Abstract
The number of genetic lesions necessary to generate leukemia in humans is unknown, but it is possible that certain specific abnormalities, eg, fusion genes, known to be associated with acute and chronic leukemia are produced relatively frequently in human cells but require other events to occur before the leukemia becomes manifest. We investigated this possibility by studying peripheral blood leukocytes from normal individuals and various hematopoietic cell lines for the presence and expression of the p210 and the p190 types of the BCR-ABL gene associated with chronic myeloid leukemia (CML) and acute lymphoblastic leukemia. We used two-step reverse transcriptase-polymerase chain reaction (RT-PCR) assays in which batches of 108 cells per sample were tested in 40 replicate reactions. We estimate that this assay is 1.5 logs more sensitive than the two-step RT-PCR assays that we use routinely to assess minimal residual disease. BCR-ABL fusion gene transcripts of various configurations were found in circulating leukocytes from 12 of the 16 healthy adults analyzed. Transcripts with an e1a2 junction (p190 BCR-ABL) were present in 11 and p210-type transcripts with b2a2 and/or b3a2 junctions were detected in 4 individuals. The same RT-PCR assays in non-CML cell lines showed the presence of classical or aberrant p210-type mRNA in 3 of 7 lines and of p190-type transcripts in all 7 lines of hematopoietic origin (HL60, KG1, U937, Kasumi, Jurkat, JVM13, and JVM25), whereas the NIH3T3 murine fibroblast line was reproducibly negative for these fusion genes. These findings confirm and extend previous reports on the detection of leukemia-associated genes in normal leukocytes and suggest that certain fusion genes are generated relatively frequently in hematopoietic cells, but only infrequently do the cells acquire the additional changes necessary to produce leukemia in humans. Although there is only a small probability that such innocent BCR-ABL–carrying leukocytes are detected by conventional RT-PCR assays, they may be the source of some sporadically positive tests in leukemia patients in long-term remission.
© 1998 by The American Society of Hematology.
THE DEMONSTRATION OF the t(9;22)(q34;q11) chromosomal translocation and/or of the BCR-ABL fusion gene in hematopoietic cells from patients with clinical features of a chronic myeloproliferative disease usually establishes the diagnosis of chronic myeloid leukemia (CML). Likewise, the presence of BCR-ABL in the blast cell population of patients with acute leukemia is generally accepted as the molecular basis of Philadelphia (Ph)-positive acute lymphoblastic leukemia (ALL) or, more rarely, acute myeloblastic leukemia (AML). In CML, the breakpoint in the BCR gene nearly always occurs in the major breakpoint cluster region (M-bcr), and the resultant BCR-ABL mRNA molecules with a b2a2 or a b3a2 junction encode a p210BCR-ABL fusion protein. In two thirds of Ph+ ALLs, the BCR breakpoint falls in the minor breakpoint cluster region (m-bcr) and the hybrid BCR-ABL transcript contains an e1a2 junction and is translated as a p190BCR-ABL product.1 The fact that both BCR-ABL fusion proteins can induce a CML-like or an acute lymphoproliferative disease in murine models2-4 provides strong evidence of their fundamental causal role of these leukemias in humans.
It is estimated that patients with leukemia have a total burden of more than 1012 malignant cells in the body at presentation,5 and the overwhelming majority of their circulating leukocytes harbor the fusion gene. The kinetics of expansion of the BCR-ABL–containing clone during the preclinical stage are not known, because this clone is usually identified only when the leukemia becomes overt. There is some indirect evidence that the formation of the BCR-ABL hybrid gene may not be the initial or the sole event in the development of CML. For example, studies of allelic biochemical markers in Epstein-Barr virus (EBV)-transformed B cells from chronic-phase CML show that most clones expressing the same markers as the malignant clone have a high frequency of cytogenetic abnormalities; however, they may not contain the t(9;22) translocation.6 Similarly, we and others7-9have observed a number of patients who, after treatment with interferon-α, achieve a complete cytogenetic response, as defined by the disappearance of Ph+ metaphases, but show the emergence of another clone with a different chromosomal abnormality. These cases may reflect the existence of a pool of secondary, weaker clonal mutations in the background of chronic-phase CML, which only becomes visible when the predominant t(9;22)-positive clone is suppressed. Overall, the data suggest that the pathogenesis of CML, like that of most types of cancer, is multistep.
The idea that BCR-ABL, and perhaps other fusion genes, may be formed inconsequently in hematopoietic cells, ie, without necessarily leading to a selective growth advantage, was strengthened recently by two studies. The first was the report by Biernaux et al10 on the detection of hybrid BCR-ABL mRNA transcripts in leukocytes from approximately 30% of normal adults. The second piece of evidence comes from our observation that various types of leukemia-associated fusion genes are generated at low frequency in some hematopoietic cell lines in standard culture conditions in the absence of specific mutagenic stimuli.11 We have now extended this line of investigation by analyzing peripheral blood leukocytes from healthy individuals as well as a series of Ph− hematopoietic cell lines for the presence of BCR-ABL fusion transcripts of the CML (p210) and/or the ALL (p190) types. We show here that a low background of BCR-ABL hybrid gene formation can be actually detected in leukocytes from the majority of normal adults and in non-CML cell lines, with a strong predominance for the p190 type of fusion.
MATERIALS AND METHODS
Blood samples.
Sixteen healthy subjects were studied, a group composed of 7 male and 9 female laboratory workers aged 23 to 46 years of age (mean ± SD = 33 ± 7 years of age). A single donation of 100 to 120 mL peripheral blood was obtained from each individual after informed consent was obtained, and the samples were immediately coded to ensure anonymity of the donors. The blood specimen was subjected to red blood cell lysis,12 and the isolated white blood cells (WBCs) were washed twice in phosphate-buffered saline (PBS) and lysed in a guanidinium thiocyanate (GTC) solution.13 Multiple aliquots of GTC lysates corresponding each to 107 WBCs were stored at −80°C until processed for RNA extraction.
Cell lines.
Seven human hematopoietic cell lines and one murine fibroblast line (NIH-3T3) were analyzed in this study. The former group was composed of KG1, HL60 and Kasumi (AML lines), U937 (histiocytic lymphoma), Jurkat (T-ALL), JVM13 (B-prolymphocytic leukemia), and JVM25 (EBV-immortalized normal lymphoblastoid line). Stocks from all these cell lines were analyzed by conventional cytogenetics and fluorescence in situ hybridization (FISH) with BCR and ABL probes14 to confirm the absence of a t(9;22). Cells from exponentially growing cultures of these lines were harvested, washed in PBS, and lysed in GTC for storage as described above.
Reverse transcriptase-polymerase chain reaction (RT-PCR) assay.
RNA extraction and reverse transcription were performed as previously reported.13,15 The 40 μL cDNA synthesis from each 107 cell aliquot was diluted in distilled H2O to 80 μL, and 1 μL was tested for quality control in a one-step PCR amplification of normal ABL sequences.16 The remaining 79 μL cDNA from each of 10 × 107 cell aliquot was then divided into 4 × 100 μL PCR tests for the first-step amplification of a given fusion gene transcript, followed by a second-step (nested) amplification of 1 μL first-step products. Separate amplifications of p210- and p190-type BCR-ABL transcripts were performed with primers, reaction composition, and thermocycling conditions as previously described17(Fig 1). Positive controls for these PCRs were diluted cDNA preparations from the BV173 (for p210 BCR-ABL) and the SD1 (for p190 BCR-ABL) cell lines, previously standardized in our single-test diagnostic protocols for reproducible detection of 1 leukemia cell in 105 to 106 nonhematopoietic cells (murine fibroblasts).
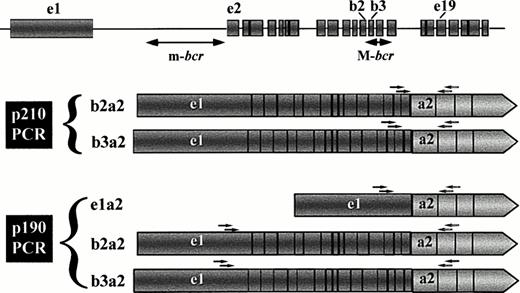
Schematic representation of the types of leukemia-associated BCR-ABL transcripts that can be amplified in the two PCR protocols. Primers for first-step and second (nested)-step amplifications are indicated as arrows over the exon regions corresponding to their sequences. The upper diagram shows the BCR gene structure according to Chissoe et al29 and the major and minor breakpoint cluster regions (M-bcr and m-bcr, respectively). Because of the position of the BCR primers, the p210 PCR assay can only detect transcripts derived from breaks in M-bcr, whereas the p190 PCR assay is able to amplify fusion transcripts arising from breakpoints in either m-bcr or M-bcr (ie, from alternative splicing of the primary M-bcr derived message).
Schematic representation of the types of leukemia-associated BCR-ABL transcripts that can be amplified in the two PCR protocols. Primers for first-step and second (nested)-step amplifications are indicated as arrows over the exon regions corresponding to their sequences. The upper diagram shows the BCR gene structure according to Chissoe et al29 and the major and minor breakpoint cluster regions (M-bcr and m-bcr, respectively). Because of the position of the BCR primers, the p210 PCR assay can only detect transcripts derived from breaks in M-bcr, whereas the p190 PCR assay is able to amplify fusion transcripts arising from breakpoints in either m-bcr or M-bcr (ie, from alternative splicing of the primary M-bcr derived message).
Each batch of PCR tests contained 40 tubes of test cDNAs from 10 cell aliquots, 1 tube with the positive control cDNA, and 4 negative controls derived from the RNA extraction, cDNA synthesis, and first-step and second-step PCR blanks. Extensive precautions were taken to avoid the risk of PCR contamination. The six stages of cell preparation, RNA extraction, cDNA synthesis, first-step PCR, second-step PCR, and electrophoresis of PCR products were each performed in six separate laboratory rooms in dedicated laminar air-flow cabinets. Mixes of reagents for cDNA synthesis and PCR amplification were prepared in bulk and stored as single-use aliquots with dedicated pipettes and plugged tips, in a PCR-free laboratory, by personnel not involved with handling cells and/or actual PCR procedures. Aliquots from all mixes were extensively tested on mock 45-tube PCR assays containing no DNA template (H2O blanks) before and at various intervals during the period of testing of actual cellular material and were always found to be contamination-free. A murine cell line (NIH-3T3) representing a source of cellular RNA from which amplification of a human BCR-ABL message could definitely not occur was used as a reliable negative control for contamination.
Five microliters of the second-step PCR products were electrophoresed through ethidium-bromide stained agarose gels, visualized, and photographed under UV light. In view of the extensive and comprehensive negative controls included for every procedure in this study, a sample was scored as positive if any of the 40 tests produced an amplified product. For confirmation of the BCR-ABL nature of these products, representative fragments of each individual size were gel-purified and sequenced by conventional methods.
RESULTS
In this study, we investigated the possibility that BCR-ABL fusion genes are generated at low frequency in vivo and/or in vitro in rare cells from within a t(9;22)-negative cell population. To detect these postulated rare events, we analyzed the total amount of cDNA synthesized from 108 normal WBCs or from Ph− cell lines in 40 replicate PCR tests for the presence of p210- or p190-type fusion mRNA transcripts (Fig 2). The results of these tests are presented in Tables 1 and2.
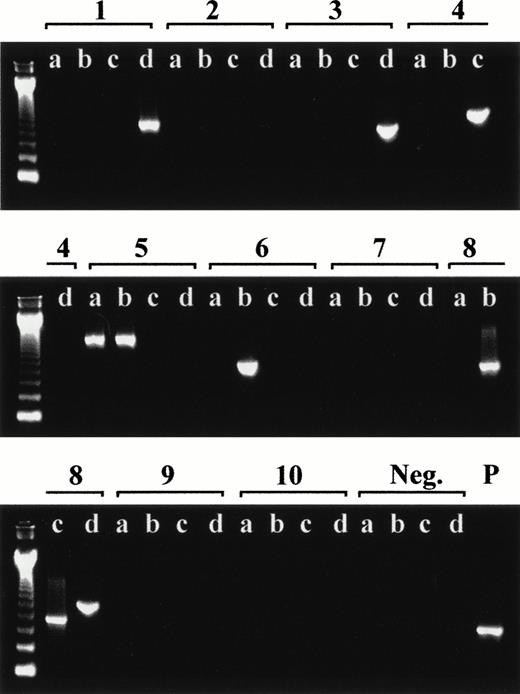
Representative RT-PCR screening for p190 BCR-ABL transcripts in leukocytes from a healthy adult. The ethidium-bromide–stained agarose gels show quadruplicate PCRs (labeled a, b, c, and d) for each cDNA aliquot (labeled 1 through 10) including 1 of the 4 negative (Neg.) controls and 1 positive (P) control specimen (SD1 cell line). The first lane on each gel is a 123-bp DNA ladder marker.
Representative RT-PCR screening for p190 BCR-ABL transcripts in leukocytes from a healthy adult. The ethidium-bromide–stained agarose gels show quadruplicate PCRs (labeled a, b, c, and d) for each cDNA aliquot (labeled 1 through 10) including 1 of the 4 negative (Neg.) controls and 1 positive (P) control specimen (SD1 cell line). The first lane on each gel is a 123-bp DNA ladder marker.
Detection of BCR-ABL Transcripts in Circulating Leukocytes From Healthy Adults
| Normal Donor No. . | Sex/Age . | p210 BCR-ABL . | p190 BCR-ABL . | ||
|---|---|---|---|---|---|
| Positive cDNA Sample-150 (of 10) . | Positive PCR Test (of 40) . | Positive cDNA Sample-150 (of 10) . | Positive PCR Test (of 40) . | ||
| 1 | M/34 | 0 | 0 | 6 | 9 |
| 2 | M/28 | NT | NT | 3 | 6 |
| 3 | M/27 | 3 | 7 | 4 | 5 |
| 4 | F/32 | 0 | 0 | 2 | 2 |
| 5 | F/28 | 0 | 0 | 3 | 4 |
| 6 | F/26 | 1 | 1 | 0 | 0 |
| 7 | F/23 | 0 | 0 | 0 | 0 |
| 8 | F/35 | 0 | 0 | 4 | 4 |
| 9 | F/46 | 0 | 0 | 0 | 0 |
| 10 | F/27 | 0 | 0 | 1 | 1 |
| 11 | M/43 | 3 | 3 | 2 | 2 |
| 12 | M/40 | 0 | 0 | 2 | 3 |
| 13 | M/32 | 0 | 0 | 0 | 0 |
| 14 | F/32 | 0 | 0 | 0 | 0 |
| 15 | M/35 | 2 | 4 | 7 | 18 |
| 16 | F/33 | 0 | 0 | 3 | 4 |
| Normal Donor No. . | Sex/Age . | p210 BCR-ABL . | p190 BCR-ABL . | ||
|---|---|---|---|---|---|
| Positive cDNA Sample-150 (of 10) . | Positive PCR Test (of 40) . | Positive cDNA Sample-150 (of 10) . | Positive PCR Test (of 40) . | ||
| 1 | M/34 | 0 | 0 | 6 | 9 |
| 2 | M/28 | NT | NT | 3 | 6 |
| 3 | M/27 | 3 | 7 | 4 | 5 |
| 4 | F/32 | 0 | 0 | 2 | 2 |
| 5 | F/28 | 0 | 0 | 3 | 4 |
| 6 | F/26 | 1 | 1 | 0 | 0 |
| 7 | F/23 | 0 | 0 | 0 | 0 |
| 8 | F/35 | 0 | 0 | 4 | 4 |
| 9 | F/46 | 0 | 0 | 0 | 0 |
| 10 | F/27 | 0 | 0 | 1 | 1 |
| 11 | M/43 | 3 | 3 | 2 | 2 |
| 12 | M/40 | 0 | 0 | 2 | 3 |
| 13 | M/32 | 0 | 0 | 0 | 0 |
| 14 | F/32 | 0 | 0 | 0 | 0 |
| 15 | M/35 | 2 | 4 | 7 | 18 |
| 16 | F/33 | 0 | 0 | 3 | 4 |
Abbreviation: NT, not tested.
cDNA synthesized from 107 cells.
Detection of BCR-ABL Transcripts in Primary Normal Leukocytes and in Cell Lines: Summary of Results
| Source of Cells . | Characteristics . | No. Positive/ No. Tested . | |
|---|---|---|---|
| p210 BCR-ABL . | p190 BCR-ABL . | ||
| Normal adults (PB WBC) | 7 male and 9 female, age = 33 ± 7 yr | 4/15 (27%) | 11/16 (69%) |
| Hematopoietic cell lines (Ph−) | AML, histiocytic NHL, T-ALL, B-PLL, normal EBV-transformed B cells | 3/7 (43%) | 7/7 (100%) |
| Murine fibroblast line | NIH-3T3 (4 batches) | 0/2 (0%) | 0/4 (0%) |
| Source of Cells . | Characteristics . | No. Positive/ No. Tested . | |
|---|---|---|---|
| p210 BCR-ABL . | p190 BCR-ABL . | ||
| Normal adults (PB WBC) | 7 male and 9 female, age = 33 ± 7 yr | 4/15 (27%) | 11/16 (69%) |
| Hematopoietic cell lines (Ph−) | AML, histiocytic NHL, T-ALL, B-PLL, normal EBV-transformed B cells | 3/7 (43%) | 7/7 (100%) |
| Murine fibroblast line | NIH-3T3 (4 batches) | 0/2 (0%) | 0/4 (0%) |
Abbreviations: PB WBCs, peripheral blood white blood cells; AML, acute myeloblastic leukemia; ALL, acute lymphoblastic leukemia; PLL, prolymphocytic leukemia.
BCR-ABL fusion transcripts of the p210 type were detected in leukocytes from 4 of 15 (27%) normal individuals and in 3 (HL60, KG1, and Jurkat) of the 7 hematopoietic cell lines tested, but never in the NIH-3T3 murine fibroblast line. In 3 individuals and in the Jurkat cell line, 2 to 3 of the 10 cell aliquots yielded positive bands in more than 1 of the 4 PCR replicates; in the fourth healthy subject, as well as in HL60 and KG1, amplification products were observed in only 1 of 10 cell aliquots. Fusion transcripts with classical b2a2 and/or b3a2 junction were detected in all 4 positive individuals, and in 2 of these, additional hybrid molecules with unusual junctions, such as b4a2, b5a2, and e17a2, were also coexpressed. Jurkat cells exhibited b3a2 transcripts, whereas in KG1 and HL60, PCR products of larger sizes were found, representing mRNA molecules with variable fragments of ABL intron 1a inserted between BCR exon b5 and ABL exon a2 (Fig 3A).
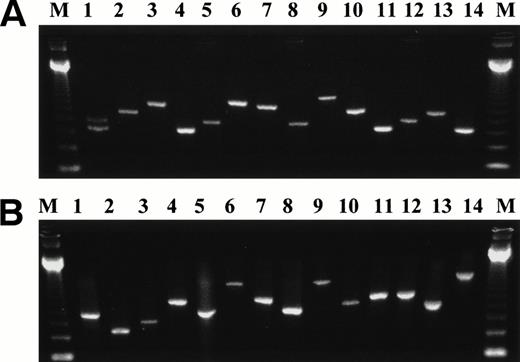
Representative second-step RT-PCR products of the different types of BCR-ABL fusion transcripts detected in leukocytes from normal individuals and non-CML cell lines. The BCR-ABL junction is represented by a double-headed arrow (↔). (A) p210-type transcripts: 1 = BCR ex.b2 ↔ ABL ex.a2 and BCR ex.b3 ↔ ABL ex.a2; 2 = BCR ex.b4 ↔ ABL ex.a2; 3 = BCR ex.b5 ↔ ABL ex.a2; 4 = BCR ex.b2 ↔ ABL ex.a2; 5 = BCR ex.b3 ↔ ABL ex.a2; 6 = BCR ex. e17 ↔ ABL ex.a2; 7 = BCR ex.b5 ↔ ABL ex.a2; 8 = BCR ex.b3 ↔ ABL ex.a2; 9 = BCR ex.b5 ↔ 153 bp of ABL intron 1a + ex.a2; 10 = BCR ex.b5 ↔ ex.a2; 11 = BCR ex.b2 ↔ ABL ex.a2; 12 = BCR ex.b3 ↔ ABL ex.a2; 13 = BCR ex.b4 ↔ ABL ex.a2; 14 = BCR ex.b2 ↔ ABL ex.a2. (B) p190-type transcripts: 1 = BCR ex.e1 ↔ ABL ex.a2; 2 = BCR ex.e1 + 56 bp of intron 1 ↔ ABL ex.a3; 3 = BCR ex.e2 ↔ ABL ex.a3; 4 = BCR ex.5 ↔ ABL ex.a3; 5 = BCR ex.e1 ↔ ABL ex.a2; 6 = BCR ex.e4 ↔ ABL ex.a2; 7 = BCR ex.e1 + 108 bp of intron 1 ↔ ABL ex.a2; 8 = BCR ex.e1 ↔ ABL ex.a2; 9 = BCR ex.e4 ↔ ABL ex.a2; 10 = BCR ex.e1 + 46 bp of intron 1 ↔ ABL ex.a2; 11 = BCR ex.e2 ↔ ABL ex.a2; 12 = BCR ex.e1 + 172 bp of intron 1 ↔ ABL ex.a2; 13 = BCR ex.e1 ↔ ABL ex.a2; 14 = BCR ex.e1 + 246 bp of intron 1 ↔ 213 bp of ABL intron 2 + ex.a3.
Representative second-step RT-PCR products of the different types of BCR-ABL fusion transcripts detected in leukocytes from normal individuals and non-CML cell lines. The BCR-ABL junction is represented by a double-headed arrow (↔). (A) p210-type transcripts: 1 = BCR ex.b2 ↔ ABL ex.a2 and BCR ex.b3 ↔ ABL ex.a2; 2 = BCR ex.b4 ↔ ABL ex.a2; 3 = BCR ex.b5 ↔ ABL ex.a2; 4 = BCR ex.b2 ↔ ABL ex.a2; 5 = BCR ex.b3 ↔ ABL ex.a2; 6 = BCR ex. e17 ↔ ABL ex.a2; 7 = BCR ex.b5 ↔ ABL ex.a2; 8 = BCR ex.b3 ↔ ABL ex.a2; 9 = BCR ex.b5 ↔ 153 bp of ABL intron 1a + ex.a2; 10 = BCR ex.b5 ↔ ex.a2; 11 = BCR ex.b2 ↔ ABL ex.a2; 12 = BCR ex.b3 ↔ ABL ex.a2; 13 = BCR ex.b4 ↔ ABL ex.a2; 14 = BCR ex.b2 ↔ ABL ex.a2. (B) p190-type transcripts: 1 = BCR ex.e1 ↔ ABL ex.a2; 2 = BCR ex.e1 + 56 bp of intron 1 ↔ ABL ex.a3; 3 = BCR ex.e2 ↔ ABL ex.a3; 4 = BCR ex.5 ↔ ABL ex.a3; 5 = BCR ex.e1 ↔ ABL ex.a2; 6 = BCR ex.e4 ↔ ABL ex.a2; 7 = BCR ex.e1 + 108 bp of intron 1 ↔ ABL ex.a2; 8 = BCR ex.e1 ↔ ABL ex.a2; 9 = BCR ex.e4 ↔ ABL ex.a2; 10 = BCR ex.e1 + 46 bp of intron 1 ↔ ABL ex.a2; 11 = BCR ex.e2 ↔ ABL ex.a2; 12 = BCR ex.e1 + 172 bp of intron 1 ↔ ABL ex.a2; 13 = BCR ex.e1 ↔ ABL ex.a2; 14 = BCR ex.e1 + 246 bp of intron 1 ↔ 213 bp of ABL intron 2 + ex.a3.
RT-PCR tests with primers designed to amplify predominantly the p190 type of BCR-ABL transcripts yielded positive results in 11 of 16 (69%) of the healthy subjects and in all 7 hematopoietic cell lines. Only 1 of the 4 individuals positive for the p210 type of transcripts did not show coexpression of the p190 type of message. Four batches of NIH-3T3 murine fibroblasts tested on different occasions during this study were reproducibly negative in a total of 160 PCRs. In the normal adults testing positive, the number of cell aliquots with detectable products varied from 1 to 7 of 10 and in approximately one fourth of these cell aliquots amplified fragments were obtained in at least 2 of the 4 replicate PCRs. The 7 hematopoietic cell lines showed 1 to 9 cDNA aliquots with p190 amplification products, almost invariably in replicate PCR tests. The majority (74%) of the transcripts had the expected e1a2 junction, but in 7 samples larger fragments were also observed. These were shown upon sequencing to represent BCR-ABL transcripts with atypical junctions such as e2a2, e2a3, e1a3, e4a2, and e5a3 and fusions containing BCR and/or ABL intronic sequences in various rearrangements between e1 and a2 (Fig 3B).
DISCUSSION
In simplistic terms, the two equations CML = BCR-ABL and BCR-ABL = CML (or Ph+ ALL) are generally valid. The first means that, with rare and questionable exceptions,18 all patients with bona fide CML have a BCR-ABL rearrangement in their malignant cell clone. The second implies that the finding of a BCR-ABL fusion gene in hematopoietic cells from an individual is a sign of impending or overt CML (or Ph+ ALL, AML, etc). Similar equations could in fact be proposed for other types of leukemias that are now known to be consistently associated with specific types of fusion genes.
The results of the present study and those of Biernaux et al10 provide a different perspective on the second equation. Thus, we show here that BCR-ABL hybrid genes are formed and transcribed in leukocytes from greater than two thirds of healthy adults. Fusion transcripts with b2a2 or b3a2 junctions, the characteristic configurations of CML, were detected in 27% of the subjects tested, a frequency very similar to that found by Biernaux et al10 in their analysis of a larger adult population. Moreover, when we used an additional PCR protocol with primers that enable the amplification of the hybrid mRNA molecules typically found in Ph+ ALL, ie, those with an e1a2 junction, BCR-ABL fusion transcripts with this structure were detected in the majority (69%) of normal individuals. Similar findings were observed in the screening of 7 non-CML hematopoietic cell lines, all of which expressed one or more types of BCR-ABL hybrid transcripts.
The difference in the frequency of detection of p210 and p190 type of transcripts has several possible explanations. The most likely is that BCR-ABL fusion genes derived from an m-bcr breakpoint, from which only e1a2 chimeric mRNA can be transcribed, are generated more frequently both in vivo (normal leukocytes) and in vitro (cell lines) than those derived from M-bcr breakpoints. Such a bias at the primary stage of genetic recombination before the influence of growth advantage selection processes has been also observed upon exposure of hematopoietic cell lines to high-dose ionizing radiation, where the formation of some but not other forms of leukemia-associated fusion genes can be induced.11 Alternatively, in some or all normal individuals and non-CML cell lines the BCR-ABL gene might be derived from a M-bcr breakpoint and might produce both b2a2/b3a2 and e1a2 mRNA molecules by alternative splicing of the primary RNA transcript (Fig 1). In this scenario, the higher frequency of cases expressing only e1a2 type of transcripts would be due either to an unknown mechanism preferentially favoring the e1a2 form of splicing or to a technical artifact resulting from possible different sensitivities of the two PCR assays. We think the latter is unlikely, because extensive semiquantitative PCR titrations of the individual p210 and p190 protocols have shown them to have a comparable level of sensitivity.17
Taken together, our present findings confirm and extend those from Biernaux et al10 and the reported observations of BCL2-IgH fusion transcripts in leukocytes from healthy subjects19,20and provide strong evidence that the mere generation of specific chromosomal translocations and gene fusions is not sufficient to cause a malignant process. These data have important biological and clinical implications. From the point of view of leukemogenesis, they suggest that such forms of illegitimate genetic recombination occur regularly in hematopoietic precursors and in cultured cell lines as a consequence of an inherent, basal level of genomic instability. To be successful in producing a leukemic phenotype, these fundamental errors of DNA replication and repair have to fulfill at least two criteria: (1) the fusion gene structure must allow for the production of a functional protein with direct or indirect oncogenic properties and (2) the chromosomal translocation must occur in a relatively early precursor cell with self-renewal capacity. In other words, only the combination of a correct fusion gene in the correct primitive hematopoietic progenitor has a potential selective advantage and can become successful as an expanding clone. It is also possible that for some types of leukemia, of which CML may be one,21 additional mutations need to be accumulated in the affected cell clone before a true malignant expansion can occur. It is likely therefore that the BCR-ABL genes detected in the circulating leukocytes from healthy individuals do not reflect incipient leukemia, because they were generated in relatively mature, harmless progenitors from which the derived clones are eventually lost through normal cell differentiation and death. An alternative possibility is that in the overwhelming majority of normal persons the immune system is able to recognize and to eliminate the BCR-ABL expressing cells shortly after their generation and initial period of replication, preventing their uncontrolled expansion as a malignant clone. It is also interesting to note that several of the BCR-ABL transcripts detected in normal leukocytes and in the non-CML cell lines have an aberrant structure due to wrong junctions between BCR and ABL exons or to the insertion of intervening sequences; such genes would encode truncated BCR-ABL proteins that would presumably not be able to promote cell growth advantage. Because they are nonleukemogenic, only rarely are these aberrant BCR-ABL transcripts found in CML or ALL patient samples,22 23 and in these instances they are always secondary to the main, correctly spliced type of message.
From the clinical point of view, the detection of BCR-ABL transcripts in healthy subjects could in principle raise concern as to the significance and validity of the evaluation of minimal residual disease in CML and Ph+ ALL by PCR assays. To dispel these doubts, it is important to consider the difference in sensitivity between the RT-PCR assays optimized for detection and follow-up of residual leukemia in patients’ specimens and the assay designed for the present study. In the p210 and p190 RT-PCR protocols used in our laboratory for the analysis of clinical samples,17 the RNA content from approximately 2.5 × 106 WBCs is analyzed by one single PCR test. In contrast, the modified strategy we used for screening normal individuals required the analysis of the whole RNA content from 108 WBCs in 40 replicate PCR tests to score a given sample as positive or negative. Because the sensitivity of the two types of assays at the level of individual reactions is identical (same amount of cDNA per reaction and same primers and reaction conditions), the overall sensitivity of the modified assay can be estimated as 40 times higher than that of the clinical protocol. On the basis of our results, the prevalence of BCR-ABL carrying leukocytes in the blood of healthy individuals is probably around less than 1 to 10 in 108 WBCs, indicating that an average 70-kg normal adult, with a mean blood volume of 5.6 L and 5 to 10 × 109 WBCs/L, may have around 300 to 6,000 circulating leukocytes with a t(9;22) and possibly an unknown number in the bone marrow. It is not known whether these BCR-ABL–positive cells arise independently a large number of times in each individual or whether they are part of a single clone that has undergone a considerable although limited degree of expansion. If the latter, such a clone would have to double from 8 to 13 times to produce 256 to 8,192 cells, ie, the range of t(9;22)-positive cells estimated to be present in the circulation of healthy adults. This would imply that tests for residual disease in patients’ samples may have a 1 in 40 chance of being false positive, ie, of detecting this low background of leukemia-unrelated BCR-ABL–containing leukocytes. Such a low rate of false positivity would not be of practical significance, because the clinical value of residual leukemia assessment relies on its pattern of evolution during a sequential quantitative PCR analysis after treatment (bone marrow transplantation and α-interferon), rather than on isolated qualitative PCR test results.24,25 Even for patients monitored by qualitative RT-PCR assays after allogeneic bone marrow transplantation, the minimum criterion for therapeutic intervention in the form of donor lymphocyte infusions is the finding of two sequential positive tests.26 The chance of two consecutive false positive assays occurring would be 1/40 × 1/40 = 1/1,600, a probability too low to be of any practical concern. However, it is possible that the chance detection of nonmalignant BCR-ABL–positive leukocytes may underlie the phenomenon of apparent transient or intermittent molecular relapse observed in some CML patients who remain in clinical remission after bone marrow transplantation.27 28
Supported in part by grants from the Leukaemia Research Fund (UK) and the Dr Mildred Scheel-Stiftung fur Krebsforshung (Germany).
Address reprint requests to Junia V. Melo, MD, PhD, Department of Haematology-ICSTM, Hammersmith Hospital, Ducane Road, London W12 0NN, UK; e-mail: jmelo@rpms.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal