IG-13, a rabbit pan-Myc antiserum generated by immunizing with a bacterially produced full-length human c-Myc protein (I. Guzhova, L.-G. Larsson, unpublished data, April 1993), 5042, a rabbit antiserum specific for chicken v- and c-Myc (kindly provided by Dr S. Hann and Dr R.N. Eisenman) and C-33 monoclonal pan-Myc antibodies (Santa Cruz Biotechnology [SCB], Santa Cruz, CA) were used. The C-33 MoAb was conjugated with 10 μg biotin-X-NHS (Calbiochem-Novabiochem, La Jolla, CA) per milligram antibody in 0.1 mol/L NaHCO3 pH 7.4, 0.1 mol/L NaCl for 1 hour at room temperature (RT) followed by dialysis in phosphate-buffered saline (PBS). For analysis of Mad1 and Max, 266-4 rabbit Mad1 antiserum,33 C-19 rabbit Mad1 antiserum (SCB), 91-4 rabbit Max antiserum,33 and C-17 rabbit Max antiserum (SCB) were employed.
Transfections and assays for reporter activity.
For electroporation, 2 × 107 cells in 0.5 mL of RPMI-1640 were mixed with the DNA in a 0.4 cm electroporation cuvette (Bio-Rad, Laboratories, Richmond, VA), incubated for 5 minutes at RT and electroporated at 960 μF, 300 V in a Bio-Rad gene pulser. After 10 minutes on ice, the cells were suspended in RPMI-1640, 10% FCS at a concentration 0.3 × 106 cells/mL and cultured for 16 to 24 hours. For differentiation experiments, the transfected cells were split into aliquots, which were treated with the differentiation agents as described above. The harvested cells were washed in PBS, suspended in 1 mL of TEN (40 mmol/L Tris pH 7.5, 1 mmol/L ethylenediamine tetraacetate (EDTA), 150 mmol/L NaCl) and left on ice for 10 minutes. After centrifugation, the cells were resuspended in 200 μL of 0.25 mol/L Tris pH 7.8, lysed by freeze thawing, and then the lysate was clarified by centrifugation. For CAT-assays an equal amount of protein per sample was incubated with 1 μCi 14C-chloramphenicol (Sigma), 0.2 mg/mL butyryl-CoA (Sigma) in 100 μL at 37°C for 1 hour. After extraction with mixed xylenes (Sigma) the amount of acetylated 14C-labeled chloramphenicol was determined by scintillation counting. For luciferase assays, cells were lysed in a buffer containing 25 mmol/L Tris pH 7.8, 2 mmol/L EDTA, 10% glycerol, 1% Triton X-100, and 2 mmol/L DTT for 10 minutes on ice. Extracts were mixed with assay buffer (25 mmol/L glycyl glycine, 15 mmol/L MgSO4, 5 mmol/L adenosine triphosphate), then 50 μL of 25 μmol/L luciferin was injected and the light emission was measured for 3 seconds by using a Lumat LB 9501/16 Berthold luminometer (EG&G Berthold, Bad Wildbad, Germany). For β-gal assays extracts were mixed with 300 μL of Z buffer (60 mmol/L Na2HPO4, 40 mmol/L NaH2PO4, 10 mmol/L KCl, 1 mmol/L MgSO4) after which 100 μL of O-nitrophenyl β-D-galactopyranoside (ONPG; Sigma) (4 mg/mL in Z buffer) was added. The solution was then incubated at 37°C until the yellow color appeared after which the reaction was stopped with 250 μL of 1 mol/L Na2CO3. The absorbance was measured at 414 nm in a Titertek Multiscan MCC/340.
The following DNA constructs were used in the transient transfections: PrT-CAT contains a 3.1 kb fragment including 250 bp of the α-prothymosine promoter, exon 1, intron 1, and part of exon 2 of the α-prothymosine gene linked to a CAT reporter gene. The E-box situated in the first intron is replaced by two Gal4 binding sites in the GalmE-PrT-CAT construct.44 The two constructs were kindly provided by Dr M. Eilers. m4mintk-Luc and mintk-Luc contains a minimal tk promoter (−32/+51) in front of a luciferase reporter gene with or without a tetramer of the CMD oligonucleotide upstream of the tk promoter.45 pCMV-myc contains a full length c-myccDNA downstream of the cytomegalovirus (CMV)-promoter in the construct pEQ176P2. In pCMV-MycΔBR, the c-myc cDNA insert of pSPMycΔBR,7 lacking the basic region, was cloned into the CMV expression vector pCB6. CMV-Luc contains the luciferase gene driven by a CMV promoter. hubactp/lacZ was used to normalize the transfection efficiency and contains 4 kb of the human β-actin promoter linked to a lacZ reporter gene,46 kindly provided by Dr U. Lendahl. For stable transfections, 35 μg of linearized m4mintk-Luc DNA was cotransfected with 5 μg of pRSV-hygr, which was used as a selectable marker. Hygromycin (400 μg/mL) (Calbiochem Inc, San Diego, CA) was added 2 days after transfection and resistant clones were picked 3 weeks after transfection, expanded, and analyzed for the presence of the integrated construct by luciferase assays and polymerase chain reaction analysis.
DNA-binding assays.
The solid phase DNA-binding assay (SODA) was performed as described previously.33 Briefly, cells were lysed under low stringency conditions in L-buffer and equal amounts of protein were immunoprecipitated with antibody. The washed samples were incubated with 1 ng of 32P-labeled oligonucleotide in 30 μL of gelshift buffer containing 100 ng of salmon sperm DNA for 25 minutes at room temperature.33 After washing the amount of bound oligonucleotide was measured in a scintillation counter. The oligonucleotides used were CMD (5′-TCAGACCACGTGGTCGGG), which contains an optimal Myc/Max binding site (underlined) and CMM (5′-TCAGACCAGCTGGTCGGG) containing a mutated Myc/Max binding site (underlined).
Two-dimensional gel electrophoresis.
35S-labeled proteins were immunoprecipitated, collected, and washed as described above and incubated in 50 μL of 20 mmol/L Tris pH 8, 1 mmol/L EDTA, 0.3% SDS, 1% DTT at 95°C for 10 minutes and then 50 μL of 2D-sample buffer (9 mol/L Urea, 2% 2-mercaptoethanol, 2% Pharmalyte 3-10, 0.5% Triton X-100, BFB) was added. The samples were separated in the first dimension by isoelectric focusing with immobiline drystrips pH 3-10 (Pharmacia Biotech, Uppsala, Sweden) in a horizontal Multiphor II system (Pharmacia Biotech) followed by SDS-PAGE in the second dimension. For blocking experiments, the IG-13 pan-Myc antiserum was incubated with 10 μg of recombinant c-Myc protein for 30 minutes at RT. Phosphatase treatment was performed by incubating the washed immunoprecipitates with 150 U of λ-phosphatase (Biolabs, Beverly, MA) for 30 minutes at 30°C.
RESULTS
TPA + IFN-γ costimulation restores differentiation and growth arrest in v-Myc expressing U-937 human monoblasts.
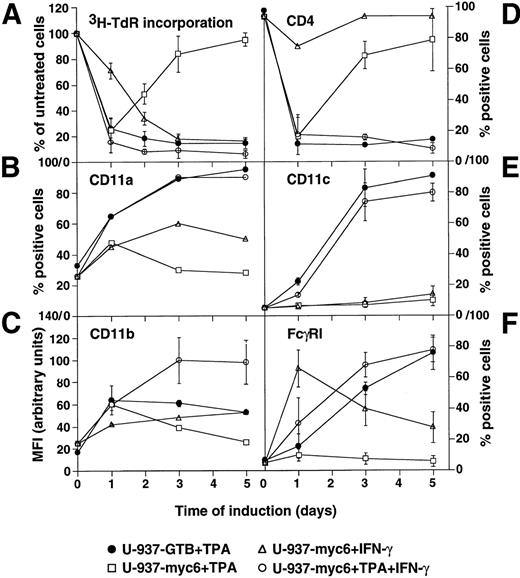
Growth and differentiation of v-Myc–expressing U-937-myc-6 and parental U-937-GTB cells in response to TPA, IFN-γ, or the combination of TPA + IFN-γ were determined by analysis of3H-TdR-incorporation and the expression of a number of differentiation-related surface antigens, respectively (Fig1). The incorporation of 3H-TdR declined during the first day of TPA-induced differentiation in both cell lines, but proliferation was gradually regained the following days in the v-Myc–expressing cells (Fig 1A). However, TPA + IFN-γ costimulation prevented the late increase in3H-TdR-incorporation in agreement with our previous report.39 IFN-γ alone reduced DNA synthesis with slower kinetics.
Growth and differentiation of v-Myc expressing U-937-myc-6 and parental U-937-GTB cells. Cells were induced by TPA, IFN-γ, or TPA + IFN-γ and analyzed at the indicated time points. (A) 3H-TdR incorporation. Cells were labeled with 10 μCi of 3H-TdR for 1 hour. (B) CD11a, (C) CD11b, (D) CD4, (E) CD11c, and (F) FcγRI expression. The surface antigen expression was measured by fluorescence-activated cell sorter analysis by using specific antibodies.
Growth and differentiation of v-Myc expressing U-937-myc-6 and parental U-937-GTB cells. Cells were induced by TPA, IFN-γ, or TPA + IFN-γ and analyzed at the indicated time points. (A) 3H-TdR incorporation. Cells were labeled with 10 μCi of 3H-TdR for 1 hour. (B) CD11a, (C) CD11b, (D) CD4, (E) CD11c, and (F) FcγRI expression. The surface antigen expression was measured by fluorescence-activated cell sorter analysis by using specific antibodies.
In contrast to the U-937-GTB cells, TPA-induced changes in the expression of the early differentiation markers CD11a, CD11b, and CD4 occurred only transiently in U-937-myc-6 cells, whereas the later markers CD11c and FcγRI were completely inhibited, indicating that the differentiation process was initiated normally but was subsequently aborted in parallel with resumed cell growth (Fig 1B through F). Costimulation with TPA + IFN-γ restored the expected changes in the expression of all five differentiation markers analyzed. IFN-γ alone influenced transiently the expression of some of the surface antigens, whereas others were unaffected.
In summary, TPA-induced differentiation and growth arrest, although initiated normally, was aborted in the U-937-myc-6 cells but was restored by TPA + IFN-γ costimulation. Because v-Myc is continuously expressed in these cells39 (see below), this result raises the question how TPA + IFN-γ costimulation might override the Myc-induced block of differentiation.
v-Myc is continuously synthesized and the expression of Mad1 is not restored in TPA + IFN-γ–treated U-937-myc-6 cells.
To investigate the synthesis of Mad1, c-Myc and v-Myc, and Max, the respective proteins were immunoprecipitated from35S-labeled U-937-myc-6 and U-937-GTB cell extracts with specific antibodies. In agreement with previous reports, the synthesis of Mad1 increased in response to TPA in U-937-GTB cells30,31 33 but not in U-937-myc-6 cells (Fig2A), indicating that the continued presence of v-Myc directly or indirectly influenced the expression of Mad1. The synthesis of v-Myc increased somewhat in the U-937-myc-6 cells, whereas the c-Myc synthesis was reduced after TPA stimulation in both cell lines. No major TPA-induced changes were observed in the expression of the p21 and p22 Max proteins. From these findings, one possibility is that TPA + IFN-γ might re-establish differentiation in v-Myc expressing U-937 cells by restoring the expression of Mad1 or by increasing the expression of other members of the Mad/Mnt family.
Myc, Mad, and Max protein synthesis in U-937-myc-6 and U-937-GTB cells. (A) U-937-myc-6 and U-937-GTB cells were treated with TPA for 3 days before 35S-methionine labeling. High stringency lysates were immunoprecipitated with pan-Myc (IG-13), Mad1 (C-19), and Max (C17) antibodies and analyzed by SDS-PAGE. (B) Time course of Myc, Mad1, and Max proteins synthesis during stimulation of U-937-myc-6 cells. TPA, IFN-γ, or TPA + IFN-γ were added for the time indicated and the proteins were analyzed as in (A).
Myc, Mad, and Max protein synthesis in U-937-myc-6 and U-937-GTB cells. (A) U-937-myc-6 and U-937-GTB cells were treated with TPA for 3 days before 35S-methionine labeling. High stringency lysates were immunoprecipitated with pan-Myc (IG-13), Mad1 (C-19), and Max (C17) antibodies and analyzed by SDS-PAGE. (B) Time course of Myc, Mad1, and Max proteins synthesis during stimulation of U-937-myc-6 cells. TPA, IFN-γ, or TPA + IFN-γ were added for the time indicated and the proteins were analyzed as in (A).
A detailed kinetic study showed that the synthesis of Mad1 increased during the first 4 hours in TPA- and TPA + IFN-γ–treated cells (Fig2B), but in contrast to the U-937-GTB cells (Fig 2A), returned to basal levels by 12 hours after both treatments. Northern blot analysis ofmad1 mRNA showed a similar pattern of expression (data not shown). The expression of mxi1 mRNA, which normally increases late after TPA treatment in U-937 cells,31 remained unchanged after TPA, IFN-γ, or TPA + IFN-γ treatment of U-937-myc-6 cells (data not shown). Furthermore, costimulation with IFN-γ did not increase the expression of mad3, mad4 or mntmRNA (data not shown). v-Myc was continuously synthesized, whereas synthesis of the endogenous c-Myc protein gradually decreased after TPA and TPA + IFN-γ treatment, but was less affected by IFN-γ alone (Fig 2B). The different treatments induced some transient changes in the synthesis ratio of the two Max proteins p21 and p22. Thus, we concluded that terminal differentiation induced by TPA + IFN-γ cannot be explained by a restored expression of Mad1 or by an increased mRNA expression of other mad/mnt-family genes.
TPA + IFN-γ inhibits Myc-regulated E-box–dependent reporter-gene activity.
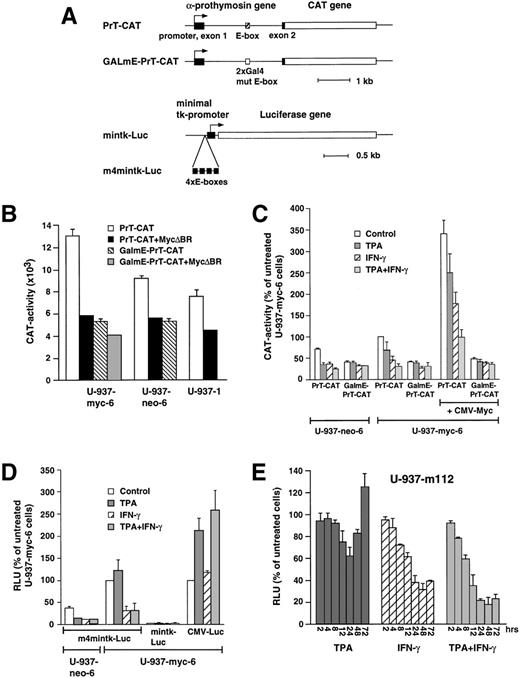
Because TPA + IFN-γ treatment neither reduced the synthesis of v-Myc nor increased the expression of Mad1 or other mad/mnt-family members we hypothesized that TPA + IFN-γ might affect the activity of Myc or Mad through post-translational mechanisms or alternatively bypass the Myc/Max/Mad network by affecting their downstream targets. We addressed this question by using the Myc-regulated α-prothymosine promoter and an artificial Myc-responsive promoter to determine whether the transactivating capacity of Myc could be modulated in response to TPA + IFN-γ. The Myc-responsive promoter/reporter gene constructs44,45 47 are depicted in Fig3A.
Regulation of E-box–dependent promoter/reporter activity during induced differentiation of U-937 cells. (A) Schematic presentation of the reporter constructs. A detailed description of the constructs is given in Materials and Methods. (B) Repression of basal E-box–dependent PrT-CAT activity by a dominant negative c-Myc mutant. 20 × 106 cells of the indicated U-937 clones were electroporated with 20 μg of Pr-T-CAT or GalmE-PrT-CAT with or without 30 μg of pCMV-myc▵BR. CAT activity was determined 16 hours after transfection. CAT or luciferase activities (B through D) were normalized to β-gal activity by cotransfection with 20 μg hubactp/lacZ. (C) PrT-CAT activity during differentiation of U-937 clones. PrT-CAT or GalmE-PrT-CAT were electroporated with or without 5 μg of pCMV-myc as described in (B). Cells from separate electroporations were pooled and divided into aliquots, which were treated with the inducers indicated. (D) m4mintk-Luc activity during differentiation of U-937 clones. The experiment was performed as in (C). As a reference, mintk-Luc, lacking E-boxes and pCMV-Luc, which contains the luciferase gene driven by a CMV-promoter, was used. (E) Kinetics of m4mintk-Luc activity during differentiation of U-937-m112 cells. U-937-m112 is a subclone of U-937-myc-6 containing a stably integrated m4mintk-Luc construct. The cells were induced by TPA, IFN-γ, or the TPA + IFN-γ for the indicated time points and assayed for luciferase activity. The results (C through E) are presented as percentage of untreated U-937-myc-6 cells. (B-E) The data of at least three independent experiments are summarized.
Regulation of E-box–dependent promoter/reporter activity during induced differentiation of U-937 cells. (A) Schematic presentation of the reporter constructs. A detailed description of the constructs is given in Materials and Methods. (B) Repression of basal E-box–dependent PrT-CAT activity by a dominant negative c-Myc mutant. 20 × 106 cells of the indicated U-937 clones were electroporated with 20 μg of Pr-T-CAT or GalmE-PrT-CAT with or without 30 μg of pCMV-myc▵BR. CAT activity was determined 16 hours after transfection. CAT or luciferase activities (B through D) were normalized to β-gal activity by cotransfection with 20 μg hubactp/lacZ. (C) PrT-CAT activity during differentiation of U-937 clones. PrT-CAT or GalmE-PrT-CAT were electroporated with or without 5 μg of pCMV-myc as described in (B). Cells from separate electroporations were pooled and divided into aliquots, which were treated with the inducers indicated. (D) m4mintk-Luc activity during differentiation of U-937 clones. The experiment was performed as in (C). As a reference, mintk-Luc, lacking E-boxes and pCMV-Luc, which contains the luciferase gene driven by a CMV-promoter, was used. (E) Kinetics of m4mintk-Luc activity during differentiation of U-937-m112 cells. U-937-m112 is a subclone of U-937-myc-6 containing a stably integrated m4mintk-Luc construct. The cells were induced by TPA, IFN-γ, or the TPA + IFN-γ for the indicated time points and assayed for luciferase activity. The results (C through E) are presented as percentage of untreated U-937-myc-6 cells. (B-E) The data of at least three independent experiments are summarized.
Electroporation of the α-prothymosine promoter constructs PrT-CAT and GalmE-PrT-CAT (the latter lacking the Myc-responsive E-box) into three U-937 clones showed that the PrT-CAT activity was somewhat higher in the v-Myc–expressing U-937-myc-6 cells than in the U-937-1 or U-937-neo-6 control cells and that the activity of GalmE-PrT-CAT was reduced to 40% of that of PrT-CAT (Fig 3B). To estimate the contribution of Myc in these transactivations, a dominant negative c-Myc mutant lacking the basic region (pCMV-mycΔBR) was cotransfected. In all three cell lines, this resulted in repression of the basal activity of PrT-CAT to a level that corresponded to the level of activity obtained with GalmE-PrT-CAT (Fig 3B). Cotransfection of pCMV-mycΔBR had little effect on GalmE-PrT-CAT. These results indicate that Myc is the main contributor of E-box–dependent transcription of the α-prothymosine promoter in U-937 cells, which is in agreement with the conclusion drawn previously by using other cell types.44
Treatment of U-937-neo-6 cells lacking v-myc with TPA, IFN-γ and TPA + IFN-γ all reduced PrT-CAT activity, but had little effect on GalmE-PrT-CAT (Fig 3C). In U-937-myc-6 cells, TPA did not reduce the activity of PrT-CAT to the same extent as in U-937-neo-6 cells, but IFN-γ and in particular TPA + IFN-γ were efficient inhibitors. To further establish that TPA + IFN-γ inhibited the transactivating properties of c-Myc, PrT-CAT, and GalmE-PrT-CAT were cotransfected with a c–Myc-expression vector. pCMV-myc stimulated PrT-CAT activity 3 to 4-fold, but did not affect GalmE-PrT-CAT. IFN-γ alone and in particular TPA + IFN-γ inhibited pCMV–myc-induced reporter activity, whereas TPA alone resulted in little inhibition (Fig 3C). GalmE-PrT-CAT was unaffected under these conditions.
Similar results were obtained with an additional Myc-inducible promoter/reporter construct, m4mintk-Luc, driven by four copies of a Myc:Max E-box binding site upstream of a minimal tk promoter. The m4mintk-Luc reporter activity was 2 to 3-fold higher in U-937-myc-6 cells as compared with U-937-neo-6 cells (Fig 3D), presumably because of their constitutive expression of v-Myc, whereas the activity of the mintk-Luc construct, lacking the E-boxes, was low. TPA, IFN-γ, and TPA + IFN-γ all reduced the m4mintk-Luc activity in U-937-neo-6 to 30% to 40% of untreated cells. In contrast, the luciferase activity was unaffected or even increased in TPA-treated U-937-myc-6 cells, whereas both IFN-γ and TPA + IFN-γ reduced the activity to 30% of untreated cells (Fig 3D). The activity of mintk-Luc was not affected under these conditions. It is relevant to point out that in addition to Myc other E-box–binding transcription factors may contribute to the activity of m4mintk-Luc. To rule out that the treatments led to a general inhibition of transactivation, the activity of a CMV-promoter/reporter construct (pCMV-Luc) was assayed. The activity of pCMV-Luc (Fig 3D) and other promoters, such as a Rous Sarcoma Virus long terminal repeat and a long herpes simplex virus tk-promoter (data not shown) were rather enhanced by TPA and TPA + IFN-γ treatment and were not affected by IFN-γ alone.
To assess, whether the treatments would affect m4mintk-Luc activity in a chromatin environment, a U-937-myc-6 clone with stable integration of the m4mintk-Luc construct (U-937-m112) was used. Kinetic experiments showed that TPA treatment led to a reduced reporter activity during the first day of treatment, which increased again and had even exceeded that of untreated cells at 72 hours. In contrast, TPA + IFN-γ treatment led to a permanent inhibition of the reporter activity, which was down to 20% of control values by 24 hours. IFN-γ alone inhibited the reporter activity with a somewhat slower kinetic and to a lesser extent as the costimulation with TPA. Taken together these experiments suggest that TPA + IFN-γ, in particular, but also IFN-γ treatment alone inhibits transactivation by v-Myc and c-Myc.
TPA + IFN-γ costimulation decreases the DNA-binding activity of Myc.
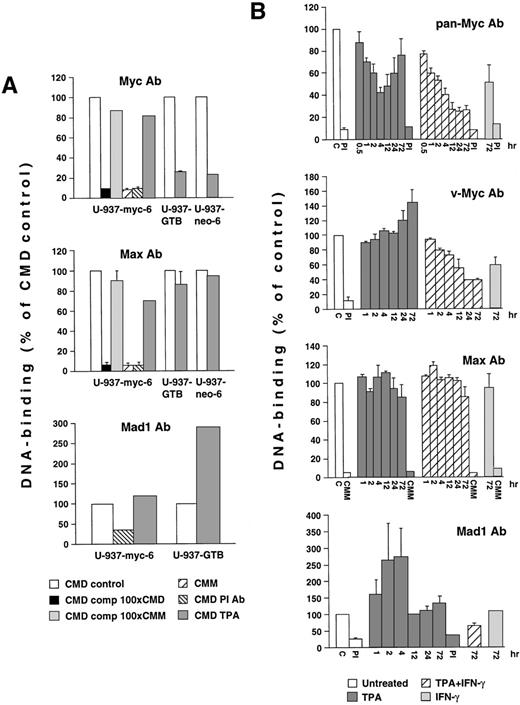
Several alternative mechanistic explanations can be envisioned for the ability of TPA + IFN-γ and IFN-γ to inhibit v-Myc and c-Myc activity as measured by transactivation assays. One possible explanation is that Myc is competed out from its DNA-binding sites by Mad/Mnt-family proteins or other E-box binding transcription factors or alternatively that TPA + IFN-γ directly regulate the function of the transactivation, DNA binding or heterodimerization domains of Myc. Because the activity of a c-Myc–transactivation domain/Gal4 fusion protein did not seem to be affected by TPA + IFN-γ in transient transactivation assays using reporter constructs driven by Gal4 DNA binding sites (data not shown), we measured the capacity of native Myc, Max and Mad1-complexes to bind specifically to DNA in response to TPA + IFN-γ treatment. For this purpose, we used SODA (solid-phase DNA-binding assay), which we recently developed based on partial purification of native Myc/Max/Mad complexes.33Unfortunately, the conventional electrophoretic mobility shift assay can not be applied for these types of studies as previously discussed.33
Low stringency immunoprecipitations of Myc-, Mad1-, and Max-complexes from v-Myc-expressing U-937 and control (U-937-GTB and U-937-neo-6) cells were performed by using antisera that do not interfere with dimerization or DNA binding. The immunocomplexes were incubated with labeled oligonucleotides containing a Myc/Max binding site (CMD) or a mutated version of CMM as described.33 Figure4A shows that α-Max or α-Myc immunocomplexes bound specifically to the CMD oligonucleotide, because the binding was competed with unlabeled CMD but not CMM oligonucleotides and only background levels were detected with the preimmune serum or labeled CMM. The DNA-binding activity of α-Myc immunoprecipitates decreased after TPA-induced differentiation of U-937-GTB and U-937-neo-6 control clones, but remained high in v-Myc expressing U-937-myc-6 cells. Smaller variations in DNA binding were seen in the α-Max immunoprecipitations. The DNA binding of Mad1 immunocomplexes was low in untreated cells but increased after TPA-stimulation of U-937-GTB but not in U-937-myc-6 cells. The DNA-binding activity measured seemed to correlate with the expression levels of the respective proteins.
Analysis of the DNA-binding activity of Myc, Mad1, and Max complexes during induced differentiation of U-937 cells. (A) Lysates from untreated U-937-myc-6, U-937-neo-6, and U-937-GTB cells or cells induced by TPA for 3 days were subjected to SODA.33The lysates were immunoprecipitated under low stringency conditions by using specific antisera and incubated with 32P-labeled CMD oligonucleotide, containing a Myc/Mac binding site or a mutated version (CMM) after which the amount of bound oligonucleotide was measured. The specificity was determined by competition with an excess of cold CMD or CMM oligonucleotide and by using preimmune (PI) serum. (B) Kinetic study of the DNA binding of total Myc-, v-Myc-, and total Max-containing and Mad1-containing complexes during differentiation of U-937-myc-6 cells. The cells were induced with TPA, IFN-γ, or the TPA + IFN-γ for the indicated time and subjected to SODA as in (A). The antisera used in (A) and (B) were IG-13 (anti-pan-Myc), 5042 (anti-chicken Myc), 266-4 (anti-Mad1) or 91-4 (anti-Max). The results are presented as a percentage of untreated cells. The error bars represent the standard deviations of the means from at least three independent experiments.
Analysis of the DNA-binding activity of Myc, Mad1, and Max complexes during induced differentiation of U-937 cells. (A) Lysates from untreated U-937-myc-6, U-937-neo-6, and U-937-GTB cells or cells induced by TPA for 3 days were subjected to SODA.33The lysates were immunoprecipitated under low stringency conditions by using specific antisera and incubated with 32P-labeled CMD oligonucleotide, containing a Myc/Mac binding site or a mutated version (CMM) after which the amount of bound oligonucleotide was measured. The specificity was determined by competition with an excess of cold CMD or CMM oligonucleotide and by using preimmune (PI) serum. (B) Kinetic study of the DNA binding of total Myc-, v-Myc-, and total Max-containing and Mad1-containing complexes during differentiation of U-937-myc-6 cells. The cells were induced with TPA, IFN-γ, or the TPA + IFN-γ for the indicated time and subjected to SODA as in (A). The antisera used in (A) and (B) were IG-13 (anti-pan-Myc), 5042 (anti-chicken Myc), 266-4 (anti-Mad1) or 91-4 (anti-Max). The results are presented as a percentage of untreated cells. The error bars represent the standard deviations of the means from at least three independent experiments.
A kinetic analysis of the DNA binding in response to TPA, TPA + IFN-γ, and IFN-γ treatment in U-937-myc-6 cells using pan-Myc antiserum showed that the DNA-binding of total v-Myc + c-Myc immunocomplexes rapidly declined after TPA treatment but increased again at 24 hours, reaching 75% of the value of untreated cells by 72 hours (Fig 4B). In TPA + IFN-γ–treated cells the total Myc DNA binding continued to decline past 4 hours and reached a minimum by 24 hours and thereafter. IFN-γ alone also reduced Myc DNA binding but to a lesser extent. Experiments using a v-Myc-specific antiserum showed that the v-Myc DNA-binding activity did not change substantially early after TPA stimulation but increased at later time points. In TPA + IFN-γ–treated cells, however, the v-Myc DNA binding gradually decreased with the lowest values at 24 and 72 hours. The v-Myc DNA binding was to a lesser extent reduced also after treatment with IFN-γ alone.
Only slight variations in the DNA-binding of α-Max immunocomplexes were observed in response to any of the treatments (Fig 4B). These complexes, which contain at least Myc and Mad1 (Fig5B and ref 33), presumably represent the sum of all different Max-containing heterodimers as well as of Max:Max homodimers. In contrast, the DNA binding of the Mad1-containing immunocomplexes increased transiently within 1 hour after TPA treatment with maximal activity 4 hours after stimulation but returned to control levels at late time points (Fig 4B). IFN-γ and TPA + IFN-γ treatment for 72 hours did not increase the binding above the basal level. In summary, the inhibition of Myc-regulated, E–box-dependent transactivation is paralleled by a decrease in Myc-specific DNA binding in response to TPA + IFN-γ treatment in U-937-myc-6 cells.
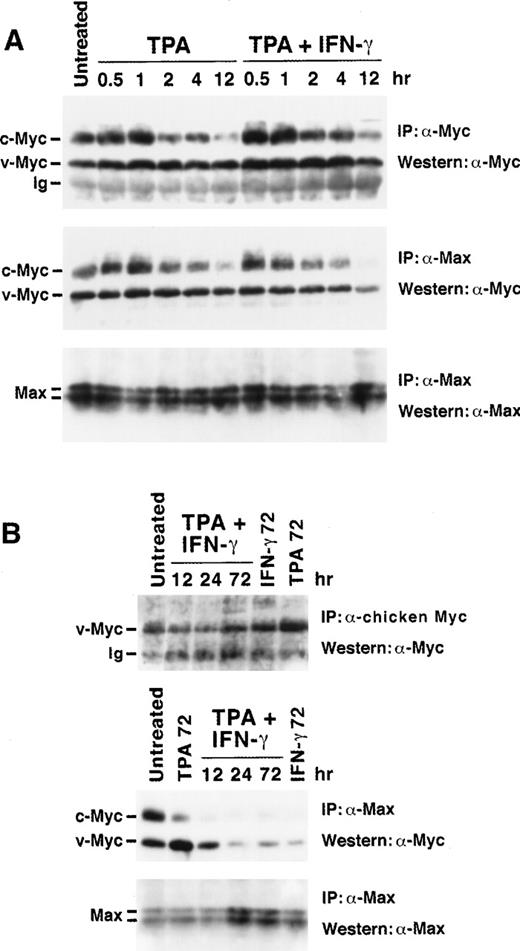
Western blot analysis of total Myc and Myc:Max complexes during induced differentiation of U-937-myc-6 cells. (A) Early kinetics of TPA- and TPA + IFN-γ–stimulation for the times indicated. (B) Late kinetics of TPA- and/or IFN-γ–stimulation. Upper panels: analysis of total Myc expression. (A) v- plus c-Myc and (B) v-Myc were immunoprecipitated from low stringency lysates by rabbit pan-Myc antibodies (IG-13) or rabbit chicken Myc-specific antibodies (5042), respectively, followed by Western analysis using monoclonal pan-Myc antibodies (C-33). Middle and lower panels: analysis of Myc:Max complexes and Max expression, respectively. Aliquots of the same lysates as in the upper panels were immunoprecipitated by using C17 Max antibodies followed by Western analysis of coimmunoprecipitated Myc (middle panel) and of Max (lower panel). Equal amounts of protein were immunoprecipitated for each lane. The figure shows representative results from 1 of 4 independent experiments.
Western blot analysis of total Myc and Myc:Max complexes during induced differentiation of U-937-myc-6 cells. (A) Early kinetics of TPA- and TPA + IFN-γ–stimulation for the times indicated. (B) Late kinetics of TPA- and/or IFN-γ–stimulation. Upper panels: analysis of total Myc expression. (A) v- plus c-Myc and (B) v-Myc were immunoprecipitated from low stringency lysates by rabbit pan-Myc antibodies (IG-13) or rabbit chicken Myc-specific antibodies (5042), respectively, followed by Western analysis using monoclonal pan-Myc antibodies (C-33). Middle and lower panels: analysis of Myc:Max complexes and Max expression, respectively. Aliquots of the same lysates as in the upper panels were immunoprecipitated by using C17 Max antibodies followed by Western analysis of coimmunoprecipitated Myc (middle panel) and of Max (lower panel). Equal amounts of protein were immunoprecipitated for each lane. The figure shows representative results from 1 of 4 independent experiments.
TPA + IFN-γ costimulation reduces the amount of Myc:Max complexes.
The results above suggest that the inhibition of Myc activity by TPA + IFN-γ is at least, in part, because of an effect on the DNA-binding capacity of Myc itself rather than increased competition from Mad1 or other E–box-binding transcription factors at the Myc/Max binding site or interference with the transactivation domain of Myc. The reduced DNA-binding activity of Myc-containing complexes could be due to a reduced steady state level of v-Myc, to an inhibition of Myc DNA binding per se, or to reduced Myc:Max complex formation or stability. To address these questions we measured the steady state levels of v-Myc, c-Myc, and Max, and the capacity of Myc and Mad1 to heterodimerize with Max in response to treatment.
Coimmunoprecipitation studies were performed under low stringency conditions followed by Western blot analysis of coimmunoprecipitated proteins. The total amount of v-Myc, c-Myc, and Max immunoprecipitated with chicken Myc-specific, pan-Myc, and Max antiserum, respectively, were compared with the amounts of Myc coimmunoprecipitated with Max antiserum (Fig 5). The amount of v-Myc:Max complexes (middle panel) and the total level of v-Myc (upper panel) was relatively stable during the first 4 hours of TPA and TPA + IFN-γ stimulation (Fig 5A). Thereafter the amount of v-Myc:Max complexes declined significantly after TPA + IFN-γ treatment reaching a minimum at 24 hours (six-fold reduction), whereas only a slight reduction in total v-Myc steady state level was observed (Fig 5B). IFN-γ alone also reduced the amount of v-Myc:Max as compared with total v-Myc. The reduction of v-Myc:Max complexes in response to TPA + IFN-γ or IFN-γ treatments was not caused by decreased steady state levels of Max as shown in the lower panels of Figs 5A and B. In contrast, both the steady state level of v-Myc and the amount of v-Myc:Max complexes increased 2 to 2.5-fold after 72 hours of TPA treatment. The steady state levels of c-Myc declined after TPA + IFN-γ or TPA treatment but not significantly after IFN-γ treatment as expected from the synthesis rates of c-Myc (Figs 2 and 5; data not shown). Few c-Myc:Max complexes were observed in TPA + IFN-γ and IFN-γ treated cells as compared with TPA-treated cells at 72 hours post induction (Fig 5B, middle panel). In conclusion, these results suggest that a potentially important consequence of the treatment with TPA + IFN-γ and with IFN-γ alone is the reduction in the fraction of total v-Myc and c-Myc, which is complexed with Max.
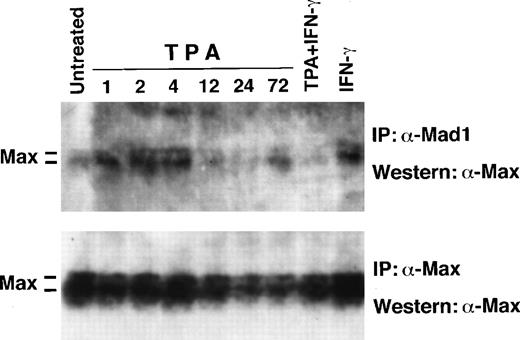
The amount of Max coimmunoprecipitated with Mad1 increased transiently after 1 to 4 hours of TPA induction, but then returned to basal levels (Fig 6), essentially reflecting the synthesis of Mad1 (Fig 2). No increase in the amount of Max in complex with Mad1 was observed late after TPA + IFN-γ treatment in agreement with the unchanged synthesis of Mad1. These findings argue against an increase in Mad1 protein due to stabilization or increased Mad1:Max affinity. The apparent increase in Max and Mad1:Max after IFN-γ treatment was because of overloading.
Western blot analysis of total Max and of Mad1:Max complexes during induced differentiation of U-937-myc-6 cells. U-937-myc-6 cells were induced with TPA, IFN-γ, or TPA + IFN-γ for the times indicated. (Upper panel) Max was coimmunoprecipitated from low stringency lysates using C19 Mad1 antibodies followed by Western blot analysis of Max by using C17 Max antibodies (Lower panel). Max was immunoprecipitated from aliquots of the same lysates with C17 Max antibodies followed by immunoblot analysis using the same antibodies.
Western blot analysis of total Max and of Mad1:Max complexes during induced differentiation of U-937-myc-6 cells. U-937-myc-6 cells were induced with TPA, IFN-γ, or TPA + IFN-γ for the times indicated. (Upper panel) Max was coimmunoprecipitated from low stringency lysates using C19 Mad1 antibodies followed by Western blot analysis of Max by using C17 Max antibodies (Lower panel). Max was immunoprecipitated from aliquots of the same lysates with C17 Max antibodies followed by immunoblot analysis using the same antibodies.
TPA + IFN-γ costimulation results in modification of Myc.
The data presented above suggest that the inhibition of Myc-induced transactivation and of the DNA-binding activity of Myc by TPA + IFN-γ is because of reduced Myc:Max heterodimerization. Destabilization of Myc:Max heterodimers could be the result of TPA + IFN-γ–induced unidentified proteins competing for Myc or Max, or direct modification(s) of Myc or Max by for instance phosphorylation/dephosphorylation.
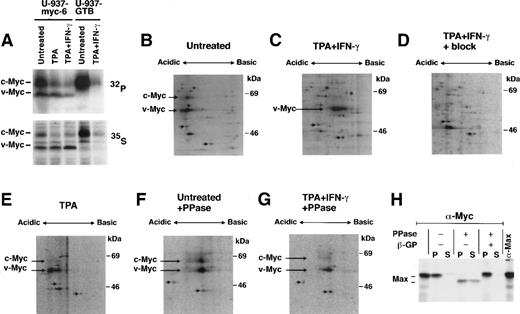
To assess whether the overall level of phosphorylation of v-Myc and c-Myc changed in response to TPA + IFN-γ treatment in U-937-myc-6 and U-937-GTB cells, aliquots of untreated and induced cultures were in vivo labeled with 35S-methionine and32P-orthophosphate in parallel, whereafter the Myc proteins were immunoprecipitated and analyzed by SDS-PAGE. Fig7A shows that the relative intensity of the32P-labeled v-Myc and c-Myc proteins were significantly reduced as compared to the corresponding 35S-labeled proteins in TPA + IFN-γ treated, but not in untreated or TPA-treated cells, indicating dephosphorylation of the Myc proteins in response to the former treatment. To substantiate this further we analyzed immunoprecipitated 35S-labeled Myc proteins from untreated and treated cells by isoelectric focusing followed by SDS-PAGE (Fig 7B-G). In response to TPA + IFN-γ v-Myc (long arrow) shifted towards the basic side as compared to marker spots (short arrows) (Fig 7C). To a lesser extent, IFN-γ alone also induced a shift toward a more basic pI (data not shown), whereas TPA did not (Fig 7E). A shift toward a basic isoelectric position might be caused by dephosphorylation. Therefore, we treated immunoprecipitated Myc from control and TPA + IFN-γ–treated cells with phosphatase and analyzed the products on 2D gels. The different Myc isoforms collapsed into one major species that migrated at a pI similar to the most basic isoform after TPA + IFN-γ treatment (Fig 7F and G). Together these findings suggest that Myc proteins are dephosphorylated on stimulation with TPA + IFN-γ. Because of the moderate level of Myc in these cells we have, unfortunately, not been able to obtain enough32P-incorporation into the proteins to perform 2D phosphopeptide analysis to establish which phosphorylation sites are affected in response to TPA + IFN-γ-stimulation. Presently, we are investigating whether costimulation with IFN-γ affects Myc function in a similar way in other cell types in which such studies may be easier to perform.
TPA + IFN-γ costimulation induces modification of Myc. (A) Aliquots of untreated or stimulated U-937-myc-6 and U-937-GTB cells were in vivo labeled with 32P-orthophosphate (upper panel) or 35S-methionine (lower panel) as indicated whereafter cell lysates were immunoprecipitated with IG-13 pan-Myc antiserum. (B through G) 35S-labeled lysates of untreated (B and F), TPA + IFN-γ- (C, D, and G) or TPA stimulated (E) U-937-myc-6 cells were immunoprecipitated with IG-13 pan-Myc antibodies and analyzed by 2D-gel electrophoresis as described in Materials and Methods. In (D) the antibody was blocked by incubation with recombinant c-Myc protein. In (F) and (G) the immunoprecipitates were phosphatase treated. The positions of v-Myc and c-Myc and of two unspecific spots are indicated by long and short arrows, respectively. (H) BK3A cells were labeled with 35S-methionine, lysed in L-buffer and Myc:Max complexes immunoprecipitated using Myc-specific antibodies (5042). Treatment with alkaline phosphatase (PPase) in the presence or absence of β-glycerophosphate (β-GP) was performed as indicated. Max extracted in L-buffer (S) or bound to Myc (P) was analyzed by SDS-PAGE and fluorography.
TPA + IFN-γ costimulation induces modification of Myc. (A) Aliquots of untreated or stimulated U-937-myc-6 and U-937-GTB cells were in vivo labeled with 32P-orthophosphate (upper panel) or 35S-methionine (lower panel) as indicated whereafter cell lysates were immunoprecipitated with IG-13 pan-Myc antiserum. (B through G) 35S-labeled lysates of untreated (B and F), TPA + IFN-γ- (C, D, and G) or TPA stimulated (E) U-937-myc-6 cells were immunoprecipitated with IG-13 pan-Myc antibodies and analyzed by 2D-gel electrophoresis as described in Materials and Methods. In (D) the antibody was blocked by incubation with recombinant c-Myc protein. In (F) and (G) the immunoprecipitates were phosphatase treated. The positions of v-Myc and c-Myc and of two unspecific spots are indicated by long and short arrows, respectively. (H) BK3A cells were labeled with 35S-methionine, lysed in L-buffer and Myc:Max complexes immunoprecipitated using Myc-specific antibodies (5042). Treatment with alkaline phosphatase (PPase) in the presence or absence of β-glycerophosphate (β-GP) was performed as indicated. Max extracted in L-buffer (S) or bound to Myc (P) was analyzed by SDS-PAGE and fluorography.
We hypothesized that this dephosphorylation might be the basis for the reduced interaction with Max and, thus, be the trigger for the inhibition of Myc-specific DNA binding and transactivation. To evaluate whether dephosphorylation might affect Myc:Max dimer stability, low stringency Myc immunoprecipitates of 35S-labeled cell lysates were phosphatase treated and the amount of Max that was either bound to Myc or free determined. After phosphatase treatment, Max had an increased mobility (because of dephosphorylation of C-terminal sites; B.L., unpublished data, October 1994) and a substantial portion was in the supernatant (Fig 7H). No release from Myc was observed after mock incubation or when the activity of the phosphatase was blocked. Although the dephosphorylation of Myc observed in vivo is only partial and no dephosphorylation of Max was seen (data not shown), these findings are consistent with the results discussed above; dephosphorylation of Myc in Myc:Max heterodimers leads to a decreased stability of the complex.
DISCUSSION
Constitutive expression of v-Myc blocks induced differentiation of human U-937 monoblasts similar to other cellular differentiation systems.42,48-53 However, costimulation with IFN-γ and TPA restores terminal differentiation and G1 cell-cycle arrest despite continuous expression of v-Myc39; Figs 1 and2). These findings suggest that the combination of TPA with IFN-γ can interfere with the growth promoting and differentiation blocking activities of Myc. We envisioned that these signals could counteract Myc by at least three different mechanisms. First, by increased expression or activity of members of the mad-family. Second, by acting on Myc directly through inhibition of its activity or through affecting its steady state level by a posttranslational mechanism. Third, by acting downstream of Myc; for instance, through independent regulation of Myc target genes.
Our results show that the induced expression of Mad1 and the subsequent increased formation of Mad1:Max heterodimers and associated DNA-binding activity observed normally during TPA-induced differentiation of U-937 cells30,31 33 occurred only transiently in v-Myc expressing cells (Figs 2, 4, and 6). This indicates a direct or indirect role of Myc in the regulation of the mad1 gene. Restored differentiation by TPA + IFN-γ was, however, not accompanied by an elevated level of Mad1 nor of its heterodimers with Max or its DNA-binding activity (Figs 2, 4, and 6). Furthermore, costimulation with IFN-γ did not increase the expression of mxi1,mad3, mad4 or mnt mRNA (data not shown). Together, the results argue against a model were TPA + IFN-γ restore differentiation and growth arrest by using the Mad/Mnt family to counteract Myc activity. Apparently, terminal differentiation of U-937 cells can proceed with only basal level of mad-family gene expression under these conditions. However, we cannot exclude post-transcriptional and/or posttranslational regulation of Mad/Mnt proteins, which potentially could contribute to restoring differentiation on TPA + IFN-γ treatment. It is also possible that Mad1 could play a role during the early phase of TPA- and TPA + IFN-γ–induced differentiation and growth inhibition, during which Mad1 is transiently induced.
Although we have no evidence supporting an involvement of themad/mnt-family, our results suggest that TPA + IFN-γ–induced signals restore differentiation and growth arrest by interfering directly with the activity of Myc proteins. We base this conclusion on three observations: (1) The Myc-regulated, E-box–dependent activity of two different promoter/reporter gene constructs is inhibited by TPA + IFN-γ, in particular, but also by IFN-γ alone (Figs 3C-E); (2) The DNA-binding activity of v-Myc and of total Myc (v-Myc + c-Myc) is reduced after TPA + IFN-γ treatment and to a lesser extent by IFN-γ treatment (Fig 4); (3) The fraction of total v-Myc, which interacts with Max, declines in response to TPA + IFN-γ and IFN-γ treatment (Fig 5).
The kinetics and the magnitude of the reduction in v-Myc:Max heterodimers in response to TPA + IFN-γ is similar to the kinetics and the magnitude of reduction in DNA-binding activity of Myc and of the inhibition of E-box–dependent promoter/reporter activity (Figs 3E,4, and 5). Because DNA binding and transactivation by Myc are dependent on heterodimerization with Max, it seems likely that the Myc:Max interaction is the main target of the TPA + IFN-γ-induced negative signals. Reduced Myc:Max heterodimerization could potentially be the result of increased competition for Max by other Mad/Mnt proteins than Mad1. Because no antibodies of high enough quality directed against these proteins were available, we were unable to address this question. However, we find this explanation unlikely because the expression of their respective genes remains unaltered. Our preliminary data suggest that these proteins are not abundant (F.B., L.-G.L., B.L., unpublished data, May 1998) and that the amount of Max in the cells does not seem to be limiting. Another possibility is that TPA + IFN-γ induce unidentified proteins binding Myc and replacing Max. Although this is an attractive hypothesis, we have at present no evidence supporting this theory. A third possibility, which we favor, is that Myc:Max heterodimerization is affected through modification(s) of Myc.
Our results show that TPA + IFN-γ–induced and IFN-γ–induced inhibition of Myc activity is correlated with direct modification of v-Myc and c-Myc as determined by their shift toward more basic isoelectric points in response to these treatments. Because phosphatase treatment of Myc immunoprecipitates from control and stimulated cells led to a similar pI it is likely that TPA + IFN-γ and IFN-γ treatments result in dephosphorylation of Myc, a conclusion supported also by the reduced intensity of 32P-labeled as compared with 35S-labeled v-Myc and c-Myc proteins after TPA + IFN-γ treatment. Both Myc and Max are phosphorylated by protein kinase CK2 near their basic regions.54-56 Mutations of these sites in Max have been reported to affect the DNA-binding activity of Max:Max homodimers.55,56 In addition, Myc contains CK2 phosphorylation sites in its central acidic region.54 This region seems to be dispensable for transformation of fibroblasts, but was reported to be essential for transformation of monocytic cells.57 A cluster of phosphorylation sites in the N-terminus of c-Myc has been suggested to be targeted by CDK/cyclin A/p107, MAPK and GSK3.58-62 Neither the CK2 phosphorylation sites of c-Myc and Max nor the N-terminal phosphorylation sites of c-Myc have, however, been implicated in regulation of the steady state of Myc:Max heterodimers, although this issue has not been specifically addressed previously. However, our results show that phosphatase treatment of Myc:Max complexes led to a release of Max (Fig 7H), providing direct evidence that the stability of Myc:Max heterodimers is sensitive to the phosphorylation status of Myc. At present, it is unclear whether the phosphorylation status of Max is relevant for the effects described above. We think this is rather unlikely because no evidence for altered phosphorylation in response to TPA + IFN-γ treatment has been obtained (data not shown). The phosphorylation site(s) of Myc that are targets for TPA + IFN-γ–induced dephosphorylation are not known presently and therefore their potential role for the inhibitory action of these signals and the phosphatase(s) involved remain to be elucidated.
Stimulation with IFN-γ in combination with TPA or IFN-γ alone arrests v-Myc–expressing U-937 cells in the G1 phase of the cell cycle and seems to affect v-Myc activity through the same mechanism, yet only the combination with TPA results in differentiation of the cells. The likely explanation for this is that downregulation or inactivation of Myc is a prerequisite but is not sufficient for differentiation of these cells and that TPA provides the necessary Myc-independent signals for the process to initiate.
In conclusion, our results suggest that TPA and IFN-γ costimulation restores differentiation and growth arrest in v-Myc-transformed U-937 cells by inhibiting Myc function. This most likely occurs primarily through destabilization of Myc:Max heterodimers, which is accompanied by and possibly involves modification of v-Myc and c-Myc by dephosphorylation. We can, however, not exclude that these signals may have other additional effects on growth and differentiation acting for instance downstream of Myc. Our conclusion challenges the generally held view that the Myc/Max/Mad network is essentially regulated through pretranslational control of the expression of its components. We suggest that a biologically relevant post-translation level of regulation of v-Myc and c-Myc is also in operation and that this level of regulation is modulated by external signals. This may be an additional safeguard mechanism to ensure inactivation of Myc function during differentiation and potentially other cellular processes as well as a mechanism to counteract Myc activity in response to oncogenic events resulting in deregulated Myc expression. Although the players involved in this regulation are yet to be identified, we believe that the present findings may form a basis for new ways to antagonize the activity of Myc in addition to the Mad/Mnt proteins. Therefore, it will be important in the future to determine whether the regulation of Myc function described here for monocytic cells is also valid in other malignant and normal hematopoietic as well as in other cell types. The identification of such alternative pathways of counteracting Myc are important both for the understanding of the function of the Myc-network but may potentially also be relevant for the treatment of hematopoietic and other tumors with deregulated Myc expression.
ACKNOWLEDGMENT
We thank Drs M. Eilers, R. Eisenman, S. Hann, and U. Lendahl for reagents. We also thank Dr G.R. Adolf, Ernst-Boehringer Institute, Vienna, for generous gifts of IFN-γ.
F.B. and S.W. contributed equally to this report.
Supported by grants from the Swedish Cancer Society, the Children Cancer Foundation of Sweden, the Lovisa & Thielman, the Selander and the Wiberg Foundations to L-G.L. and the Deutsche Forschungsgemeinschaft, the Deutsche Krebshilfe and the Fonds der Chemischen Industrie to B.L.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Lars-Gunnar Larsson, Uppsala Genetic Center, Department of Plant Biology, Swedish University of Agricultural Sciences, Box 7080, S-750 07 Sweden; e-mail: Lars-Gunnar.Larsson@vbiol.slu.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal