Stage-specific expression of the human β-like globin genes is controlled by interactions between regulatory elements near the individual genes and additional elements located upstream in the Locus Control Region (LCR). Elucidation of the mechanisms that govern these interactions could suggest strategies to reactivate fetal (γ) or embryonic (ɛ) genes in individuals with severe hemoglobinopathies. This study extends an earlier analysis of a transgenic construct, HS3ɛγ, testing: (A) the effect of substitution of HS2 for HS3 on stage-specific expression of the ɛ and γ genes and, (B) the role of an evolutionarily conserved YY1 binding site in transcriptional regulation of the γ gene. The data show that both HS3ɛγ and HS2ɛγ can individually support embryonic expression of ɛ and fetal expression of Aγ. Thus, the cis regulators of distinct expression patterns for ɛ and γ are likely to reside near the genes, rather than in specific hypersensitive sites of the LCR. Alterations in Aγ expression patterns observed in transgenic lines carrying a construct with a mutation in a conserved YY1 binding site at −1086 indicate that this site might function to facilitate active transcription of the γ gene in fetal life.
THE FIVE FUNCTIONAL β-LIKE globin genes are arranged on human chromosome 11 in an order (5′ ε-Gγ-Aγ-δ-β 3′) that parallels their pattern of expression during development. The ε gene is expressed in early embryonic life and tightly repressed at 6 to 8 weeks of gestation, concomitant with full activation of the two γ genes. At birth, a second transcriptional switch results in the silencing of the γ genes and the activation of the adult δ and β genes. Debilitating diseases result if this developmental program is not completed (failure to activate the β gene causes β thalassemia) or if the adult β gene is defective (a single point mutation results in a protein structural alteration that causes sickle cell anemia). One potential approach to the therapy of both of these diseases is based on reactivation of the silenced γ (or possibly ε) gene; but a thorough understanding of the specific molecular components that control ε and γ gene expression is required to achieve this goal.
Prevailing models of globin gene regulation embrace the notion that globin gene expression requires communication between the Locus Control Region, or LCR (a regulatory region spanning 22 kb upstream from the ε gene), and elements of the individual promoters.1Elegant studies using in situ hybridization with intronic probes have shown that only one promoter is active at a time; expression “flip-flops” between genes in a dynamic way.2 Because the activity of each promoter is facilitated by the LCR, the flip-flopping is thought to reflect a competition between promoters for interaction with the LCR such that the half-life of the promoter:LCR interaction dictates the relative level of expression of each gene.1 2
The mechanisms by which promoter:LCR communication governs hemoglobin switching are unknown and various models have been proposed, including: a looping model, in which distally located LCR elements contact promoters directly3; a binary model, in which the role of the LCR is to control chromatin structure such that transcription is either on or off4,5; and a tracking model, which proposes that intergenic transcripts that extend through the locus act to deliver Pol II or a portion of the necessary transcriptional machinery directly to the individual promoters.6 7
Although debate continues as to which of these models provides the most accurate mechanistic basis for hemoglobin switching, two practical questions can be addressed: (1) Are the distinct developmental expression patterns exhibited by each globin gene (eg, embryonic expression of ε and fetal expression of γ) determined by the LCR or by gene proximal elements, or both? (2) Which specific sequence elements within the promoters and/or LCR are important for the generation of these patterns? The work presented here addresses both of these issues.
First, we tested the ability of HS2 or HS3 alone to direct distinct expression patterns of a human ε gene (embryonic) and a linked human γ gene (embryonic and fetal) in transgenic mice. Previous studies had investigated function of these HS sites linked to γ and β genes,8 or to Gγ , Aγ , δ, and β genes.9 Here, we wished to analyze ε and γ expression in the absence of competition from β. Four independent transgenic lines were created using a construct, HS2εγ, in which a 1.9 kb fragment spanning HS2 is linked to the human ε and γ genes. Stage-specific ε and γ transgene expression patterns in these mice were compared with patterns observed in mice created earlier carrying a related construct, HS3εγ,10 in which the 1.9 kb region of HS3 replaces HS2. Though subtle differences in transgene expression profiles were observed, both HS2 and HS3 supported distinct expression patterns for ε (embryonic expression followed by silencing in fetal life) and γ (peak expression in fetal life and silencing thereafter). Taken together with the previous literature, these results indicate that gene-proximal elements are involved in the control of stage-specificity of the ε and γ globin genes.
Second, in a previous study, phylogenetic footprinting analysis in conjunction with transient transfection studies suggested that an evolutionarily conserved YY1 binding site located at −1086 bp upstream from the γ gene might be one of the cis elements important for γ gene silencing in adult life.11 YY1 is a ubiquitously distributed transcription factor of the zinc finger class that can act as both an activator and as a repressor, and in some cases, mediates a switch between activation and repression of gene expression (reviewed in Shi, Lee, and Galvin12). To study the role of this site in stage-specific regulation of the γ gene, a debilitating mutation was introduced into the −1086 YY1 binding site in the HS3εγ construct and five independent transgenic lines were generated. Contrary to expectation, examination of γ expression patterns in these mice suggested that this site might be involved in the activation of the γ gene in fetal life.
MATERIALS AND METHODS
Construction and mutagenesis of plasmid vectors.
The plasmid HS3εγ was described earlier,10 and contains a 1.9 kb HindIII fragment (HUMHBB 3267-5172) encompassing the core of HS3; a 3.8 kb EcoRI fragment (HUMHBB 7482-21251) containing the entire human ε gene with 2 kb of 5′ flanking sequence and 300 bp of 3′ flanking sequence; and two tandemHindIII fragments (3.3 and 0.7 kb, HUMHBB 38085-42135) containing the entire human Aγ gene with 1.2 kb of 5′ flanking sequence and 1.1 kb of 3′ flanking sequence, including the Aγ enhancer region.13 The ε and Aγ fragments of HS2εγ are identical to HS3εγ, but a 1.9 kb KpnI/PvuII fragment (GenBank entry HUMHBB 7765-9653) containing human LCR DNase I-hypersensitive site 2 (HS2) replaces HS3.
To alter the YY1 binding site at −1086 bp upstream from γ in the plasmid HS3εγ, the Unique Site Elimination (U.S.E.) method14 was used with slight modifications. An oligonucleotide (5′CTGGTGAGTATTCAACCAAGTC-3′) spanning a ScaI site in the ampicillin resistance gene of the vector pBlueScript II SK(−) was used as the selection oligonucleotide; this oligonucleotide encodes a mutation of theScaI site (underlined) that interferes with ScaI digestion but does not alter ampicillin resistance. An additional sense strand oligonucleotide containing a mutation at the −1086 YY1 binding site was used as the mutagenic primer: 5′-TAATGGTCCAAgcgGTCAGAAACAGCACTG-3′, in which the lower case letters indicate the site of a 3 bp mutation. The mutagenic and selection primers were annealed to the plasmid HS3εγ (in a 10:1 ratio of mutagenic primer to selection primer) and primer extension and ligation were accomplished as described previously.14 The resultant plasmid DNA was transformed into the repair-deficient strain BMH 71-18 (mutS). Bulk plasmid DNA isolated from an overnight culture grown in the presence of ampicillin (50 μg/mL) was transformed into DH5α. Bulk DNA isolated from this second overnight culture (with ampicillin) was digested with ScaI, and retransformed into DH5α. Plasmid DNA was isolated from single colonies appearing after growth on ampicillin-containing agar plates and the successful introduction of the ScaI mutation was tested by digestion withScaI. Undigested plasmids were sequenced to confirm that the YY1 mutation had also been introduced. In addition, to confirm that unwanted additional mutations had not been randomly introduced into key regulatory regions, the entire γ promoter (−1350 to +1) as well as the core of HS3 (HUMHBB 4214-5172) were sequenced in the mutant plasmid. No sequence alterations other than the targeted YY1 mutation were found.
Creation of transgenic mice.
Inserts of all three plasmids were released by digestion withKpnI and NotI. Insert fragments of 9,814 bp (HS2εγ) or 9,875 bp (HS3εγ and HS3εγ−1086mut) were gel isolated, dissolved in injection buffer (10 μmol/L Tris-HCl, pH 7.4; 2 μmol/L EDTA, pH 8.0), and microinjected into F2hybrid zygotes from C57BL/6J X SJL/J parents. Microinjections were performed by the University of Michigan Transgenic Mouse Core. Transgenic founders were detected by polymerase chain reaction (PCR) of tail DNA using primers specific for each construct. A sense strand oligonucleotide (5′-GTGAATCAAATATTTATCTTGCAGGTGGCCT-3′, HUMHBB 8972-9002) located in HS2, and an antisense strand oligonucleotide (5′-CAAATTGTTATTATTCCAGGCCACTGAATT-3′, HUMHBB 17573-17602) located in the distal ε promoter, were used as primers for the HS2εγ transgene. These primers amplify a 817 bp product. For the HS3εγ and HS3εγ−1086muttransgene, a sense strand oligonucleotide in HS3 (5′-AGCTGCTGCAGTCAAAGTCGAATGCAGCTG-3′, HUMHBB 5123-5152), and an antisense strand oligonucleotide in the distal ε promoter (5′-TCCATCCATTTCTACCATTTCTTTCTCCTA-3′, HUMHBB 17916-17945), were used. These primers amplify a 1,156 bp product. PCR reactions were performed for 35 cycles (94°C, 30 seconds; 60°C, 90 seconds; 72°C, 120 seconds). Founders testing positive by PCR were bred to obtain F1 males. For each founder line, two or three different F1 males were bred to obtain embryos and fetuses (F2) for analysis.
Southern blot analysis.
Fluorometrically quantitated transgenic mouse genomic DNA (8 to 10 μg) isolated from the tails of F1 males was digested withXhoI (for HS2εγ lines) or NsiI (for HS3εγ and HS3εγ−1086mut lines), fractionated on a 0.7% agarose gel, and transferred to nylon membranes. The blots were subsequently probed with a 753 bp HindIII fragment containing the Aγ enhancer sequence (HUMHBB 41383 to 42135). Copy numbers were determined by phosophoImager analysis of Southern blots after comparison to standards generated by dilution of plasmid constructs into nontransgenic mouse DNA.
RNA isolation and S1 nuclease assay.
To determine the pattern of transgene expression during development, 6- to 8-week old CD1 female mice (Charles River Laboratories, Wilmington, MA) were mated with F1 or F2transgenic males. The morning on which the copulatory plug was detected was designated day 0. On the following days, tissues were removed for analysis: day 10 (yolk sac), day 12 (yolk sac and fetal liver), day 14 (fetal liver), day 16 (fetal liver) and 4-weeks after birth (peripheral blood). In some cases, the day-18 fetal liver or adult (>6 months) blood was also examined. Tissues were dissected, snap frozen in liquid nitrogen, and stored at −80°C. Other tissues from the same embryo, fetus, or adult were collected simultaneously and genotyped with the PCR assay. Total RNA was purified from transgenic samples using TRIzol (GIBCO BRL, Gaithersburg, MD) according to manufacturers directions. RNA pellets were dissolved in diethyl pyrocarbonate (DEPC)-treated water, and quantitated spectrophotometrically.
For S1 analysis, probes were labeled with T4 DNA polynucleotide kinase and used in the hybridization. The probes for human ε, human Aγ, mouse ζ , and mouse βh1 were described previously.15,16The mouse α (mα) probe consisted of an NheI-BamHI genomic fragment that protects 179 bases of mα exon 2 (GenBankV00714, coordinates 630 to 808). Typically, 1.0 μg of RNA from yolk sacs or fetal livers or 250 ng of RNA from the peripheral blood of 4-week old pups was used in S1 nuclease protection assays, performed as described previously.16 For each RNA sample, assays were performed by multiplex analysis, using both ζ+α+ε and ζ+α+γ probe sets. Additional hybridizations were performed in which the amount of RNA was tripled to confirm that probes were not limiting in the assay. Protected bands were scanned and quantitated by phosphoImager analysis (Molecular Image, Bio-Rad, Hercules, CA).
RESULTS
Transgene expression patterns during development.
F1 or F2 males from four independent founder lines for HS2εγ, four founder lines for HS3εγ, and five founder lines for HS3εγ−1086M were bred; embryos or fetuses were collected for analysis at five developmental time points. In most cases, three or more independent embryos or fetuses were analyzed for each time point. The collective data are displayed in Table 1 (HS3εγ), Table 2 (HS2εγ), and Table 3(HS3εγ−1086M), in which the expression levels of human ε and Aγ transgenes are presented as the percentage of combined mα and ζ mRNA levels in the yolk sac, or the percentage of mα level in the fetal liver and peripheral blood. The data are corrected for copy number. A portion of the data in Table 1 (data for 10- and 12-day yolk sac, 12- and 14-day liver) was reported earlier.10
HS3ɛγ Lines: Copy Number Corrected ɛ and γ Expression Levels
| Line (F0) . | Copy Number . | Sample . | ε Expression Level* . | ε Silencing Ratio† . | γ Expression Level* . | γ Silencing Fetal/Adult‡ . |
|---|---|---|---|---|---|---|
| 150 | 2 | d10 yolk sac | 28 ± 9 (2) | 23 | 41 ± 20 (2) | 11.5 |
| d12 yolk sac | 9.4 ± 1.8 (3) | 19 ± 2 (3) | ||||
| d12 fetal liver | 3.7 ± 1.6 (3) | 32 ± 7 (3) | ||||
| d14 fetal liver | 1.6 ± 0.3 (2) | 138 ± 36 (3) | ||||
| d16 fetal liver | 1.2 ± 0.8 (3) | 117 ± 41 (3) | ||||
| 4-week blood | 1.2 ± 0.5 (2) | 12 ± 2 (2) | ||||
| 155 | 7 | d10 yolk sac | 5.0 ± 3.0 (3) | 7.0 | 9.2 ± 1.3 (3) | 8.0 |
| d12 yolk sac | 5.6 ± 2.3 (3) | 11 ± 4 (4) | ||||
| d12 fetal liver | 0.54 ± 0.01 (2) | 5.8 ± 2.1 (3) | ||||
| d14 fetal liver | 0.25 ± 0.06 (3) | 12 ± 9 (3) | ||||
| d16 fetal liver | 0.30 ± 0.17 (3) | 7.6 ± 1.6 (3) | ||||
| 4-week blood | 0.8 ± 0.05 (2) | 1.5 ± 0.5 (3) | ||||
| 2564 | 10 | d10 yolk sac | 21 ± 3 (3) | 140 | 25 ± 4 (3) | 7.4 |
| d12 yolk sac | 9.5 ± 2.5 (3) | 55 ± 22 (3) | ||||
| d12 fetal liver | 1.1 ± 0.5 (3) | 55 ± 29 (3) | ||||
| d14 fetal liver | 0.72 ± 0.12 (3) | 126 ± 59 (4) | ||||
| d16 fetal liver | 0.34 ± 0.04 (2) | 69 ± 44 (2) | ||||
| 4-week blood | 0.15 ± 0.01 (3) | 17 ± 3 (3) | ||||
| 2649 | 25 | d10 yolk sac | 3.2 ± 1.2 (2) | 77 | 6.8 ± 2.5 (2) | 46 |
| d12 yolk sac | 5.8 ± 0.6 (3) | 29 ± 13 (3) | ||||
| d12 fetal liver | 0.34 ± 0.12 (3) | 12 ± 3 (3) | ||||
| d14 fetal liver | 0.33 ± 0.14 (3) | 17 ± 8 (3) | ||||
| d16 fetal liver | 0.15 ± 0.05 (3) | 7.8 ± 4.3 (3) | ||||
| 4-week blood | 0.075 ± 0.041 (3) | 0.37 ± 0.23 (3) |
| Line (F0) . | Copy Number . | Sample . | ε Expression Level* . | ε Silencing Ratio† . | γ Expression Level* . | γ Silencing Fetal/Adult‡ . |
|---|---|---|---|---|---|---|
| 150 | 2 | d10 yolk sac | 28 ± 9 (2) | 23 | 41 ± 20 (2) | 11.5 |
| d12 yolk sac | 9.4 ± 1.8 (3) | 19 ± 2 (3) | ||||
| d12 fetal liver | 3.7 ± 1.6 (3) | 32 ± 7 (3) | ||||
| d14 fetal liver | 1.6 ± 0.3 (2) | 138 ± 36 (3) | ||||
| d16 fetal liver | 1.2 ± 0.8 (3) | 117 ± 41 (3) | ||||
| 4-week blood | 1.2 ± 0.5 (2) | 12 ± 2 (2) | ||||
| 155 | 7 | d10 yolk sac | 5.0 ± 3.0 (3) | 7.0 | 9.2 ± 1.3 (3) | 8.0 |
| d12 yolk sac | 5.6 ± 2.3 (3) | 11 ± 4 (4) | ||||
| d12 fetal liver | 0.54 ± 0.01 (2) | 5.8 ± 2.1 (3) | ||||
| d14 fetal liver | 0.25 ± 0.06 (3) | 12 ± 9 (3) | ||||
| d16 fetal liver | 0.30 ± 0.17 (3) | 7.6 ± 1.6 (3) | ||||
| 4-week blood | 0.8 ± 0.05 (2) | 1.5 ± 0.5 (3) | ||||
| 2564 | 10 | d10 yolk sac | 21 ± 3 (3) | 140 | 25 ± 4 (3) | 7.4 |
| d12 yolk sac | 9.5 ± 2.5 (3) | 55 ± 22 (3) | ||||
| d12 fetal liver | 1.1 ± 0.5 (3) | 55 ± 29 (3) | ||||
| d14 fetal liver | 0.72 ± 0.12 (3) | 126 ± 59 (4) | ||||
| d16 fetal liver | 0.34 ± 0.04 (2) | 69 ± 44 (2) | ||||
| 4-week blood | 0.15 ± 0.01 (3) | 17 ± 3 (3) | ||||
| 2649 | 25 | d10 yolk sac | 3.2 ± 1.2 (2) | 77 | 6.8 ± 2.5 (2) | 46 |
| d12 yolk sac | 5.8 ± 0.6 (3) | 29 ± 13 (3) | ||||
| d12 fetal liver | 0.34 ± 0.12 (3) | 12 ± 3 (3) | ||||
| d14 fetal liver | 0.33 ± 0.14 (3) | 17 ± 8 (3) | ||||
| d16 fetal liver | 0.15 ± 0.05 (3) | 7.8 ± 4.3 (3) | ||||
| 4-week blood | 0.075 ± 0.041 (3) | 0.37 ± 0.23 (3) |
(Human ε or γ)/(mouse α/4) + (mouse ζ/2)/copy. Mean ± standard deviation (number of independent samples assayed).
Calculated as: Highest expression level in embryonic samples/expression level in adult sample.
Calculated as: Highest expression level in fetal samples/expression level in adult sample.
HS2ɛγ Lines: Copy Number Corrected ɛ and γ Expression Levels
| Line (F0) . | Copy Number . | Sample . | ε Expression Level* . | ε Silencing Ratio† . | γ Expression Level* . | γ Silencing Ratio‡ . |
|---|---|---|---|---|---|---|
| 166 | 18 | d10 yolk sac | 4.1 ± 0.2 (3) | 9.5 | 11 ± 9 (3) | 2.0 |
| d12 yolk sac | 3.1 ± 1.1 (3) | 13 ± 7 (3) | ||||
| d12 fetal liver | 1.0 ± 0.4 (3) | 8.4 ± 4.5 (2) | ||||
| d14 fetal liver | 0.55 ± 0.19 (3) | 7.0 ± 0.4 (2) | ||||
| d16 fetal liver | 0.67 ± 0.28 (3) | 5.9 ± 3.0 (3) | ||||
| 4-week blood | 0.43 ± 0.11 (2) | 4.4 ± 1.7 (2) | ||||
| 186 | ∼300 | d10 yolk sac | 0.08 ± 0.06 (3) | 6 | 0.3 ± 0.1 (3) | 58 |
| d12 yolk sac | 0.12 ± 0.01 (3) | 2.1 ± 1.3 (3) | ||||
| d12 fetal liver | 0.064 ± 0.043 (3) | 4.9 ± 3.0 (3) | ||||
| d14 fetal liver | 0.052 ± 0.029 (3) | 3.5 ± 2.2 (3) | ||||
| d16 fetal liver | 0.015 ± 0.001 (3) | 1.2 ± 1.0 (3) | ||||
| 4-week blood | 0.02 ± 0.002 (3) | 0.06 ± 0.03 (3) | ||||
| 187 | 10 | d10 yolk sac | 5.0 ± 2.6 (3) | 35 | 23 ± 11 (3) | 9.5 |
| d12 yolk sac | 11 ± 1 (2) | 121 ± 106 (2) | ||||
| d12 fetal liver | 1.7 ± 0.8 (2) | 106 ± 9 (2) | ||||
| d14 fetal liver | 0.68 ± 0.25 (3) | 104 ± 54 (3) | ||||
| d16 fetal liver | 0.15 ± 0.12 (2) | 70 ± 31 (2) | ||||
| 4-week blood | 0.31 ± 0.22 (3) | 11 ± 1 (3) | ||||
| 192 | 25 | d10 yolk sac | 1.3 ± 1.2 (3) | 8.8 | 8.7 ± 3.2 (3) | 163 |
| d12 yolk sac | 1.4 ± 0.7 (3) | 13 ± 3 (2) | ||||
| d12 fetal liver | 0.35 ± 0.30 (3) | 15 ± 3 (2) | ||||
| d14 fetal liver | 0.14 ± 0.05 (3) | 44 ± 34 (3) | ||||
| d16 fetal liver | 0.17 ± 0.07 (3) | 12 ± 3 (3) | ||||
| 4-week blood | 0.16 ± 0.10 (3) | 0.27 ± 0.09 (3) |
| Line (F0) . | Copy Number . | Sample . | ε Expression Level* . | ε Silencing Ratio† . | γ Expression Level* . | γ Silencing Ratio‡ . |
|---|---|---|---|---|---|---|
| 166 | 18 | d10 yolk sac | 4.1 ± 0.2 (3) | 9.5 | 11 ± 9 (3) | 2.0 |
| d12 yolk sac | 3.1 ± 1.1 (3) | 13 ± 7 (3) | ||||
| d12 fetal liver | 1.0 ± 0.4 (3) | 8.4 ± 4.5 (2) | ||||
| d14 fetal liver | 0.55 ± 0.19 (3) | 7.0 ± 0.4 (2) | ||||
| d16 fetal liver | 0.67 ± 0.28 (3) | 5.9 ± 3.0 (3) | ||||
| 4-week blood | 0.43 ± 0.11 (2) | 4.4 ± 1.7 (2) | ||||
| 186 | ∼300 | d10 yolk sac | 0.08 ± 0.06 (3) | 6 | 0.3 ± 0.1 (3) | 58 |
| d12 yolk sac | 0.12 ± 0.01 (3) | 2.1 ± 1.3 (3) | ||||
| d12 fetal liver | 0.064 ± 0.043 (3) | 4.9 ± 3.0 (3) | ||||
| d14 fetal liver | 0.052 ± 0.029 (3) | 3.5 ± 2.2 (3) | ||||
| d16 fetal liver | 0.015 ± 0.001 (3) | 1.2 ± 1.0 (3) | ||||
| 4-week blood | 0.02 ± 0.002 (3) | 0.06 ± 0.03 (3) | ||||
| 187 | 10 | d10 yolk sac | 5.0 ± 2.6 (3) | 35 | 23 ± 11 (3) | 9.5 |
| d12 yolk sac | 11 ± 1 (2) | 121 ± 106 (2) | ||||
| d12 fetal liver | 1.7 ± 0.8 (2) | 106 ± 9 (2) | ||||
| d14 fetal liver | 0.68 ± 0.25 (3) | 104 ± 54 (3) | ||||
| d16 fetal liver | 0.15 ± 0.12 (2) | 70 ± 31 (2) | ||||
| 4-week blood | 0.31 ± 0.22 (3) | 11 ± 1 (3) | ||||
| 192 | 25 | d10 yolk sac | 1.3 ± 1.2 (3) | 8.8 | 8.7 ± 3.2 (3) | 163 |
| d12 yolk sac | 1.4 ± 0.7 (3) | 13 ± 3 (2) | ||||
| d12 fetal liver | 0.35 ± 0.30 (3) | 15 ± 3 (2) | ||||
| d14 fetal liver | 0.14 ± 0.05 (3) | 44 ± 34 (3) | ||||
| d16 fetal liver | 0.17 ± 0.07 (3) | 12 ± 3 (3) | ||||
| 4-week blood | 0.16 ± 0.10 (3) | 0.27 ± 0.09 (3) |
HS3ɛ−1086Mγ Lines: Copy Number Corrected ɛ and γ Expression Levels
| Line (F0) . | Copy Number . | Sample . | ε Expression Level3-150 . | ε Silencing Ratio3-151 . | γ Expression Level3-150 . | γ Silencing Fetal/Adult3-152 . |
|---|---|---|---|---|---|---|
| 4403 | 2 | d10 yolk sac | 0.82 ± 0.34 (3) | 4.2 | 5.5 ± 4.6 (3) | 0.5 |
| d12 yolk sac | 1.1 ± 0.1 (3) | 7.3 ± 0.8 (3) | ||||
| d12 fetal liver | 0.22 ± 0.02 (3) | 2.3 ± 0.2 (2) | ||||
| d14 fetal liver | 0.19 ± 0.03 (3) | 3.2 ± 2.9 (3) | ||||
| d16 fetal liver | 0.24 ± 0.08 (2) | 5.9 ± 0.9 (2) | ||||
| 4-week blood | 0.26 ± 0.08 (3) | 6.1 ± 1.6 (3) | ||||
| 4456 | 32 | d10 yolk sac | 0.46 ± 0.23 (3) | 1.6 | 0.63 ± 0.12 (2) | 0.4 |
| d12 yolk sac | 0.5 ± 0.19 (3) | 0.82 ± 0.27 (3) | ||||
| d12 fetal liver | 0.31 ± 0.01 (2) | 0.18 ± 0.06 (2) | ||||
| d14 fetal liver | 0.30 ± 0.11 (3) | 0.21 ± 0.07 (2) | ||||
| d16 fetal liver | 0.31 ± 0.05 (3) | 0.02 ± 0.01 (3) | ||||
| 4-week blood | 0.32 ± 0.04 (3) | 0.52 ± 0.29 (3) | ||||
| 4463 | 10 | d10 yolk sac | 2.6 ± 2.0 (3) | 13.8 | 8.4 ± 7.2 (3) | 9.3 |
| d12 yolk sac | 5.1 ± 1.6 (3) | 20 ± 10 (3) | ||||
| d12 fetal liver | 1.2 ± 0.06 (3) | 6.7 ± 4.4 (3) | ||||
| d14 fetal liver | 0.42 ± 0.14 (3) | 9.3 ± 7.7 (4) | ||||
| d16 fetal liver | 0.28 ± 0.03 (3) | 5.3 ± 4.5 (2) | ||||
| 4-week blood | 0.37 ± 0.01 (2) | 1.0 ± 0.02 (3) | ||||
| 4485 | 33 | d10 yolk sac | 12 ± 5.0 (3) | 6.8 | 18 ± 10 (3) | 0.3 |
| d12 yolk sac | 17 ± 5.0 (3) | 33 ± 22 (3) | ||||
| d12 fetal liver | 5.1 ± 4.1 (3) | 12 ± 2 (2) | ||||
| d14 fetal liver | 2.0 ± 0.7 (3) | 6.5 ± 0.9 (3) | ||||
| d16 fetal liver | 2.5 ± 0.8 (3) | 5.0 ± 0.1 (3) | ||||
| 4-week blood | 2.5 ± 0.5 (3) | 22 ± 9 (3) | ||||
| 4491 | 16 | d10 yolk sac | 0.9 ± 0.1 (3) | 10 | 5.5 ± 1.0 (3) | 4.3 |
| d12 yolk sac | 1.2 ± 0.3 (3) | 11 ± 2.0 (3) | ||||
| d12 fetal liver | 0.21 ± 0.05 (6) | 6.3 ± 1.5 (6) | ||||
| d14 fetal liver | 0.30 ± 0.04 (2) | 7.3 ± 0.4 (2) | ||||
| d16 fetal liver | 0.21 ± 0.07 (3) | 5.4 ± 1.0 (3) | ||||
| 4-week blood | 0.12 ± 0.03 (3) | 1.7 ± 0.7 (3) |
| Line (F0) . | Copy Number . | Sample . | ε Expression Level3-150 . | ε Silencing Ratio3-151 . | γ Expression Level3-150 . | γ Silencing Fetal/Adult3-152 . |
|---|---|---|---|---|---|---|
| 4403 | 2 | d10 yolk sac | 0.82 ± 0.34 (3) | 4.2 | 5.5 ± 4.6 (3) | 0.5 |
| d12 yolk sac | 1.1 ± 0.1 (3) | 7.3 ± 0.8 (3) | ||||
| d12 fetal liver | 0.22 ± 0.02 (3) | 2.3 ± 0.2 (2) | ||||
| d14 fetal liver | 0.19 ± 0.03 (3) | 3.2 ± 2.9 (3) | ||||
| d16 fetal liver | 0.24 ± 0.08 (2) | 5.9 ± 0.9 (2) | ||||
| 4-week blood | 0.26 ± 0.08 (3) | 6.1 ± 1.6 (3) | ||||
| 4456 | 32 | d10 yolk sac | 0.46 ± 0.23 (3) | 1.6 | 0.63 ± 0.12 (2) | 0.4 |
| d12 yolk sac | 0.5 ± 0.19 (3) | 0.82 ± 0.27 (3) | ||||
| d12 fetal liver | 0.31 ± 0.01 (2) | 0.18 ± 0.06 (2) | ||||
| d14 fetal liver | 0.30 ± 0.11 (3) | 0.21 ± 0.07 (2) | ||||
| d16 fetal liver | 0.31 ± 0.05 (3) | 0.02 ± 0.01 (3) | ||||
| 4-week blood | 0.32 ± 0.04 (3) | 0.52 ± 0.29 (3) | ||||
| 4463 | 10 | d10 yolk sac | 2.6 ± 2.0 (3) | 13.8 | 8.4 ± 7.2 (3) | 9.3 |
| d12 yolk sac | 5.1 ± 1.6 (3) | 20 ± 10 (3) | ||||
| d12 fetal liver | 1.2 ± 0.06 (3) | 6.7 ± 4.4 (3) | ||||
| d14 fetal liver | 0.42 ± 0.14 (3) | 9.3 ± 7.7 (4) | ||||
| d16 fetal liver | 0.28 ± 0.03 (3) | 5.3 ± 4.5 (2) | ||||
| 4-week blood | 0.37 ± 0.01 (2) | 1.0 ± 0.02 (3) | ||||
| 4485 | 33 | d10 yolk sac | 12 ± 5.0 (3) | 6.8 | 18 ± 10 (3) | 0.3 |
| d12 yolk sac | 17 ± 5.0 (3) | 33 ± 22 (3) | ||||
| d12 fetal liver | 5.1 ± 4.1 (3) | 12 ± 2 (2) | ||||
| d14 fetal liver | 2.0 ± 0.7 (3) | 6.5 ± 0.9 (3) | ||||
| d16 fetal liver | 2.5 ± 0.8 (3) | 5.0 ± 0.1 (3) | ||||
| 4-week blood | 2.5 ± 0.5 (3) | 22 ± 9 (3) | ||||
| 4491 | 16 | d10 yolk sac | 0.9 ± 0.1 (3) | 10 | 5.5 ± 1.0 (3) | 4.3 |
| d12 yolk sac | 1.2 ± 0.3 (3) | 11 ± 2.0 (3) | ||||
| d12 fetal liver | 0.21 ± 0.05 (6) | 6.3 ± 1.5 (6) | ||||
| d14 fetal liver | 0.30 ± 0.04 (2) | 7.3 ± 0.4 (2) | ||||
| d16 fetal liver | 0.21 ± 0.07 (3) | 5.4 ± 1.0 (3) | ||||
| 4-week blood | 0.12 ± 0.03 (3) | 1.7 ± 0.7 (3) |
None of the constructs exhibited copy-number–dependent expression of the human transgenes (Tables 1 to 3). Rather, line-to-line variation was observed in expression levels, most likely attributable to position effects. Therefore, to permit comparison of patterns of transgene expression during development, the data in Tables 1 to 3 were normalized for each transgene. The point of maximal transgene (ε or γ) expression for each line was taken as 100% and all other points were expressed relative to this value. The expression patterns for ε and γ transgenes in all 13 lines are compared in Fig 1.
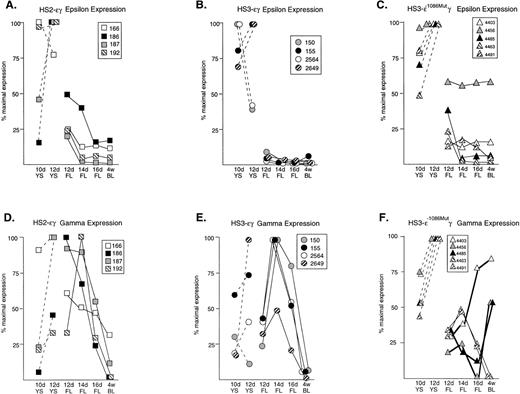
Normalized expression patterns of the ɛ and Aγ human transgenes in mice carrying HS2ɛγ, HS3ɛγ, and HS3ɛγ−1086Mut constructs. (A) HS2ɛγ: ɛ transgene expression patterns; (B) HS3ɛγ: ɛ transgene expression patterns; (C) HS3ɛγ−1086Mut: ɛ transgene expression patterns; (D) HS2ɛγ: γ transgene expression patterns; (E) HS3ɛγ: γ transgene expression patterns; (F) HS3ɛγ−1086Mut: γ transgene expression patterns. For all lines, expression data are expressed relative to the peak (maximal) expression level, which is taken as 100%. Dashed lines represent yolk sac samples; solid lines represent fetal liver and postnatal samples. The identity of each transgenic line is given by the key provided in the box accompanying each set of curves. For (F) γ expression curves for the three transgenic lines that exhibit a phenotype after deletion of the −1086 bp YY1 site are shown with bold lines.
Normalized expression patterns of the ɛ and Aγ human transgenes in mice carrying HS2ɛγ, HS3ɛγ, and HS3ɛγ−1086Mut constructs. (A) HS2ɛγ: ɛ transgene expression patterns; (B) HS3ɛγ: ɛ transgene expression patterns; (C) HS3ɛγ−1086Mut: ɛ transgene expression patterns; (D) HS2ɛγ: γ transgene expression patterns; (E) HS3ɛγ: γ transgene expression patterns; (F) HS3ɛγ−1086Mut: γ transgene expression patterns. For all lines, expression data are expressed relative to the peak (maximal) expression level, which is taken as 100%. Dashed lines represent yolk sac samples; solid lines represent fetal liver and postnatal samples. The identity of each transgenic line is given by the key provided in the box accompanying each set of curves. For (F) γ expression curves for the three transgenic lines that exhibit a phenotype after deletion of the −1086 bp YY1 site are shown with bold lines.
Embryonic expression of the ε gene in transgenic mice carrying HS2εγ and HS3εγ constructs.
Embryonic expression and postembryonic silencing of the ε transgene occurred in mice carrying both HS3εγ and HS2εγ constructs, as can be appreciated in the S1 analyses shown in Fig 2A and 2B. The raw data in Tables 1 and2 confirm that both HS2 and HS3 support efficient human ε transgene expression in 10- and 12-day yolk sacs. In fact, two of the HS3εγ lines (150 and 2564) exhibit high ε expression levels (28% and 21% of mα-like chains, respectively); these levels are similar to those observed when a yeast artificial chromosome (YAC) containing the entire human β-like cluster was injected into transgenic mice.17 18 All other HS3εγ and HS2εγ lines expressed ε at levels between 1.4% and 11% of mα-like chains. One HS2εγ line (186) with a particularly high number of transgene copies exhibited much lower absolute levels of human ε expression, 0.12% of mα-like chains (Table 2).
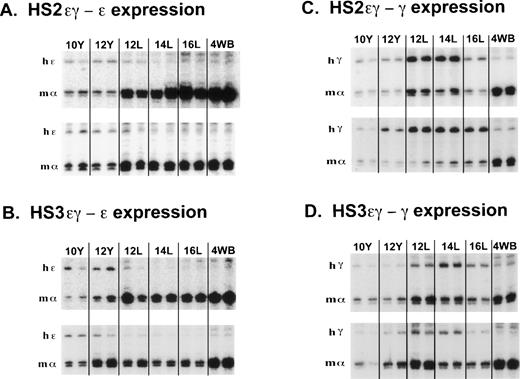
S1 nuclease protection assay of human ɛ and Aγ transgene expression during mouse development in mice carrying HS2ɛγ and HS3ɛγ constructs. (A) Human ɛ transgene expression data from HS2ɛγ lines 187 (top panel) and 166 (bottom panel). Phosphorimager scans of S1 protection assays analyzing human ɛ transgene (hɛ) and mouse gene (m) expression are shown from two embryos or fetuses at each time point. The band protected by the mouse ζ probe is not shown but was included in the calculations in Table 2. Y = yolk sac; L = fetal liver; 4WB = blood from 4-week old pups. (B) Human ɛ transgene expression data from HS3ɛγ lines 2649 (top panel) and 2564 (bottom panel). The band protected by the mouse ζ probe is not shown but was included in the calculations in Table 1. (C) Human Aγ transgene expression in HS2ɛγ lines. Samples are from line 192 (top panel) and 186 (bottom panel). (D) Human Aγ transgene expression in HS3ɛγ lines. Samples are from line 150 (top panel) and 155 (bottom panel).
S1 nuclease protection assay of human ɛ and Aγ transgene expression during mouse development in mice carrying HS2ɛγ and HS3ɛγ constructs. (A) Human ɛ transgene expression data from HS2ɛγ lines 187 (top panel) and 166 (bottom panel). Phosphorimager scans of S1 protection assays analyzing human ɛ transgene (hɛ) and mouse gene (m) expression are shown from two embryos or fetuses at each time point. The band protected by the mouse ζ probe is not shown but was included in the calculations in Table 2. Y = yolk sac; L = fetal liver; 4WB = blood from 4-week old pups. (B) Human ɛ transgene expression data from HS3ɛγ lines 2649 (top panel) and 2564 (bottom panel). The band protected by the mouse ζ probe is not shown but was included in the calculations in Table 1. (C) Human Aγ transgene expression in HS2ɛγ lines. Samples are from line 192 (top panel) and 186 (bottom panel). (D) Human Aγ transgene expression in HS3ɛγ lines. Samples are from line 150 (top panel) and 155 (bottom panel).
Figure 1 (A and B) compares the pattern of ε transgene expression in four HS2 εγ lines (1A) and four HS3εγ lines (1B). In each of these eight independent lines, all data points for fetal and adult ε expression fall below the levels observed in 10- and 12-day yolk sac, indicating silencing of the ε gene in fetal life. Silencing (downregulation from peak ε expression level to adult expression level) was consistent in all lines, ranging from six- to 35-fold in HS2εγ lines (Table 2) and seven- to 140-fold in HS3εγ lines (Table 1). However, for both constructs, ε gene silencing was not complete; ε gene expression persisted at low levels beyond the embryonic stage into fetal and adult stages (Fig 2A and 2B). Residual expression levels of ε in 4-week old animals were similar for the two constructs, ranging from 1.2% to 0.075% of mα chains for HS3εγ mice (Table 1) and 0.43% to 0.02% of mα chains for HS2εγ mice (Table 2).
Aγ expression in mouse fetal liver and postnatal silencing of Aγ expression in mice carrying HS2εγ and HS3εγ constructs.
The S1 analysis of Aγ transgene expression in two lines of mice carrying each type of construct is shown in Fig 2C (HS2εγ) and 2D (HS3εγ). A peak of expression in fetal life can be discerned for all four lines shown here. The peak of fetal expression in the various HS2εγ lines occurred at 12 or 14 days, and mRNA levels fell thereafter (Fig 1D). For HS3εγ, the pattern of Aγ expression during development was remarkably consistent (Fig 1E) despite differences in absolute expression level among the lines (Table 1). In all HS3εγ lines, the Aγ transgene was expressed efficiently (and at levels greater than ε gene expression, relative to mα chains, Table 1) in 10- and 12-day embryonic yolk sacs; fetal expression levels peaked at 14 days, and subsequently fell in 16-day liver and postnatal samples (Fig 1E). Overall silencing of the Aγ gene (fold reduction in transcript level from peak prenatal point to the 4-week postnatal point) in HS2εγ lines varied from three- to 163-fold (Table 2), whereas in HS3εγ lines, overall Aγ silencing efficacy varied between 7.4- and 78-fold (Table 1). Interestingly, for all eight of the lines examined, this degree of silencing is significantly greater than was observed using constructs containing the μLCR linked to the same γ fragment in the absence of the ε gene (Stamatoyannopoulos, Clegg, and Li,19 construct μLCR-3.9: three lines examined exhibit silencing ratios of 1.7-, 1.2-, and 1.2-fold). Thus, either the presence of the ε gene itself, or the increased distance between the LCR element(s) and the γ gene may result in more effective γ silencing.
Mutagenesis of the YY1 binding site at −1086 of the Aγ gene.
A 3 bp mutation that severely reduces YY1 binding to a high affinity, evolutionarily conserved YY1 binding site at −1086 bp upstream from γ was previously described.11 The same mutation was introduced into the HS3εγ backbone and this mutant construct (HS3εγ−1086M) was used to generate five independent transgenic founder lines. Compiled S1 nuclease protection data for ε and Aγ transgene expression in these five lines are provided in Table 3. As with the parent construct (HS3εγ), apparent position effects cause line-to-line variation in the absolute level of transgene expression in mice carrying HS3εγ−1086M. When the data are normalized (all points expressed as percent of maximal expression level), the shape of the resulting expression curves for the ε gene are not remarkably different from those of the wild-type construct, in that expression is highest in embryonic life and falls thereafter (Fig 1C). However, all lines, particularly 4456, show a reduced ε silencing ratio; these ratios range from 1.6- to 13.8-fold in HS3εγ−1086M mice as compared with seven- to 140-fold in HS3εγ mice (Table 3).
For the Aγ transgene, a striking difference in expression pattern is observed in mice carrying the mutant construct (Fig 1F) compared with the wild-type parent construct (Fig 1E). As discussed above, mice carrying wild-type HS3εγ exhibit a consistent peak in expression in the 14-day fetal liver. In contrast, three of the five lines carrying the mutant construct (4403, 4456, 4485) show no such 14-day peak. In fact, levels of Aγ in fetal life for these lines are lower than those seen either in the 10-day yolk sac or in adult life, resulting in an unusual U-shaped expression curve that to our knowledge has not been previously observed in transgenic animals carrying the human Aγ gene. For the remaining two mutant lines, the expression curve is similar to that observed for wild-type line 2649.
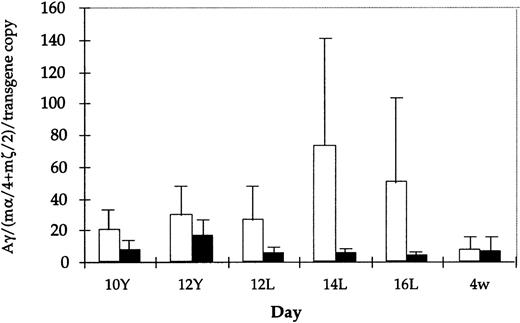
Absolute values for Aγ transgene expression in the 14- and 16-day fetal liver are also lower in mice carrying HS3 εγ−1086M compared with those carrying the wild-type construct (for example, at 14 days, wild-type mice exhibit absolute levels of 138%, 126%, 17%, and 12% of mα chains per copy, whereas mutant mice have absolute levels of 9.3%, 7.3%, 6.5%, 3.2%, and 0.21%). Though these two data sets are nonoverlapping, the differences between them are not statistically significant. However, it is interesting to note that the apparent reduction in expression level is observed only in the fetal period (Fig3), because Aγ expression in embryonic life and in adult life is comparable in wild-type and mutant constructs. Together, these findings suggest that the −1086 YY1 binding site may play a role in directing the high level of Aγ transgene expression during the mouse fetal stage.
Comparison of Aγ transgene expression level (mRNA) among all HS3ɛγ and HS3ɛγ−1086Mut lines. Average mRNA level (corrected for copy number and expressed relative to m-like mRNA level) is shown for all lines carrying HS3ɛγ (white bars) and for HS3ɛγ−1086Mut (black bars). Brackets indicate standard error of the mean. None of the differences shown are statistically significant.
Comparison of Aγ transgene expression level (mRNA) among all HS3ɛγ and HS3ɛγ−1086Mut lines. Average mRNA level (corrected for copy number and expressed relative to m-like mRNA level) is shown for all lines carrying HS3ɛγ (white bars) and for HS3ɛγ−1086Mut (black bars). Brackets indicate standard error of the mean. None of the differences shown are statistically significant.
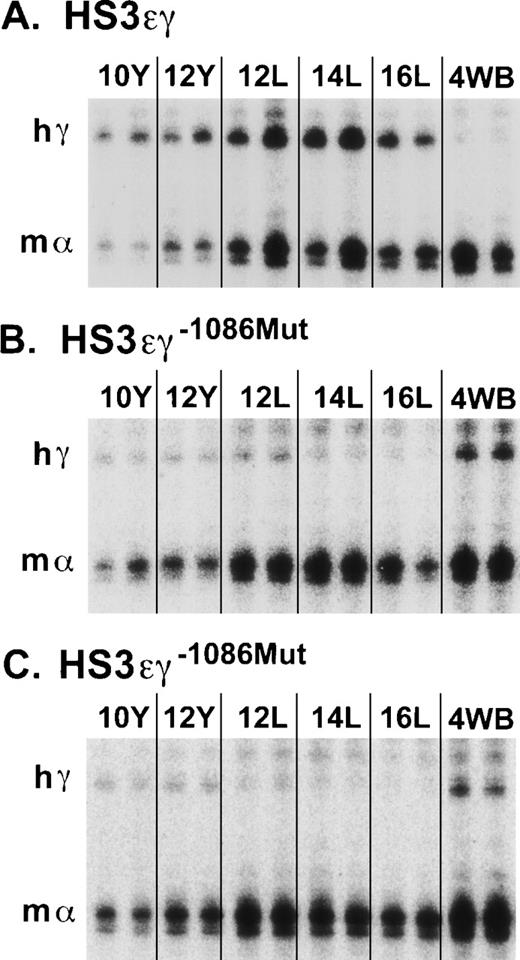
The striking difference in the appearance of the overall pattern of γ transgene expression in mutant lines 4485 and 4456 compared with that of wild-type line 2649 is shown in Fig 4. In a subset of lines, Aγ transgene expression was also examined in adults more than 6-months old. No further change in Aγ expression levels was observed in older animals in either the wild-type or the mutant lines (data not shown).
S1 nuclease protection assay of human Aγ transgene expression in HS3ɛγ and HS3ɛγ−1086Mut lines. (A) Human Aγ transgene (hγ) and mouse gene (m) expression are shown from two embryos or fetuses at each time point for the wild-type HS3ɛγ construct carried by line 2649. The band protected by the mouse ζ probe is not shown but was included in the calculations in Table 1. (B) Human Aγ transgene (hγ) and m expression in animals from HS3ɛγ−1086Mut line 4485. (C) Human Aγ transgene (hγ) and m expression in animals from HS3ɛγ−1086Mut line 4456. Y = yolk sac; L = fetal liver; 4WB = blood from 4-week-old pups.
S1 nuclease protection assay of human Aγ transgene expression in HS3ɛγ and HS3ɛγ−1086Mut lines. (A) Human Aγ transgene (hγ) and mouse gene (m) expression are shown from two embryos or fetuses at each time point for the wild-type HS3ɛγ construct carried by line 2649. The band protected by the mouse ζ probe is not shown but was included in the calculations in Table 1. (B) Human Aγ transgene (hγ) and m expression in animals from HS3ɛγ−1086Mut line 4485. (C) Human Aγ transgene (hγ) and m expression in animals from HS3ɛγ−1086Mut line 4456. Y = yolk sac; L = fetal liver; 4WB = blood from 4-week-old pups.
DISCUSSION
The human ε and γ genes exhibit distinct patterns of stage-specific expression, both in vivo during human development and when expressed as transgenes in the mouse. ε is exclusively embryonic and silenced thereafter, whereas γ is primarily a fetal gene (though it is also expressed in embryonic life in humans and in transgenic mice20-22). Three different models can explain the generation of these distinct stage-specific expression patterns: (1) these patterns may be controlled entirely by the LCR: for example, the LCR may adopt a different conformation in each stage that is suited for more effective communication with a specific promoter; (2) control of stage-specificity may be exclusively gene-proximal, individual LCR enhancers could be required for amplification of the signals specified by the genes but LCR sequences might not contribute to stage-specificity; and (3) both gene proximal and LCR sequences may hold stage-specific information. The data described in this report are consistent with the second alternative above, that gene-proximal signals must exist, because distinct stage-specific expression patterns for both ε (embryonic) and Aγ (fetal) genes are maintained regardless of whether the genes are linked to HS2 or to HS3. However, the third alternative is not excluded if HS2 and HS3 both contain redundant signals controlling these distinct patterns.
Two previous reports also support the existence of gene-linked developmental signals. First, in transgenic mice carrying human Gγ-Aγ-δ-β genes without any linked LCR sequences, stage-specific regulation of γ and β transgenes is maintained.23 Second, in earlier work from our laboratory, two variants of the HS3εγ construct, in which the γ gene fragment was derived either from the human (human γ is expressed fetally) or the galago (galagos, like other nonanthropoid mammals, express γ in embryonic life), were used to create transgenic mice.10 In multiple founder lines, the galago γ gene was expressed exclusively in the embryo, whereas the human γ gene was expressed fetally. Because the two constructs were identical except for the 4.0 kb fragment containing the γ gene, sequence differences within that fragment (not within HS3) must specify embryonic versus fetal expression of γ.
Whereas the results of the present work as well as the two studies cited above indicate that gene proximal signals mediate stage specificity, other work has stressed the importance of the LCR in developmental control. For example, Fraser et al8 linked individual hypersensitive sites to a cosmid containing the Gγ, Aγ, δ, and β genes, and observed that different hypersensitive sites drive different developmental patterns of γ and β expression. Similar conclusions were reached using constructs carrying HS2 or HS3 linked to γ and β.9 More recently, Navas et al18 deleted the core of HS3 in an otherwise intact human β-locus YAC and observed that HS3 is essential for specific parts of the switching program. Embryonic expression of ε (but not γ) was disrupted, and fetal expression of γ was diminished. Collectively, these investigations indicate that HS2 and HS3 differ in their ability to communicate with ε and γ.
How is this conclusion reconciled with data presented here, which show that both HS2 and HS3 can independently support embryonic ε and fetal γ expression? In this context, it is important to consider the attributes of the different constructs used in each study. For example, the results observed in the Fraser, et al8 study could indicate the existence of stage-specific elements within individual hypersensitive sites. Alternatively, because the construct used in that study contained both γ and β and because these genes compete for LCR interaction, the results could also reflect stage-specific differences in the outcome of the competition (eg, in the fetal stage, when both genes are present, HS3 prefers to interact with γ rather than β; but HS2 prefers β over γ in the same stage). The fact that we observe that HS2 and HS3 can each drive γ in fetal life in the absence of β in fact indicates that this alternative explanation most likely accounts for the results of that study. In addition, we found that HS3 and HS2 can each support an embryonic ε expression pattern; this had not been previously tested.
The Navas et al18 study was done using a YAC containing the entire human β-globin locus. Though such large constructs (or deletions engineered into the endogenous mouse locus) represent the best possible substrate for such experiments, it is noteworthy that the five published investigations analyzing HS3 deletions in the context of an otherwise intact mouse or human locus show rather disparate phenotypes, ranging from minimal effects24 to a major reduction in expression of all globin genes at all stages25,26 to effects on specific genes at specific stages.17,18 It has been speculated the size of the HS3 deletion may determine the phenotype.17,18 24 Removal of only the HS3 core may result in more debilitating effects via a dominant negative mechanism (ie, small deletions may destroy an activation function, but still allow sequences surrounding the core to tether to a promoter). In contrast, larger HS3 deletions seem to produce more subtle effects, as if removal of core plus tethering sequences allows another HS to substitute for the missing HS3. Here, we show that HS2 alone, at least when present in multiple copies as was the case in all of our lines, is indeed capable of supporting both embryonic ε and fetal γ expression and is therefore capable of substituting for a deleted HS3 region; likewise, HS3 could substitute for a deleted HS2 in the regulation of ε and γ patterns.
Given the importance of gene-proximal regulatory elements in the control of ε and γ gene stage specificity, the next important goal is to identify the specific elements involved in this control. The HS3 εγ construct represents a particularly useful tool to analyze such candidate elements, because despite line to line variations in the absolute levels transgene expression, this construct exhibits very reproducible expression curves for both of these genes. Thus, it was used as the backbone in which to study the role of an evolutionarily conserved YY1 binding site (at −1086 bp upstream from γ) in the regulation of γ expression. YY1 is an interesting protein in the context of the hemoglobin switching program, because it has been shown to act as either an activator or a repressor (reviewed in Shi, Lee, and Galvin12), and in at least one system, it appears to mediate developmental regulation of gene expression.27 An important attribute of YY1 is its ability to interact with a number of basal and specialized transcription factors, as well as cofactors, including: TFIIB, TAFII55, SP1, C/EBP β, c-myc, ATF/CREB, E1A, and p300/CBP (reviewed in Shi, Lee, and Galvin12). Recently, pleiohomeotic (PHO), a member of theDrosophila polycomb group of proteins was found to encode a YY1 homologue.28 Polycomb proteins form multiprotein complexes on DNA and are necessary for the maintenance of the tissue-specific pattern of homeotic gene expression (reviewed in Pirrota29). YY1 also exists as a multiprotein complex in mammalian cells,30 and may function similarly in recruitment of a protein conglomerate to DNA. In this regard, it is noteworthy that YY1 interacts with histone acetylases such as p300/CBP31,32 as well as histone deacetylases such as the RPD3-related proteins HCAC1, HDAC2, and HDAC3,33,34 such interactions have the potential to influence local chromatin structure as well as gene activity (reviewed in Grunstein35). All of these characteristics make YY1 a compelling target for study in the context of hemoglobin switching.
Here, we found that mutation of the −1086 Aγ YY1 site did not alter the developmental expression pattern of the ε transgene, though it did appear to interfere somewhat with effective ε silencing. However, clear effects of the YY1 mutation on Aγ transgene expression were observed in three of the five mutant lines. These effects included: (1) loss of the 14-day peak of Aγ expression that was observed consistently in wild-type lines, (2) apparent reduction in absolute expression levels of the Aγ transgene that was specific to fetal life, and (3) reactivation (or rebound) of Aγ gene expression after birth in three of the five lines, resulting in a U-shaped expression curve. In the remaining two HS3 εγ−1086M lines, expression curves were similar to that observed in one of the lines carrying the wild-type construct. It is possible that some aspect of the site of integration in these two lines either masked or complemented the mutant phenotype. The “all or nothing” pattern of this phenotype is interesting in light of the possible role of YY1 in determination of chromatin structure discussed above, as chromatin mediated effects characteristically exhibit this type of pattern.36
Disappearance of the 14-day fetal peak (and the generally low absolute levels of fetal Aγ expression in the mutant lines, Fig 3) suggests that the −1086 YY1 site may function in fetal activation. However, this is contrary to results observed in previous studies using transient expression assays, in which an oligonucleotide containing the −1086 YY1 binding site linked to a minimal γ promoter acted as a repressor in fetal erythroid cell lines.11 This apparent contradiction could be explained if other sequences of the distal γ promoter, present in the constructs tested here, but absent in the minimal promoter constructs, influence the function of the −1086 site. A precedent for this exists in the human papillomavirus type-18 promoter.37 In that case, a YY1 binding site in the minimal promoter (−221 to −2) acts as a repressor, but when additional sequences of the distal promoter are added (to −824), the same site appears to be an activator. Alternatively, the discordant results could be due to chromatin effects, because the transient transfection procedure, unlike the transgenic assay, does not involve chromosomal integration of the expression plasmid. In this vein, it is interesting that recent studies of the interaction of YY1 with the immunoglobulin κ 3′ enhancer show a similar paradox: in vivo footprinting studies are consistent with a stage-specific activator role for YY138 even though earlier studies of the same site in transient assays suggested that this site mediates repression.27
Another important consideration in comparing the disparate results observed in transient assays versus transgenic mice may be the concentration of YY1 within the cell. It has been shown that YY1 expression at high levels represses reporter gene activity, but activation (or relief of repression) of the same reporter is observed when lower levels of YY1 are present.39 Because the majority of studies of YY1 function have been performed using transient assays, it will be of interest to determine whether discordant outcomes are detected in other systems when the same binding sites are retested in transgenic models.
In three of the five lines carrying the mutant construct, Aγ expression levels are greater in both embryonic and postnatal life than in fetal life, giving rise to a U-shaped expression curve. None of the lines carrying wild-type constructs exhibited this pattern; nor has this pattern been observed in previous studies of HS3-γ constructs.9,19 40 Two possibilities exist to explain the apparent increase in γ transgene expression during the transition from fetal to adult life. YY1 binding to the −1086 site could mediate bona fide transcriptional repression in adult life. If this is the case, loss of YY1 binding at this site should cause postnatal upregulation of transcriptional activity. Another possibility is that the apparent fetal activator function of the YY1 binding site at −1086 of the Aγ promoter may lose its importance after birth; that is, the Aγ transcriptional state may be no longer affected by this site. Therefore, the observed effect may represent a rebound to average wild-type postnatal Aγ transgene expression levels. The fact that overall expression levels of Aγ were similar in wild-type and mutant lines (Fig 3) is supportive of the second explanation. Thus, taken together, the data suggest that the −1086 YY1 site may be required for efficient transcription of the γ gene in fetal life, but may not be required for autonomous γ gene silencing in the postnatal period.
In light of the possible role of the −1086 YY1 site in fetal γ activation, it is interesting that the galago γ gene, which is silent in the fetal liver, contains an altered YY1 binding site at the orthologous position.41 The galago γ gene is expressed exclusively in the embryonic period, in contrast to the human γ gene, which is a fetally expressed gene. We recently showed that this change in expression pattern of the γ gene is due to cis changes in regions linked to the γ genes themselves.10 It is possible that these critical changes include the sequences near the −1086 region. Although a YY1 binding site is present in both galago and human at −1086,41 the site has a different core sequence (ACAT in the human and CCAT in the galago) and is on the opposite strand (antisense in the human and sense in the galago). In the c-fos promoter, such a reversal of a YY1 binding site caused a switch in phenotype of the site between repressor and activator.42 Work is under way to test directly whether the introduction of a human-like YY1 site into the galago promoter region would be sufficient to cause fetal activation of this embryonic γ gene.
ACKNOWLEDGMENT
We wish to thank Dr Joyce Lloyd for providing the HS2 fragment that was used in cloning, and Dr Timothy Ley for S1 probes. We are also grateful to Drs Sally Camper and Thom Saunders of the University of Michigan Transgenic Mouse Core for microinjection of constructs.
This work was supported by NIH R01 HL48802 (D.L.G.) and NIH R01 HL33940 (M.G.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal