C/EBPɛ is essential for granulocytic differentiation. We investigated the role of C/EBPɛ in the transcriptional activation of various myeloid-specific genes. We found that two C/EBPɛ isoforms, p32 and p30, possessing transcriptional activation domains were coexpressed in myeloid cells. Interestingly, isoform C/EBPɛ p30 but not p32 was differentially upregulated in NB-4 promyelocytic leukemia cells treated with retinoids. Both isoforms bound specifically to C/EBP sites in myeloid promoters. The kd for C/EBPɛ binding to the C/EBP site of the neutrophil elastase promoter was 4.2 nmol/L. In transfection assays using the nonhematopoietic cell line, CV-1, the p32 isoform activated promoters from the myeloid-specific mim-1, neutrophil elastase, and granulocyte colony-stimulating factor (G-CSF) receptor genes by 2.5-, 1.8-, and 1.6-fold, respectively. The p30 isoform lacked significant transcriptional activity, suggesting that other hematopoietic-specific factors were required for its function. Consistent with this prediction, transfections into the hematopoietic cell line Jurkat showed a 9.0- and 2.5-fold activation of the mim-1 promoter by the p32 and p30 isoforms, respectively. The additional 32 NH2-terminal residues made p32 a significantly more potent transcriptional activator than p30. T lymphoblasts (Jurkat cells) and immature myeloid cells (eg, Kcl22 cells) expressed high levels of the c-myb hematopoietic transcription factor. Cotransfection of c-myb with either the p32 or p30 isoform of C/EBPɛ in CV-1 cells cooperatively transactivated the mim-1 promoter by 20- and 16-fold, respectively, and the neutrophil elastase promoter by 10-and 7-fold, respectively. Pulldown assays showed that each C/EBPɛ isoform interacted directly with the DNA binding domain of the c-myb protein. Further studies showed that Kcl22 myeloid cells only contained active C/EBPɛ, but not C/EBP, C/EBPβ, or C/EBPδ. A mutation of the C/EBP site in the neutrophil elastase promoter markedly decreased the transactivation of the promoter in Kcl22 myeloblasts. These results demonstrate a role for C/EBPɛ in regulating myeloid promoters, such as neutrophil elastase, probably through a direct interaction with c-myb.
C/EBPε1,2 IS A RECENTLY cloned member of the CCAAT enhancer binding protein (C/EBP) family of transcriptional regulators that also includes C/EBPα,3C/EBPβ,4-9 C/EBPγ,10C/EBPδ,5,11,12 and C/EBPζ.13 All members share a highly homologous basic DNA binding region and a leucine zipper dimerization domain. Their DNA binding consensus site has been identified as TKNNGYAAK (Y = C or T, K = T or G).14 In contrast to the wide range of expression of the other C/EBP members, the expression of C/EBPε is restricted to the granulocytic and T-lymphoid lineages. In the granulocytic lineage, C/EBPε is expressed in the later stages of differentiation from the promyelocyte to the mature granulocyte.15,16 Four different C/EBPε isoforms p32, p30, p27, and p14 are generated by alternative promoter usage, alternative mRNA splicing, and alternative translation start sites.2,16 As described for other C/EBP proteins, this produces either full-length proteins (C/EBPε p32, p30, and p27) having DNA binding and transactivation domains (TAD) or a truncated protein (C/EBPε p14) that retains the DNA binding domain but lacks a TAD.17 We recently mapped the essential transactivating element of C/EBPε to amino acid residues 33-50 for p32 and 1-18 for p30.18 Interestingly, the domain with maximal transactivating capacity (residues 1-70 of C/EBPε p30) is present in both isoforms. Details about the expression of C/EBPε isoforms during myeloid differentiation are presently unknown.
A number of myeloid-specific genes contain C/EBP binding sites in their promoter regions, including those encoding the receptors for granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and macrophage colony-stimulating factor (M-CSF),19-21 and the primary granule proteins, such as myeloperoxidase (MPO), neutrophil elastase, and proteinase 3.22-24 C/EBPα, in concert with the transcription factor PU.1, transactivates a number of these myeloid genes, including those encoding the receptors for G-CSF and GM-CSF. C/EBPα transactivates the M-CSF receptor in cooperation with AML-121 and the neutrophil elastase promoter in cooperation with c-myb.25C/EBPβ (NF-IL6, LAP) is known to transactivate a number of lipopolysaccharide (LPS)-responsive genes such as interleukin-1β (IL-1β)26 and tumor necrosis factor-α27 in monocytes. Ectopic expression of C/EBPε in murine macrophage cell lines confers LPS-dependent activation of IL-6 and monocyte chemoattractant protein 1. In addition, macrophage inhibitory protein 1α (MIP-1α), MIP-1β, and the M-CSF receptor have been implicated as targets of C/EBPε.28 Transactivation studies show that the mim-1 and MPO promoters can be activated by C/EBPε in myeloid cells.2
The C/EBP proteins and myb cooperate in transcriptional activation of myeloid genes. This cooperation was first described for NFM (chicken C/EBPβ) and myb, which induced the expression of myelomonocytic-related genes (eg, the mim-1 and lysozyme genes) even in nonmyeloid cells, underlining their importance as a myeloid differentiation switch.29,30 Mammalian C/EBPβ also synergizes with v-myb in the transactivation of mim-1.31Mink et al32,33 showed that both proteins interact with each other via their DNA binding domains. Recently, the cooperation of the two transcription factors was linked to their interaction with the coactivator protein p300.32,33 The coactivators p300 and the closely related CREB binding protein (CBP) are involved in translocation t(11;22) and t(8;16), respectively, which are associated with acute myeloid leukemia.34,35 Although C/EBP proteins are presently not known to be direcly involved in leukemia-associated translocations, a direct interaction between C/EBPα and the leukemia-specific fusion protein AML1-ETO has been shown to interfere with C/EBPα-mediated transactivation of myeloid genes.36
Knock-out murine studies showed that C/EBPα, C/EBPβ, and C/EBPε are critical for normal myelopoiesis. The C/EBPα−/− mice developed a complex phenotype reflecting the wide expression pattern of C/EBPα. In the hematopoietic system, they exhibit a maturation arrest in myelopoiesis at the stage of the immature myeloblasts.37The C/EBPβ−/− mice displayed predominantly defects in macrophage function.38 The C/EBPε deletional mice have aberrations in late differentiation of both the neutrophilic and eosinophic lineages. Granulocytes of C/EBPε−/− mice showed dysplastic features and are not completely functional. These animals died between 3 and 5 months after birth due to infectious complications.39
In this study, we addressed several important questions about the structure and function of C/EBPε. Are both isoforms (p32 and p30) of C/EBPε expressed in myeloid cells? Do they differ in their ability to activate transcription in hematopoietic cells? Do C/EBPε p32 and p30 bind to and transactivate myeloid promoters such as mim-1 and neutrophil elastase? Finally, does either the C/EBPε p32 or p30 form of C/EBPε directly interact and cooperatively activate transcription with c-myb?
MATERIALS AND METHODS
Eukaryotic expression vectors and promoter-reporter gene constructs.
The construction of the C/EBPε (p30) expression vector and the amino-terminal truncation mutants were described previously.2,18 Eukaryotic expression vectors for the C/EBPε isoforms p32, p27, p14, and murine C/EBPα were a generous gift from Dr K. Xanthopoulos (Aurora Inc, San Diego, CA). The pCMV4 c-myb expression vector was kindly provided by Linda Shapiro (St Jude’s Hospital, Memphis, TN), and the HA epitope-tagged CBP expression vector was from R. Goodman (Vollum Institute, Oregon Health Sciences, University of Oregon, Portland, OR).40 Neutrophil elastase promoter-luciferase constructs were described previously.23The Mim-1 promoter (−240 to +150) luciferase construct was a kind gift from Dr A. Leutz (Max Delbrueck Center for Molecular Medicine, Berlin, Germany).30
Preparation of nuclear extracts and Western blotting.
For preparation of nuclear extracts, 5 × 106 COS-1 cells were washed three times with ice-cold phosphate-buffered saline (PBS). After the last wash, adherent cells were scraped off the dish with a rubber policeman and resuspended in 500 μL extraction buffer B (20 mmol/L HEPES, pH 7.9, 20% glycerol, 10 mmol/L NaCl, 0.2 mmol/L EDTA, 1.5 mmol/L MgCl2, 0.1% Triton X, 1 mmol/L dithiothreitol [DTT], 1 mmol/L phenylmethyl sulfonyl fluoride [PMSF], 40 μL/mL Complete [Boehringer, Indianapolis, IN]). After 15 minutes of incubation on ice, the nuclei were pelleted at 250g for 10 minutes. Nuclei were resuspended in extraction buffer B, and NaCl was added dropwise with mixing to a final concentration of 300 mmol/L NaCl. Nuclei were rocked for 60 minutes at 4°C. Samples were microcentrifuged at 12,000 RPM and supernatants were frozen at −80°C.
A total of 107 Kcl22, U937, KG-1, and NB4 cells were washed twice in ice-cold PBS and subsequently lysed in RIPA buffer supplemented with protease inhibitors. NB4 cells (5 × 105) were grown in RPMI + 10% fetal bovine serum (FBS) + 10−6 mol/L all-trans-retinoic acid for 24 hours before protein extracts were prepared in RIPA buffer.
Nuclear extracts (15 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% polyacrylamide gel. The gel was electroblotted on a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, MA). The membrane was blocked with 1% gelatin and incubated with the primary antibody (rabbit polyclonal antibody, dilution 1:1,000 to 1:2,000) and a secondary horseradish peroxidase-conjugated donkey antirabbit antibody (Amersham, Arlington Heights, IL). Detection was performed using the ECL detection kit (Amersham).
The generation of affinity-purified rabbit polyclonal C/EBPε antibody against amino acids 1-115 of C/EBPε p30 was described previously by us.2 Rabbit polyclonal antibodies to C/EBPα, C/EBPβ, C/EBPδ, and murine and human c-myb, epitope (aa 98-108) of influenza hemagglutinin protein as well as CBP were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
In vitro transcription and translation.
C/EBPε and C/EBPα genes cloned downstream of the T7 promoter in pCDNA I/III (Invitrogen, Carlsbad, CA) were transcribed with T7 RNA polymerase and in vitro translated in rabbit reticulocyte lysate using the TNT coupled Reticulocyte lysate system (Promega, Madison, WI) with35S methionine (>1,000 Ci/mmol; Amersham) as a label in the translation reaction. The procedure was performed according to the manufacturer.
Pulldown assays.
A fusion of the gene coding for maltose binding protein (MBP) and C/EBPε was constructed using the pMalc2 plasmid (New England Biolabs, Beverly, MA) as a backbone. C/EBPε cDNA (p30) was cloned in-frame downstream of the MBP sequence. MBP, MBP-C/EBPε, GST-C/EBPε 1-115 (amino-acids 1-115 of C/EBPε p30), and GST were expressed in the bacterial host BL21. GST-c-myb and truncation mutants were kindly provided by Dr Timothy Bender and expressed as previously described.41 42 Soluble protein was prepared by sonication of the bacterial culture 4 hours after induction with 100 mmol/L isoprophyl β-D thiogalactopyranoside (IPTG). Amylose resin (New England Biolabs) or glutathione sepharose beads (Pharmacia Biotech, Uppsala, Sweden) were loaded with equal amounts of the different fusion proteins. The loaded amylose resin and the glutathione sepharose beads were washed three times with column buffer (20 mmol/L Tris-Cl, pH 7.4, 150 mmol/L NaCl, 0.5 mmol/L EDTA, 0.1mmol/L DTT) and then incubated 2 hours either with COS-1 cell nuclear extract expressing either the c-myb or C/EBPε protein or with in vitro translated 35S-methionine labeled C/EBPε protein. The resin (MBP fusion) or the beads (GST fusions) were washed three times with pulldown assay buffer (150 mmol/L NaCl, 20 mmol/L Tris-Cl, pH 7.5, 0.3% NP40, 0.1 mmol/L EDTA, 1 mmol/L DTT, 1 mmol/L PMSF, 10 μg/mL leupeptin and pepstatin) and resuspended in 25 μL 1× SDS-PAGE sample buffer. The samples were boiled for 5 minutes and separated by electrophoresis on a 4% to 15% gradient SDS polyacrylamide minigel (Bio-Rad, Hercules, CA). The gel was electroblotted overnight and probed with antibody against either c-myb or C/EBPε. When using 35S-methionine–labeled proteins, the gel was directly fixed in 40% methanol/10% acetic acid; dehydrated in 10% methanol, 2.5% acetic acid, and 2.5% glycerol; dried; and exposed overnight to Kodak Biomax X-ray film (Eastman Kodak, Rochester, NY).
Electromobility shift assay (EMSA).
Double-stranded oligonucleotides (30 bp) containing the C/EBP consensus site of a specific promoter and adjacent sequences were end-labeled with 32P γATP. A standard reaction contained 1 ng labeled probe, 10 μg COS-1 nuclear extract expressing either C/EBPε or C/EBPα, 2 μg pdIdC, and 4.5 μg bovine serum albumin (BSA) in a 20 μL volume. Competing cold oligonucleotides (shown below at 10- and 100-fold molar excess) or antibodies (1 μg/μL) were added where indicated. Electrophoresis was performed on a 4% polyacrylamide gel at 30 mA. Neutrophil elastase: 5′ TCGAGGCCAGGATGGGGCAATACAACCC 3′; mutant neutrophil elastase: 5′ TCGAGGCCAGGACTCGAGGATACAACCCG 3′; G-CSF receptor: 5′ AAGGTGTTGCAATCCCAGC 3′.
Transient transfections.
For protein expression, 10 μg of eukaryotic expression vector (C/EBPε, C/EBPα, c-myb, and CBP-HA) was transfected into COS-1 cells (10-cm dish). Transfection was performed with Superfect reagent (Qiagen, Valencia, CA) over 3 hours according to the manufacturer’s protocol. Nuclear extracts were prepared after 48 hours, as described above.
For reporter gene assays, CV-1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) + 10% fetal calf serum (FCS). Approximately 5 × 105 cells/dish were plated in 60-mm dishes at 1 day before transfection. Promoter-reporter constucts (5 μg) and expression vectors (100 ng to 1 μg) as well as a control 1 μg pCMV-Gal vector (1 μg) were ethanol precipitated and resuspended in 25 μL sterile TE buffer. Transfection was performed with Superfect reagent (Qiagen) as described above. Cells were washed with PBS and fed with DMEM + 10% FCS. Protein extracts were prepared 40 hours later in reporter lysis buffer (Promega). Luciferase activity was determined according to standard procedures. β-Galactosidase activity was used to normalize transfection efficiencies.
Approximately 2.5 × 107 Kcl22 or U937 myeloid cells were transfected by electroporation at 340 V with 15 μg of reporter plasmid, 5 μg of expression plasmid, and 3 μg of β-galactosidase vector in 500 μL RPMI + 10% FCS. Cells were harvested after 16 hours.
Transfection of 2 to 3 × 106 Jurkat cells was performed with 20 μL Lipofectin reagent (GIBCO BRL, Gaithersburg, MD) according to the manufacturer’s protocol using 3 μg reporter plasmid, 330 ng expression plasmid (or 75 to 750 ng in dose-response experiments), and 1 μg pCMV-βGal vector and adjusted to 5 μg total DNA with empty expression vector. At 24 hours posttransfection, cells were activated with TPA (5 ng/mL) and calcium ionophore A21387 (125 ng/mL). At 48 hours posttransfection, cells were lysed with reporter lysis buffer (Promega) and micocentrifuged at 12,000 RPM. Supernatants were assayed for luciferase activity. To monitor the levels of the exogenously expressed C/EBPε proteins, a pellet from each sample was resuspended in RIPA buffer (50 mmol/L Tris-Cl, pH 8.0, 1% NP40, 0.5% deoxycholicacidsodiumsalt (DOC), 0.1% SDS, 300 mmol/L NaCl, Complete 40 μL/mL), rocked for 1 hour at 4°C, and microcentrifuged at 12,000 RPM. C/EBPε was immunoprecipitated with 0.5 μg affinity-purified C/EBPε antibody and collected with Protein A sepharose beads. The beads were washed three times in IP buffer (50 mmol/L Tris-Cl, pH 7.4, 150 mmol/L NaCl, 0.5% NP40) and then resuspended in SDS-PAGE sample buffer. Samples were electrophoresed on a 10% to 20% gradient polyacrylamide gel containing SDS, electroblotted, and probed with C/EBPε antibody.
RESULTS
Myeloid leukemia cell lines express two transcriptionally active C/EBPε isoforms: p32 and p30.
Two translation start sites were previously described for C/EBPε.1,2 The first generates a protein with a predicted molecular weight of 32.2 kD (p32 isoform); the second in-frame ATG generates a protein that is 32 amino acids shorter (p30 isoform). We examined myeloid leukemia cell lines for the expression of these two isoforms by Western blot analysis and found that both isoforms were present in the myeloid cell lines Kcl22, NB4, and U937 at a ratio of approximately 2:1 (p32:p30). Cell line KG-1 did not express C/EBPε (Fig 1). We previously reported the upregulation of C/EBPε during granulocytic differentiation of cell line NB-4 with retinoid treatment.43 Interestingly, we find that only the p30 isoform of C/EBPε was upregulated 24 hours after treatment of NB4 cells with 10−6 mol/L all-trans retinoic acid. The level of isoform p32 remained unchanged, indicating that both isoforms are differentially regulated (lane 7). For comparison, C/EBPε p32 (lane 1) and p30 (lane 2) proteins expressed in transfected COS-1 cells were run alongside the nuclear extracts from myeloid cell lines.
Western blot demonstrating the expression of p32 and p30 isoforms of C/EBPɛ in myeloid cell lines: Kcl22 myeloblasts (lane 3), NB4 promyelocytes (lane 4), and U937 myelomonoblasts (lane 5). The isoforms p32 and p30 were expressed at a ratio of approximately 2:1. NB4 cells treated with 10−6 mol/L ATRA for 24 hours selectively upregulate C/EBPɛ p30 (lane 7). The KG-1 early myeloblasts did not express detectable C/EBPɛ levels (lane 6). C/EBPɛ p32 (lane 1) and p30 (lane 2) cDNAs transfected and expressed in COS-1 cells were run alongside as positive controls.
Western blot demonstrating the expression of p32 and p30 isoforms of C/EBPɛ in myeloid cell lines: Kcl22 myeloblasts (lane 3), NB4 promyelocytes (lane 4), and U937 myelomonoblasts (lane 5). The isoforms p32 and p30 were expressed at a ratio of approximately 2:1. NB4 cells treated with 10−6 mol/L ATRA for 24 hours selectively upregulate C/EBPɛ p30 (lane 7). The KG-1 early myeloblasts did not express detectable C/EBPɛ levels (lane 6). C/EBPɛ p32 (lane 1) and p30 (lane 2) cDNAs transfected and expressed in COS-1 cells were run alongside as positive controls.
C/EBPε binds to C/EBP sites in myeloid promoters.
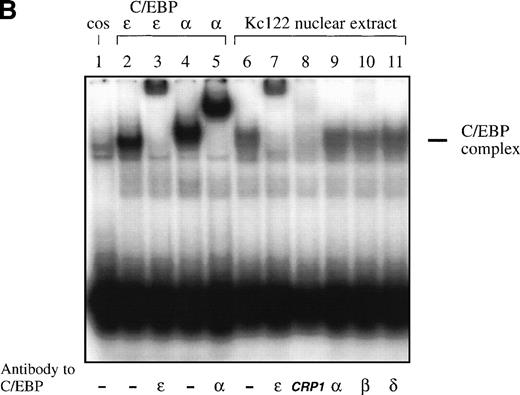
Because the DNA binding domains of C/EBP proteins are highly homologous, we hypothesized that the DNA binding site of C/EBPε is either similar or identical to the known sites of other C/EBP proteins. Double-stranded oligonucleotides representing C/EBP sites of the neutrophil elastase and G-CSF receptor promoter were used for EMSA. C/EBPε (p32/p30) expressed in COS-1 cells was able to bind to both sites and binding was competed by cold self oligonucleotide and supershifted by antibody to C/EBPε. A similar binding was seen with C/EBPα expressed in COS-1 cells (Fig 2A). Also, C/EBPε present in nuclear extracts of the myeloid cell line Kcl22 bound to the C/EBP site of the neutrophil elastase promoter (Fig2B). The complex was supershifted by the affinity-purified C/EBPε antiserum and its formation was inhibited by the commercially available antibody against CRP1 (rat homolog of C/EBPε), demonstrating that the complex contained C/EBPε (Fig 2B, lanes 7 and 8). The complex could not be supershifted by antibodies against C/EBPα, C/EBPβ, and C/EBPδ (Fig 2B, lanes 9 through 11), indicating that these proteins are not present in nuclear extracts of the Kcl22 cell line.
(A) EMSA study. Both C/EBPɛ and C/EBP bound to the C/EBP site of the promoter of the G-CSF receptor. A 32P γ-ATP–labeled, double-stranded oligonucleotide containing the C/EBP site of the promoter of the G-CSF receptor was added to lysate from cos-1 cells expressing either C/EBPɛ or C/EBP. Binding was competed by 20-fold excess of cold-self. Retarded bands were supershifted by specific antisera. (B) EMSA study with 32P γ-ATP–labeled C/EBP consensus site oligonucleotide derived from neutrophil elastase promoter. Oligonucleotides were retarded by C/EBP protein complex present in nuclear extracts of the Kcl22 cell line in characteristic location for C/EBPɛ migration. Antibodies directed against C/EBPɛ (lane 7) and CRP1 (rat C/EBPɛ) (lane 8) either supershifted or blocked binding of the complex, respectively. Antibodies to C/EBP, C/EBPβ, and C/EBPδ did not supershift or block formation of the complex (lanes 9 through 11), indicating that C/EBPɛ is the predominant C/EBP protein binding to neutrophil elastase C/EBP consensus site in Kcl22 nuclear extracts.
(A) EMSA study. Both C/EBPɛ and C/EBP bound to the C/EBP site of the promoter of the G-CSF receptor. A 32P γ-ATP–labeled, double-stranded oligonucleotide containing the C/EBP site of the promoter of the G-CSF receptor was added to lysate from cos-1 cells expressing either C/EBPɛ or C/EBP. Binding was competed by 20-fold excess of cold-self. Retarded bands were supershifted by specific antisera. (B) EMSA study with 32P γ-ATP–labeled C/EBP consensus site oligonucleotide derived from neutrophil elastase promoter. Oligonucleotides were retarded by C/EBP protein complex present in nuclear extracts of the Kcl22 cell line in characteristic location for C/EBPɛ migration. Antibodies directed against C/EBPɛ (lane 7) and CRP1 (rat C/EBPɛ) (lane 8) either supershifted or blocked binding of the complex, respectively. Antibodies to C/EBP, C/EBPβ, and C/EBPδ did not supershift or block formation of the complex (lanes 9 through 11), indicating that C/EBPɛ is the predominant C/EBP protein binding to neutrophil elastase C/EBP consensus site in Kcl22 nuclear extracts.
Because the expression of C/EBPα and C/EBPε partly overlaps during myeloid differentiation and because both proteins can bind to the same DNA motif, we studied their relative DNA binding affinities to the C/EBP binding site of the neutrophil elastase promoter. Decreasing amounts of labeled oligonucleotides representing the C/EBP site were titrated with a constant amount of COS-1 cell extract expressing the respective C/EBP protein, and the bound complexes were separated from the free probe by EMSA (Fig 3A). The amounts of bound and free probe were quantitated with an Ambis imaging system (Ambis Inc, San Diego, CA). The values were plotted as bound/free versus bound, and the results were used to determine the dissociation constant, kd (kd = −1/slope). The kd for C/EBPα was 0.65 nmol/L and the kd for C/EBPε was 4.2 nmol/L (Fig 3B and C), showing that C/EBPα had a 6.5-fold higher affinity than C/EBPε to these C/EBP binding sequences.
(A) Comparison of DNA binding affinity of C/EBPɛ (p32) and C/EBP to neutrophil elastase C/EBP site. EMSA study with32P γ-ATP–labeled C/EBP binding site oligonucleotide from the neutrophil elastase promoter and either C/EBPɛ or C/EBP expressed in COS-1 cells. The same amount of protein was incubated with decreasing concentrations of hot oligonucleotides (left to right). β emissions of the retarded bands and the free hot probe were quantitated by an AMBIS imaging system. (B) and (C) show the Scatchard analyses of the data for C/EBPɛ and C/EBP, respectively. The slope of the curves express the affinity of the protein for the DNA motif. kd: −1/slope: C/EBPɛ: 4.2 nmol/L; C/EBP: 0.65 nmol/L.
(A) Comparison of DNA binding affinity of C/EBPɛ (p32) and C/EBP to neutrophil elastase C/EBP site. EMSA study with32P γ-ATP–labeled C/EBP binding site oligonucleotide from the neutrophil elastase promoter and either C/EBPɛ or C/EBP expressed in COS-1 cells. The same amount of protein was incubated with decreasing concentrations of hot oligonucleotides (left to right). β emissions of the retarded bands and the free hot probe were quantitated by an AMBIS imaging system. (B) and (C) show the Scatchard analyses of the data for C/EBPɛ and C/EBP, respectively. The slope of the curves express the affinity of the protein for the DNA motif. kd: −1/slope: C/EBPɛ: 4.2 nmol/L; C/EBP: 0.65 nmol/L.
C/EBPε p32 is a transcriptional activator in heterologous cells.
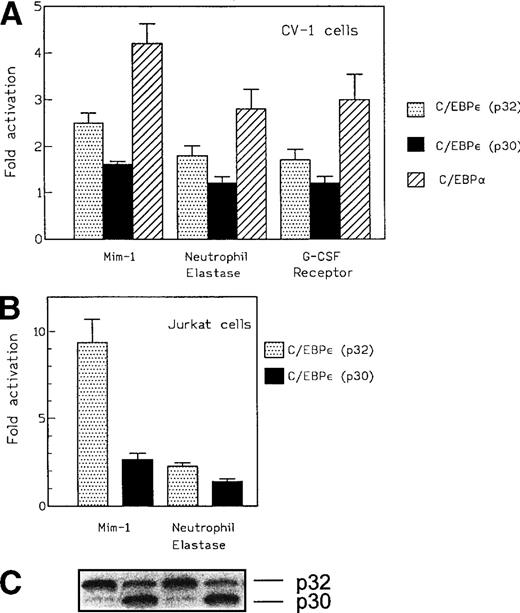
The transactivation potential of C/EBPε isoforms p32 and p30 were initially examined using the promoters of the neutrophil elastase, G-CSF receptor, and mim-1 genes in heterologous cells. Expression plasmids for either C/EBPε p32 or p30 were cotransfected with promoter-luciferase reporter constructs into CV-1 cells, which do not express endogenous C/EBP proteins. Levels of C/EBPε isoforms p32 and p30 proteins after transfection with these expression vectors were shown to be similar by Western blotting of protein extracts from transfected cells (Fig 1, lanes 1 and 2). C/EBPε isoform p32 activated transcription from the neutrophil elastase promoter by 1.8-fold compared with C/EBP site mutants of the wild-type promoters. The mim-1 promoter was activated by 2.5-fold and the G-CSF receptor promoter by 1.6-fold (Fig 4A). These results are the average of three separate experiments. In all cases, the C/EBPε isoform p30 activated the promoters less efficiently than did the p32 isoform. For comparison, the same experiments were performed with C/EBPα. In CV-1 cells, C/EBPα was more active than C/EBPε for each of the promoter-reporter gene constructs (2- to 4-fold activation; Fig 4A).
(A) Effect of C/EBPɛ isoforms p32 and p30 and C/EBP on transcription of promoter-reporter plasmids for the mim-1, neutrophil elastase, and G-CSF receptor promoters in CV-1 cells. Maximal activation was achieved with 500 ng of the expression vector. C/EBPɛ p32 activated the promoters 2.5-, 1.8-, and 1.6-fold, respectively. C/EBPɛ p30 showed only a small effect on the mim-1 promoter (1.5-fold activation). C/EBP was used as a positive control and activated the reporters approximately twofold to fourfold. For experiments using the mim-1 and neutrophil elastase promoter constructs, transactivation by the empty expression plasmid served as the baseline. (B) Effect of C/EBPɛ isoforms p32 and p30 on transcriptional activation of mim-1 and neutrophil elastase promoter in T-cell leukemia cell line Jurkat. Isoform p32 is 3.5-fold more active than p30 on the mim-1 promoter and 1.5-fold more active on the neutrophil elastase promoter. (C) Western blot demonstrates that transfected Jurkat cells expressed equal amounts of C/EBPɛ p32 and p30. (D) Dose-response curve for transcriptional activation potential of C/EBPɛ p30, p32, and C/EBP on mim-1 promoter in T-cell leukemia cell line Jurkat.
(A) Effect of C/EBPɛ isoforms p32 and p30 and C/EBP on transcription of promoter-reporter plasmids for the mim-1, neutrophil elastase, and G-CSF receptor promoters in CV-1 cells. Maximal activation was achieved with 500 ng of the expression vector. C/EBPɛ p32 activated the promoters 2.5-, 1.8-, and 1.6-fold, respectively. C/EBPɛ p30 showed only a small effect on the mim-1 promoter (1.5-fold activation). C/EBP was used as a positive control and activated the reporters approximately twofold to fourfold. For experiments using the mim-1 and neutrophil elastase promoter constructs, transactivation by the empty expression plasmid served as the baseline. (B) Effect of C/EBPɛ isoforms p32 and p30 on transcriptional activation of mim-1 and neutrophil elastase promoter in T-cell leukemia cell line Jurkat. Isoform p32 is 3.5-fold more active than p30 on the mim-1 promoter and 1.5-fold more active on the neutrophil elastase promoter. (C) Western blot demonstrates that transfected Jurkat cells expressed equal amounts of C/EBPɛ p32 and p30. (D) Dose-response curve for transcriptional activation potential of C/EBPɛ p30, p32, and C/EBP on mim-1 promoter in T-cell leukemia cell line Jurkat.
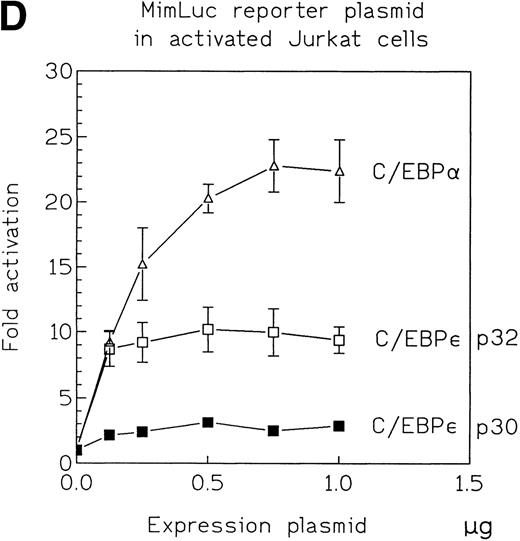
Because the overall reporter gene activity for each of the constructs was low in CV-1 cells, we used Jurkat cells, a hematopoietic cell line, to confirm a potential, concentration-independent, in vivo difference in transcriptional activation potential between C/EBPε p32 and p30. We cotransfected Jurkat cells with expression plasmids (330 ng) for either C/EBPε p32 or p30 with either the mim-1 or neutrophil elastase promoter luciferase construct. After 48 hours, we measured the reporter gene activity in cytoplasmic lysates and determined the C/EBPε expression level in the nuclear lysates (Fig 4B). Reproducibly, C/EBPε p32 was threefold to fourfold more active than p30 on the mim-1 promoter and 1.5-fold more active than p30 on the neutrophil elastase promoter. In both cases, the expression levels of p32 and p30 were similar in the transfected samples (Fig 4C). Dose-response experiments using between 125 and 1,000 ng of the expression vector and 1 μg mim-1 reporter plasmid showed that the maximal effect on reporter gene activity was reached with expression plasmid concentrations as low as 125 to 250 ng. The rank order of transactivation efficiency was C/EBPα > C/EBPε p32 > C/EBPε p30 (Fig 4D). C/EBPε p32 was more potent than C/EBPε p30 at each concentration of the expression vector.
To determine if C/EBPε could activate transcription from these promoters in myeloid cells, the activation of the neutrophil elastase was examined in Kcl22 and U937 cells. As demonstrated (Fig 2B), the only C/EBP family member binding to the neutrophil elastase promoter in Kcl22 cells was C/EBPε. The wild-type neutrophil elastase promoter construct was 7.7-fold more active than the empty vector, p19Luc, in Kcl22 cells and 5.5-fold more active in U937 cells. In both cell lines, the NE promoter with a mutation in the C/EBP site was 30% to 50% less active than the wild-type promoter. Taken together, these data demonstrate that C/EBPε can contribute to the transactivation of the neutrophil elastase promoter in myeloid cells (Fig 5).
Relative luciferase activity of neutrophil elastase promoter constructs with and without C/EBP site mutation. Constructs tested in CV-1 cells and the myeloid leukemia cell lines Kcl22 and U937. Luciferase activty of the empty vector construct p19Luc was arbitrarily set as 1. (▪) Wild-type; (▨) C/EBP site mutant.
Relative luciferase activity of neutrophil elastase promoter constructs with and without C/EBP site mutation. Constructs tested in CV-1 cells and the myeloid leukemia cell lines Kcl22 and U937. Luciferase activty of the empty vector construct p19Luc was arbitrarily set as 1. (▪) Wild-type; (▨) C/EBP site mutant.
Both C/EBPε isoforms (p32 and p30) cooperate with c-myb in the transactivation of the neutrophil elastase and mim-1 promoters.
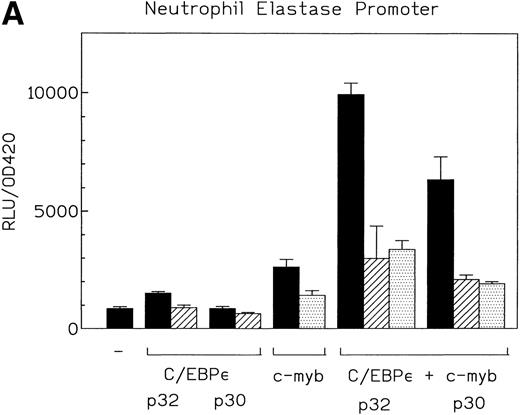
c-myb is a hematopoietic transcription factor known to cooperate with C/EBP proteins. It is highly expressed in Jurkat and Kcl22 cells (data not shown). A number of myeloid promoters have c-myb binding sites adjacent to C/EBP sites. We used the promoters from the neutrophil elastase and mim-1 genes to investigate the potential cooperation of C/EBPε p32 and p30 isoforms with c-myb. c-myb alone activated the neutrophil elastase promoter about twofold. The C/EBPε p32 isoform plus c-myb activated the neutrophil elastase promoter 10-fold in comparison to the empty expression vectors (Fig 6A). The C/EBPε p30 plus c-myb activated the same promoter sevenfold. These results indicate that both C/EBPε isoforms cooperatively activated the NE promoter with c-myb. The combination of both factors was 1.5-fold to twofold higher in activity than an additive effect. Mutation of either the C/EBP or myb sites in the neutrophil elastase promoter resulted in a loss of cooperativity between the factors (Fig 6A). This demonstrated that binding of C/EBPε to the promoter was essential for a significant transcriptional activation. The insertion of 5 bp between the C/EBP and the myb sites to rotate the relative position of the binding sites on the double helix did not change the cooperativity. Insertion of 10 bp slightly reduced cooperativity (Fig 6B).
(A) Cooperative transactivation of the neutrophil elastase promoter by c-myb and either p32 or p30 isoforms of C/EBPɛ. Expression plasmids (500 ng) or empty vectors were cotransfected with 4 μg of either the NELuc reporter or reporter plasmids mutated at either the C/EBP or myb site. Combination of C/EBPɛ and c-myb were 1.5-fold to twofold more active than the additive effect of both factors alone. Binding of both factors to their DNA motif was required for cooperation. (▪) Wild-type; (▨) C/EBP site mutant; ( ) Myb site mutant. (B) Effect of spacing between C/EBP and c-myb binding sites in neutrophil elastase promoter on transcritional activation by C/EBPɛ p30 and c-myb. Two constructs with the insertion of either 5 (NELuc +5) or 10 bp (NELuc +10) between the binding sites were used. (C) Cooperative transactivation of mim-1 promoter (−240 to +150) Luc reporter plasmid with c-myb and either the p32 or p30 isoforms of C/EBPɛ or C/EBP. Expression plasmids or empty vectors (500 ng) were cotransfected with reporter plasmid (4 μg) into CV-1 cells. Luciferase activity was assayed after 40 hours.
) Myb site mutant. (B) Effect of spacing between C/EBP and c-myb binding sites in neutrophil elastase promoter on transcritional activation by C/EBPɛ p30 and c-myb. Two constructs with the insertion of either 5 (NELuc +5) or 10 bp (NELuc +10) between the binding sites were used. (C) Cooperative transactivation of mim-1 promoter (−240 to +150) Luc reporter plasmid with c-myb and either the p32 or p30 isoforms of C/EBPɛ or C/EBP. Expression plasmids or empty vectors (500 ng) were cotransfected with reporter plasmid (4 μg) into CV-1 cells. Luciferase activity was assayed after 40 hours.
(A) Cooperative transactivation of the neutrophil elastase promoter by c-myb and either p32 or p30 isoforms of C/EBPɛ. Expression plasmids (500 ng) or empty vectors were cotransfected with 4 μg of either the NELuc reporter or reporter plasmids mutated at either the C/EBP or myb site. Combination of C/EBPɛ and c-myb were 1.5-fold to twofold more active than the additive effect of both factors alone. Binding of both factors to their DNA motif was required for cooperation. (▪) Wild-type; (▨) C/EBP site mutant; ( ) Myb site mutant. (B) Effect of spacing between C/EBP and c-myb binding sites in neutrophil elastase promoter on transcritional activation by C/EBPɛ p30 and c-myb. Two constructs with the insertion of either 5 (NELuc +5) or 10 bp (NELuc +10) between the binding sites were used. (C) Cooperative transactivation of mim-1 promoter (−240 to +150) Luc reporter plasmid with c-myb and either the p32 or p30 isoforms of C/EBPɛ or C/EBP. Expression plasmids or empty vectors (500 ng) were cotransfected with reporter plasmid (4 μg) into CV-1 cells. Luciferase activity was assayed after 40 hours.
) Myb site mutant. (B) Effect of spacing between C/EBP and c-myb binding sites in neutrophil elastase promoter on transcritional activation by C/EBPɛ p30 and c-myb. Two constructs with the insertion of either 5 (NELuc +5) or 10 bp (NELuc +10) between the binding sites were used. (C) Cooperative transactivation of mim-1 promoter (−240 to +150) Luc reporter plasmid with c-myb and either the p32 or p30 isoforms of C/EBPɛ or C/EBP. Expression plasmids or empty vectors (500 ng) were cotransfected with reporter plasmid (4 μg) into CV-1 cells. Luciferase activity was assayed after 40 hours.
A similar cooperative effect on transactivation of the mim-1 promoter was observed (Fig 6C). The C/EBPε isoform p32 plus c-myb activated the promoter 20-fold, C/EBPε p30 plus c-myb by 16-fold, and C/EBPα plus c-myb by 37-fold. Again, both C/EBPε p32 and p30 cooperated with c-myb in transcriptional activation. Together, the combination was twofold to threefold more active than a simple additive effect.
C/EBPε p32 and p30 isoforms and c-myb can interact in vitro.
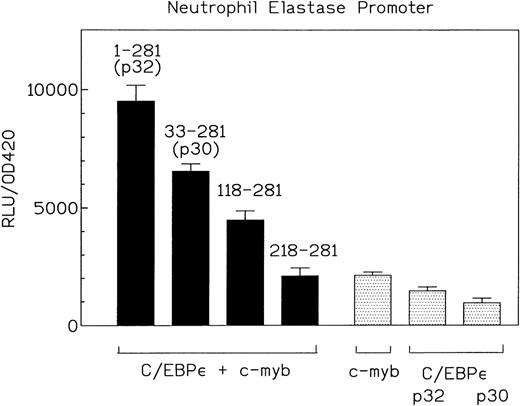
To analyze the structural and functional aspects involved in the cooperativity between C/EBPε and c-myb, we cotransfected various amino-terminal C/EBPε truncation mutants with c-myb and the neutrophil elastase promoter reporter construct (Fig 7). Suprisingly, a truncated C/EBPε protein (aa 118-281) that lacked the complete transactivation domain still cooperatively activated transcription from the promoter with c-myb, although at much lower levels than the full-length C/EBPε (Fig7). The truncation mutants alone did not transactivate the neutrophil elastase promoter (data not shown). This result implicates the carboxy-terminal half (DNA binding domain and leucine zipper) of C/EBPε in the cooperativity with c-myb.
C/EBPɛ and c-myb cooperation is mediated by the carboxy-terminal half of C/EBPɛ. Expression plasmids for C/EBPɛ p32 (aa 1-281), C/EBPɛ p30 (aa 33-281), and two amino-terminal truncation mutants of C/EBPɛ (aa 118-281 and aa 218-281) (500 ng) were cotransfected with c-myb and NELuc reporter plasmid (4 μg) into CV-1 cells. Luciferase activity was assayed after 40 hours. The C/EBPɛ 118-281 mutant lacks the transactivation domain but still cooperates with c-myb.
C/EBPɛ and c-myb cooperation is mediated by the carboxy-terminal half of C/EBPɛ. Expression plasmids for C/EBPɛ p32 (aa 1-281), C/EBPɛ p30 (aa 33-281), and two amino-terminal truncation mutants of C/EBPɛ (aa 118-281 and aa 218-281) (500 ng) were cotransfected with c-myb and NELuc reporter plasmid (4 μg) into CV-1 cells. Luciferase activity was assayed after 40 hours. The C/EBPɛ 118-281 mutant lacks the transactivation domain but still cooperates with c-myb.
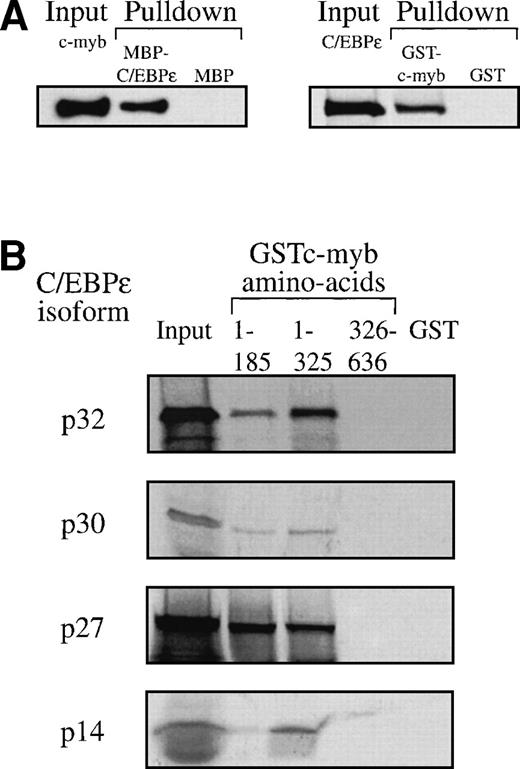
The transcriptional cooperativity between C/EBPε and c-myb could occur via either indirect or direct interactions. We tested whether both factors could physically interact using pulldown assays. c-myb was pulled down by the MBP-C/EBPε fusion, but not by MBP alone (Fig 8A). In the reciprocal experiment, C/EBPε was pulled down by GST-c-myb but not by GST alone (Fig 8A). The same result was obtained with C/EBPε expressed in COS-1 cells or in vitro synthesized, 35S-methionine labeled C/EBPε p32/p30 protein (data not shown). To define further the interacting domains, we used three GST fusion proteins representing different regions of c-myb: (1) the DNA binding domain (aa 1-185), (2) the DNA binding domain and the transactivation domain (aa 1-325), and (3) the negative regulatory domain (aa 326-636). Pulldown assays showed that all 35S-methionine–labeled C/EBPε isoforms (p32, p30, p27, and p14) interacted with the GST-c-myb fusions 1-185 and 1-325 containing the DNA binding domain of c-myb. No interaction ocurred with either the GST-c-myb fusion 326-636 or the GST control. Furthermore, interaction of c-myb with all C/EBPε isoforms indicated that the carboxy-terminal half of C/EBPε, which is common to all isoforms, is sufficient for the interaction (Fig 8B).
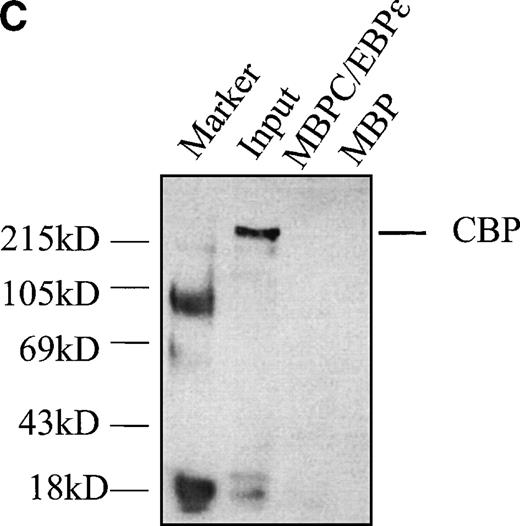
(A) C/EBPɛ physically interacts with c-myb. MBP-C/EBPɛ (p30) and GST-c-myb were used for in vitro pulldown assays. c-Myb was expressed in COS-1 cells; C/EBPɛ (p32) was35S methionine-labeled by in vitro translation in rabbit reticulocyte lysate. Pulldown of c-myb by MBP-C/EBPɛ (left); pulldown of C/EBPɛ (p32) by GSTc-myb (right). (B) Pulldown assay with three GSTc-myb truncation mutants: (a) DNA binding domain only, amino acids (aa) 1-185; (b) DNA binding domain + transactivation domain of c-myb, aa 1-325; and (c) amino-terminal, negative regulatory domain only, aa 326-636. 35S-methionine–labeled, in vitro-translated C/EBPɛ isoforms p32, p30, p27, and p14 were used as input for separate pulldown experiments. All C/EBPɛ isoforms were pulled down by GSTc-myb fusion proteins containing the DNA binding domain. (C) MBP C/EBPɛ p30 pulldown assay with CBP as input. CBP does not bind to C/EBPɛ p30 in vitro.
(A) C/EBPɛ physically interacts with c-myb. MBP-C/EBPɛ (p30) and GST-c-myb were used for in vitro pulldown assays. c-Myb was expressed in COS-1 cells; C/EBPɛ (p32) was35S methionine-labeled by in vitro translation in rabbit reticulocyte lysate. Pulldown of c-myb by MBP-C/EBPɛ (left); pulldown of C/EBPɛ (p32) by GSTc-myb (right). (B) Pulldown assay with three GSTc-myb truncation mutants: (a) DNA binding domain only, amino acids (aa) 1-185; (b) DNA binding domain + transactivation domain of c-myb, aa 1-325; and (c) amino-terminal, negative regulatory domain only, aa 326-636. 35S-methionine–labeled, in vitro-translated C/EBPɛ isoforms p32, p30, p27, and p14 were used as input for separate pulldown experiments. All C/EBPɛ isoforms were pulled down by GSTc-myb fusion proteins containing the DNA binding domain. (C) MBP C/EBPɛ p30 pulldown assay with CBP as input. CBP does not bind to C/EBPɛ p30 in vitro.
C/EBPε isoform p30 does not directly interact with coactivator protein CBP/p300 in vitro.
Transcriptional cooperativity between v-myb and C/EBPβ has been attributed to interaction with the coactivator protein, CBP/p300. To investigate a possible interaction between C/EBPε p30 and CBP/p300, we performed pulldown assays using MBP-C/EBPε as bait and CBP expressed in cos-1 cells as input using the same techniques as those described above. No in vitro interaction between CBP and C/EBPε p30 was detected (Fig 8C).
DISCUSSION
Regulation of eukaryotic transcription is a highly complex process involving chromatin accessibility, basal transcriptional machinery, and the interplay of ubiquitously expressed and lineage-restricted factors. C/EBPε has a highly restricted pattern of expression limited predominately to the granulocytic lineage.30 Therefore, it is a good candidate for the regulation of lineage-restricted target genes in granulocytic cells. Because of the high degree of homology of the DNA binding domain between C/EBPε and other C/EBP family members and published data for the rat homolog CRP1,10 we reasoned that C/EBPε can bind to the same C/EBP consensus sites. Although we demonstrated binding to the C/EBP sites of the neutrophil elastase and G-CSF receptor promoters, the binding affinity was lower for C/EBPε than for C/EBPα. Further studies are needed to clarify whether homodimers or possibly heterodimers of C/EBPε can bind with higher affinity to other DNA motifs.
The differential upregulation of C/EBPε p30 in NB4 cells treated with retinoids suggests a separate functional role for each isoform in myeloid differentiation. Although both C/EBPε p32 and p30 contain the previously mapped transactivation domain,18 our data show that they differ in their potency by which they activate target promoters. C/EBPε p32 was a significantly stronger transactivator of the mim-1 and neutrophil elastase promoters. We hypothesized that the NH2-terminal 32 amino acids may be important for interacting with additional proteins involved in the transcription of target genes of C/EBPε p32, but not p30.
The overall low transcriptional activation potential of C/EBPε in heterologous cells could be a consequence of either low affinity for the selected sites or an indication that other factors are required either for cooperation with or activation of C/EBPε in myeloid differentiation. In two model systems, we showed that a stronger transactivation can be mediated by the cooperation of C/EBPε with the hematopoietic transcription factor c-myb. For the first time, we demonstrated that a C/EBP protein can directly interact with c-myb in vitro. Interacting regions are the DNA binding domain of c-myb and the carboxy-terminal half of C/EBPε, common to all four C/EBPε isoforms. The significance of this finding is supported by our functional studies using amino-terminal truncation mutants of C/EBPε in transient transfections with c-myb which suggested an important functional role for the carboxy-terminal half of C/EBPε in the cooperation with c-myb. Mink et al32previously reported the interaction between the DNA binding domains of avian myeloblastosis virus v-myb and C/EBPβ. We conclude that the three mutations in the DNA binding domain of AMV v-myb (aa 91, 106, and 117), although important for differential binding to DNA motifs in comparison to E26 v-myb and c-myb,44 do not affect their binding to C/EBP proteins. However, the small reduction of cooperativity achieved by the insertion of 10 bp between the C/EBP and c-myb binding sites of the neutrophil elastase promoter suggests an additional mechanism. Recently, studies have shown that C/EBPβ interacted with the coactivator protein p300/CBP, which was important for the cooperation between v-myb and C/EBPβ.33 We did not observe an interaction between C/EBPε p30 and CBP in pulldown assays. Additional studies will clarify whether differences exist among C/EBPε isoforms and their interaction with coactivator proteins.
A cooperation between C/EBPε and c-myb may be relevant for target genes expressed in promyelocytes. Our data suggest a role for C/EBPε in the transcriptional regulation of the neutrophil elastase promoter. This promoter is transcriptionally regulated by members of at least three transcription factor families: C/EBP, myb, and ets factors. If multiple family members with similar binding capacity are expressed in one cell type, predicting which factor is most important in vivo is difficult. The ets family members GABP and PU.1 are both expressed in myeloid cells. Recently, studies showed that GABP is a significantly more potent activator of transcription from the neutrophil elastase promoter than PU.1, suggesting that GABP is the important ets factor in vivo.45 Also, C/EBPα and C/EBPε are coexpressed in myeloid cells expressing neutrophil elastase. Both factors can bind to the promoter and synergize with c-myb. The relative contributions of C/EBPα and C/EBPε to the expression of the neutrophil elastase gene in normal myeloid differentiation remain to be elucidated. Interestingly, forced overexpression of C/EBPε in NB4 cells increased clonal growth,2 a finding possibly related to its capacity to cooperate with c-myb, a transcription factor known to be involved in the regulation of hematopoietic cell proliferation.46
In conclusion, p32 and p30 isoforms of C/EBPε act as transcriptional activators; and their potency is markedly enhanced by c-myb. Another partner may be required for efficient transcriptional activation by C/EBPε p30 in late myeloid differentiation.
ACKNOWLEDGMENT
The authors thank Dr Kleanthis Xanthopoulos for his helpful suggestions and his generosity in providing reagents.
Supported in part by National Institutes of Health and US Army grants as well as by the C. + K. Koeffler + the Parker Hughes Fund. H.P.K. holds the Mark Goodson Chair in Oncology and is a member of the Jonsson Cancer Center.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to H. Phillip Koeffler, MD, Cedars-Sinai Medical Center, Becker Building, Room B 213, 8700 Beverly Blvd, Los Angeles, CA 90048.








This feature is available to Subscribers Only
Sign In or Create an Account Close Modal