Abstract
The current paradigm concerning the kinetics of hematopoiesis is that only the most primitive pluripotential bone marrow stem cells can support prolonged hematopoiesis whereas more differentiated, lineage-committed stem cells can only contribute to a particular lineage for a limited period of time. In this study, we present evidence that in mice, the spleen contains a long-lived myeloid-committed stem cell population(s) that continuously replenishes the mature myeloid lineage for at least 9 months. After myeloid-specific retroviral-mediated gene transfer, the exogenous gene could be detected in thioglycollate-induced macrophages and granulocytes by Southern blot analysis and by in situ polymerase chain reaction on an individual cell basis. The targeted stem cell population does not repopulate the bone marrow in secondary recipients and did not give rise to cells other than cells of the myeloid lineage. It therefore represents the first nonpluripotential stem cell population capable of replenishing a hemopoietic lineage for a long period of time. The ability to target a myeloid-specific stem cell could facilitate gene therapy of congenital disorders of the myeloid system such as lysosomal storage diseases. It also offers a unique opportunity to assess the immunologic consequences of expressing an exogenous gene of choice exclusively in the myeloid lineage.
© 1998 by The American Society of Hematology.
ALL HEMATOPOIETIC lineages including the myeloid lineage develop from pluripotential stem cells (PSC, also called primitive stem cells) which in adult mice are found in bone marrow (BM) and spleen.1 The accepted definition of a PSC is that it is capable of self-renewal and that it can differentiate into all lineages of the hematopoietic system.2-4 Thus far, a cell surface marker unique for PSC has not been identified and therefore they have not yet been purified to homogeneity. PSC are rare in BM (1 in 2,000 to 5,000 cells) and are contained within a population of BM cells with the phenotype Thy-1.1lo,Lin-,Sca-1+.2,3,5-7When this population is purified by negative selection with antibodies and complement followed by positive selection by fluorescence-activated cell sorter (FACS), it contains all BM PSC.2,3,5-7 However, the PSC constitute only 10% of that population.6 8 The rest of the cells in this population are a mix of various uncharacterized, more differentiated stem cells.
The first characterization of BM stem cells was described in the early 1960s by Till and McCulloch9 as cells that form spleen nodules containing highly replicating hematopoietic stem cells, which they termed colony-forming-units–spleen (CFU-S), 8 to 12 days postreconstitution of irradiated recipients with adult BM cells. It was later realized that the spleen nodules only contained myeloid and erythroid precursors and only have a transient capacity to replenish the myeloid and erythroid lineages.10 More recent studies have shown that only the most primitive pluripotential BM stem cells can support hematopoiesis for long periods of time.2,11-13This notion is mostly based on experiments in which the reconstitution potential of the PSC-containing subpopulation (Thy-1.1loLin-Sca-1+ cells) was compared with that of the remaining BM cells. In these experiments, only the PSC-enriched population could protect a lethally irradiated host indefinitely.2,6 7
Further support for this notion came from studies on the kinetics of hematopoiesis. Studies using retroviral-mediated tagging of BM-derived PSC with a reporter gene have shown that for a short time immediately after the reconstitution of a lethally irradiated mouse, many stem cell clones contributed to hematopoiesis. However, a few weeks later, only very few clones were contributing to the continuous replenishment of all hematopoietic lineages.14,15 These results suggested that for a short time after reconstitution, both lineage-committed as well as PSC contributed to hematopoiesis, but the more mature, lineage-committed cells are short-lived and therefore disappear within a few weeks. Similar conclusions were reached by Harrison et al.16 Using a competitive repopulation assay, these investigators showed that the appearance of all three hematopoietic lineages is highly correlated with respect to time, which they interpreted as an indication that most donor cells arise from the same PSC, and therefore, lineage-committed precursors present in the inoculum could not have contributed significantly to any of the lineages. In contrast to the finding regarding BM-derived stem cells, in this paper we describe the identification of a splenic population of very long-lived myeloid-committed stem cells that can continuously replenish the myeloid lineage. Lipopolysaccharide (LPS)-stimulated, T-cell–depleted spleen cells obtained from normal or B-cell–deficient mice were infected with a retroviral vector containing the neomycin phosphotransferase (neo) marker gene and were used to rescue lethally irradiated syngeneic recipients. In thioglycollate-induced granulocytes and macrophages, 0.5 to 1 copy/cell of the neogene could be detected by Southern blotting for at least 9 months after reconstitution. In situ polymerase chain reaction (PCR) showed the presence of the gene in greater than 80% of induced granulocytes and macrophages in the peritoneal cavity 12 months after reconstitution. The targeted stem cell in the spleen was not a self-renewing PSC because secondary, lethally irradiated recipients reconstituted with BM obtained from primary recipients never contained the exogenous gene in myeloid or any other cell type. Further indication that PSC were not targeted was the fact that the lymphoid lineage (T cells) were invariably negative. Because the average life of macrophages is only a few days,17 and that of mature granulocytes is only a few hours,18 these results indicate that the targeted lineage-committed stem cell population was the main replenishing source for the myeloid lineage for at least 9 months. Based on these results and the previous studies cited above, the splenic cells targeted in these experiments would have to be classified as myeloid precursors at an earlier differentiation stage than CFU-S cells.
MATERIALS AND METHODS
Mice.
C57Bl/6, (BALB/C × C57Bl/6)F1 and B-cell–deficient (μ knockout) C57Bl/6 male19 and female mice were purchased from the Jackson Laboratory (Bar Harbor, ME).
Retroviral vectors and virus-producing cell line.
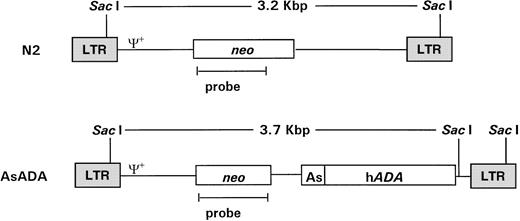
pN220 and pAsADA(Fig 1) are Moloney murine leukemia virus (MLV)-based retroviral vectors that contain the neo gene, which is expressed from the MLV long terminal repeat (LTR) promoter.pAsADA contained, in addition to the neogene, the human adenosine deaminase (ADA) gene driven by its own minimal promoter.21,22 N2 and AsADA virus-producing cells were previously described.22 Briefly, the NIH 3T3-based ecotropic murine packaging cell line GP+E-8623 were transfected with 5 μg of pN2 orpAsADA retroviral vectors using the polybrene/dimethyl sulfoxide shock method, followed by selection with G418 (0.35 mg/mL; GIBCO-BRL, Grand Island, NY). The retroviral titer for both virus-producing cell lines ranged from 1.5 to 2.0 × 107 CFU/mL.
Schematic diagram of the retroviral vectors used in gene transfer. Retroviral vectors N2 and AsADA are Moloney murine leukemia virus–based vectors. N2 contains the neo gene, expressed from the viral LTR promoter. AsADA contains neo, expressed from the viral LTR promoter, and the human ADA gene, expressed from its endogenous promoter, As. Relevant restriction enzyme sites and the distance between them are indicated. ψ+ represents the packaging signal. The 930-bp EagI-AvaI fragment in the neo coding region was used as a probe in experiments described in Figs 2 and 3.
Schematic diagram of the retroviral vectors used in gene transfer. Retroviral vectors N2 and AsADA are Moloney murine leukemia virus–based vectors. N2 contains the neo gene, expressed from the viral LTR promoter. AsADA contains neo, expressed from the viral LTR promoter, and the human ADA gene, expressed from its endogenous promoter, As. Relevant restriction enzyme sites and the distance between them are indicated. ψ+ represents the packaging signal. The 930-bp EagI-AvaI fragment in the neo coding region was used as a probe in experiments described in Figs 2 and 3.
Gene transfer.
The gene transfer protocol was performed as follows: enriched B-cell populations were prepared from the spleens of C57Bl/6, (BALB/C × C57Bl/6)F1 or B-cell–deficient (μ knockout) C57Bl/6 mice by depleting T cells with monoclonal antibody J1j, (rat IgM anti-mouse Thy-1.2),24 plus complement treatment. The remaining cells were stimulated for 24 hours with 50 μg/mL LPS and then cocultivated with a monolayer of irradiated (1,600 R) N2 or AsADA virus-producing cells in the presence of 6 μg/mL polybrene. The nonadherent cells were collected and washed 24 hours later, and 4 to 15 × 106 of the cells were injected intravenously into each lethally irradiated (1,000 R) (BALB/C × C57Bl/6)F1 recipient mouse.
Cell preparation, Southern blotting analysis, and ADA assay.
Thioglycollate was injected intraperitoneally 24 or 96 hours before the isolation of myeloid cells to induce the recruitment of granulocytes and macrophages to the peritoneum of the reconstituted recipients. Some of the peritoneal exudate cells (PEC) were fixed onto glass slides for in situ PCR and hybridization as described below. Spleen cells were isolated from the same mice from which the PEC were isolated. Part of the spleen cells were injected into the lethally irradiated secondary recipient mice and some of the spleen cells were treated with the monoclonal antibody J11d (rat IgM anti-mouse heat stable antigen)24 or J1j and complement before the extraction of genomic DNA, followed by Southern blotting or ADA assay. Southern blotting was performed according to standard procedures. For detection of the neo marker gene, 10 μg/lane of SacI-digested genomic DNA was screened using a 32P-labeled probe, anEagI-AvaI fragment from the neo coding region. To determine the clonality of the transduced population, 10 μg/lane of BamHI- or HindIII-digested genomic DNA was screened using the same probe as above.
The ADA assay was described previously.22 Briefly, 1 × 106 target cells were lysed by multiple freezing/thawing cycles, and samples were separated by electrophoresis on cellulose acetate plates. Enzyme activity was developed by an agar overlay containing adenosine, nucleoside phosphorylase, xanthine oxidase, phenazine methosulfate, and dimethylthiazol diphenylterazolium bromide.
In situ PCR and hybridization.
PEC were fixed onto glass slides with 4% paraformaldehyde, washed, and dehydrated. The cells were then permealized with proteinase K and sealed with the PCR mixture containing the neo specific primers 5′-CAGGATGATCTGGACGA and 3′-TGGATGCCGACGGATTTGCA. Cycling conditions were 94°C, 1 minute; 55°C, 1 minute; and 72°C, 1 minute 30 seconds for 30 cycles. After the PCR reaction was completed, the slides were washed and incubated with the hybridization mixture containing a 404-bp neo-specific biotinylated probe complementary to the PCR-amplified sequence excluding the primer region. Streptavidin was then added (to amplify the signal) followed by biotin-conjugated alkaline phosphatase. Color was developed using bromochloroindoyl phosphate (BCIP)/nitro blue tetrazolium (NBT).25 The cells were analyzed on a Bright-field phase, differential interference contrast microscope (Zeiss, Germany). Images were captured using a Dage CCD72 camera and Dage DSP2000 digital signal processor (Michigan City, IN) (capable of on-chip integration for low-light situations), and a Macintosh Quadra 700 (Cupertino, CA) with a Scion LG-3 frame grabber board.
RESULTS
Retrovirus-mediated gene tagging of a long-lived myeloid precursor population in the spleen.
Previously, we developed a very efficient gene transfer protocol for the introduction of exogenous genes into highly purified lymph node (LN) B and T cells for other purposes.22,26 27 We noticed that when T-cell–depleted spleen cells (rather than LN cells) were used as the target cells in this system, cells harboring the exogenous gene could also be detected in nonlymphoid tissues such as liver and lungs as well as in spleen and LN, and in some experiments, in low levels also in the thymus (data not shown). Because signals were obtained from organs that normally do not contain significant numbers of B cells, this raised the possibility that some spleen-residing stem cells were also targeted and gave rise to tissue-residing myeloid cells. To check this possibility, thioglycollate-induced granulocytes and macrophages obtained from animals reconstituted with retroviral vector–targeted BM cells were directly assayed for the presence and expression of an exogenous gene introduced by the retroviral vector.
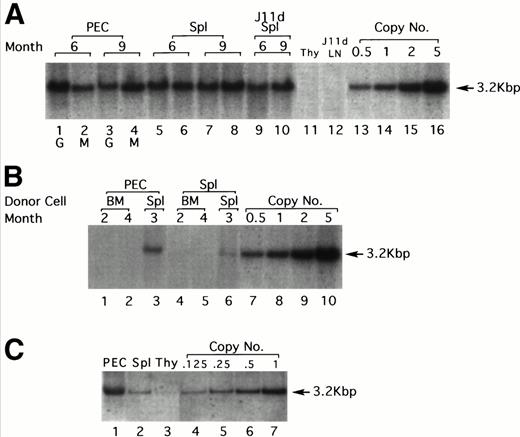
LPS-stimulated, T-cell–depleted spleen cells obtained from (BALB/c × C57Bl/6)F1 or C57Bl/6 mice were infected by cocultivation with packaging cell line producing the N2 vector virus (Fig 1) and adoptively transferred into lethally irradiated F1 recipients. Gene transduction was dependent on LPS stimulation but independent of T-cell depletion, although T-cell depletion results in a higher transduction efficiency (data not shown). A total of 30 recipient mice were injected with thioglycollate 6 to 9 months after adoptive transfer to induce granulocyte and macrophage migration into the peritoneal cavity. As assessed by α-naphthyl acetate esterase staining (macrophage-specific) versus naphthol AS-D chloroacetate esterase staining (granulocyte-specific), 24 hours after injection of thioglycollate, greater than 95% of the cells obtained from the peritoneum were granulocytes, whereas 96 hours after thioglycollate injection, greater than 95% of the cells were macrophages (data not shown). Six and nine months posttransfer, approximately 1.0 to 2.0 proviral copies/cell were detected in both peritoneal granulocytes and macrophages by Southern blotting (Fig 2A). DNA extracted from granulocytes (24 hours PEC) was analyzed in lanes 1 and 3 at 6 and 9 months postreconstitution, respectively (Fig 2A). Lanes 2 and 4 show the same analysis but for macrophages (96 hours PEC). Lanes 5 through 8 represent DNA from the spleens of the same animals. Lanes 9 and 10 represent DNA from the same spleen cells used in lanes 5 and 8, respectively, but after treatment with the monoclonal antibody J11d and complement. J11d is a cytotoxic antibody specific for the murine heat stable antigen. It removes 80% to 90% of splenic B cells but spares macrophages. The results indicate that in this experiment, approximately half of the signal (as measured by gel densitometry) obtained from the spleens was due to resident macrophages.
Gene transfer into the myeloid lineage but not pluripotential hematopoietic stem cells. (A) Southern blot analysis of PEC and spleen cells from four representative lethally irradiated mice reconstituted with 15 × 106 N2-infected T-cell–depleted spleen target cells 6 and 9 months earlier. Genomic DNA was prepared from spleen and PEC 24 and 96 hours after injection of thioglycollate and digested with the restriction enzyme SacI and probed using a neo-specific probe. Lanes 1 and 3 represent DNA from granulocytes (24 hours) and lanes 2 and 4 represent DNA from macrophages (96 hours) as indicated by the lettering below the lanes. The last four lanes represent SacI-digested pN2 plasmid DNA equivalent to 0.5, 1.0, 2.0, and 5.0 copies/cell, respectively. The middle lanes represent DNA prepared from whole spleen cells (lanes 5 through 8) or from J11d-treated spleen cells (lanes 9 and 10) obtained from the same mice used for harvesting PEC. (B) Genomic DNA extracted from PEC or spleen cells obtained from three representative secondary mice previously reconstituted with 8 × 106 N2-infected, B- and T-cell–depleted BM or spleen cells obtained from primary recipients were analyzed by Southern blotting. Lanes 1 and 2 represent DNA from macrophages (96 hours PEC) taken from mice reconstituted with BM cells from positive primary mice. Lane 3 represents DNA from macrophages (96 hours PEC) taken from a mouse reconstituted with spleen cells from a positive primary mouse. Lanes 4 through 6 represent the same mice except that the DNA was extracted from spleen rather than PEC. The last four lanes represent SacI-digested pN2 plasmid DNA equivalent to 0.5, 1.0, 2.0, and 5.0 copies/cell, respectively. (C) Genomic DNA extracted from PEC, spleen, or thymus obtained from mice reconstituted 4 months earlier with 4 × 106 N2-infected, T-cell–depleted BM taken from syngeneic, C57Bl/6 μ knockout mice. Lanes 1, 2, and 3 represent DNA from macrophages (96 hours PEC), spleen, and thymus, respectively. Lanes 4 through 7 representSacI-digested pN2 plasmid DNA equivalent to 0.125, 0.25, 0.5, and 1.0 copies/cell, respectively.
Gene transfer into the myeloid lineage but not pluripotential hematopoietic stem cells. (A) Southern blot analysis of PEC and spleen cells from four representative lethally irradiated mice reconstituted with 15 × 106 N2-infected T-cell–depleted spleen target cells 6 and 9 months earlier. Genomic DNA was prepared from spleen and PEC 24 and 96 hours after injection of thioglycollate and digested with the restriction enzyme SacI and probed using a neo-specific probe. Lanes 1 and 3 represent DNA from granulocytes (24 hours) and lanes 2 and 4 represent DNA from macrophages (96 hours) as indicated by the lettering below the lanes. The last four lanes represent SacI-digested pN2 plasmid DNA equivalent to 0.5, 1.0, 2.0, and 5.0 copies/cell, respectively. The middle lanes represent DNA prepared from whole spleen cells (lanes 5 through 8) or from J11d-treated spleen cells (lanes 9 and 10) obtained from the same mice used for harvesting PEC. (B) Genomic DNA extracted from PEC or spleen cells obtained from three representative secondary mice previously reconstituted with 8 × 106 N2-infected, B- and T-cell–depleted BM or spleen cells obtained from primary recipients were analyzed by Southern blotting. Lanes 1 and 2 represent DNA from macrophages (96 hours PEC) taken from mice reconstituted with BM cells from positive primary mice. Lane 3 represents DNA from macrophages (96 hours PEC) taken from a mouse reconstituted with spleen cells from a positive primary mouse. Lanes 4 through 6 represent the same mice except that the DNA was extracted from spleen rather than PEC. The last four lanes represent SacI-digested pN2 plasmid DNA equivalent to 0.5, 1.0, 2.0, and 5.0 copies/cell, respectively. (C) Genomic DNA extracted from PEC, spleen, or thymus obtained from mice reconstituted 4 months earlier with 4 × 106 N2-infected, T-cell–depleted BM taken from syngeneic, C57Bl/6 μ knockout mice. Lanes 1, 2, and 3 represent DNA from macrophages (96 hours PEC), spleen, and thymus, respectively. Lanes 4 through 7 representSacI-digested pN2 plasmid DNA equivalent to 0.125, 0.25, 0.5, and 1.0 copies/cell, respectively.
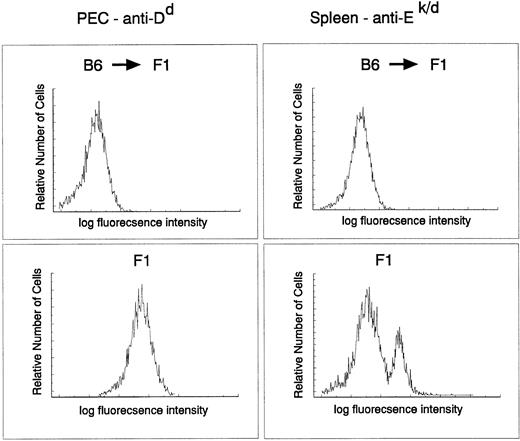
The donor origin of the hematopoietic system in the target animals was verified by flow cytometric analysis of both macrophages and B cells in recipient F1 mice reconstituted with parental C57Bl/6 target cells using major histocompatibility (MHC) class I– and II–specific antibodies. The degree of chimerism was checked directly on PEC with an anti-H-2Dd antibody and on spleen cells with MHC class II–specific anti-I-Ek,d antibodies. C57Bl/6 do not express I-E and therefore spleen cells from a C57Bl/6 → (C57Bl/6 × BALB/c)F1 should not stain with this antibody. Figure 3 clearly indicates that the myeloid and lymphoid (B cells) lineages in the reconstituted animals were derived entirely from donor stem cells (H-2b+, I-E−). Taken together, these results indicate a very efficient infection of a precursor cell population that contributes to the myeloid lineage for at least 9 months.
Myeloid and B cell in C57Bl/6 → (BALB/c × C56Bl/6)F1 chimeras are of donor origin. PEC and spleen cells obtained from C57Bl/6 → (BALB/c × C57Bl/6)F1 or (BALB/c × C57Bl/6)F1 were stained with fluorescein isothiocyanate–labeled 34-4-21S (anti-Dd) and 14-4-4 (anti-I-Ek,d) antibodies, respectively, and analyzed by FACS.
Myeloid and B cell in C57Bl/6 → (BALB/c × C56Bl/6)F1 chimeras are of donor origin. PEC and spleen cells obtained from C57Bl/6 → (BALB/c × C57Bl/6)F1 or (BALB/c × C57Bl/6)F1 were stained with fluorescein isothiocyanate–labeled 34-4-21S (anti-Dd) and 14-4-4 (anti-I-Ek,d) antibodies, respectively, and analyzed by FACS.
Pluripotential hematopoietic stem cells were not infected during gene transfer.
The next set of experiments was aimed at determining whether pluripotential hematopoietic stem cells as opposed to rather later-stage, lineage-committed progenitor cells were targeted. Two assumptions could be made if pluripotential cells were targeted in the initial inoculum. The first is that all hematopoietic lineages in the reconstituted animals should harbor the marker gene. This was not the case in any of the mice tested. Southern blots were always negative for the marker genes in B-cell–depleted LN (Fig 2A, lane 12), and in most experiments was also undetected in thymus (Fig 2A, lane 11). In some experiments, a weak signal can be detected in the thymus (Fig 2C, lane 3), which is not surprising because this organ does contain a small fraction of BM-derived macrophages. The second is that BM cells from the reconstituted animals should be able to transfer the marker gene into a secondary lethally irradiated host. To test this, BM or spleen cells from positive primary recipients were used to reconstitute secondary lethally irradiated recipients. To ensure that the cells transferred into secondary recipients were depleted of mature B cells that might contain the exogenous gene, donor BM cells were first treated with J11d and complement (to remove B cells) and also with J1j (anti–Thy-1 monoclonal antibodies, to remove mature T cells) and complement. Over 30 secondary recipients reconstituted with BM obtained from primary positive mice were examined and proviral sequences could never be detected by Southern blotting in 96 hours PEC (Fig 2B, lanes 1 and 2) and spleen (Fig 2B, lanes 4 and 5).
In contrast to BM, the spleen should contain the targeted myeloid precursor cells and therefore, in parallel, secondary irradiated animals were reconstituted with 8 × 106 B- and T-cell–depleted spleen cells obtained from positive primary reconstituted mice. In these experiments, all secondary recipients were positive for proviral sequences both in 96 hours PEC (Fig 2B, lane 3) and in spleen (Fig 2B, lane 6). The signals are weaker here (0.1 to 0.4 copies/cell for PEC) probably because only 8 × 106spleen cells were transferred; however, all six mice tested were positive. T cells isolated from spleen, LN, and thymus of these mice (secondary recipients) were invariably negative (data not shown). These results strongly suggest that pluripotential stem cells were not targeted during the first infection.
Because the infection protocol using LPS-activated, T-cell–depleted spleen cells as target cells results in the transduction of both myeloid precursors and mature B cells, the next set of experiments was designed to rule out the possibility that some of the signal is due to residual B cells. In these experiments, spleen cells obtained from B-cell–deficient mice (μ knockout mice) were used as target cells. In this case no infected B cells are transferred and the donor stem cells cannot generate any B cells; therefore, the recipient animals were totally devoid of donor origin B cells. As seen in Fig 2C, PEC obtained from such mice 4 months after cell transfer contained an average of 1 copy/cell of the exogenous gene, similar to the results obtained with normal spleen (Fig 2A). Spleen cells contained an average of 0.125 copies/cell which is, as expected, much lower than when normal spleen cells were used as target cells because in this case there are no infected B cells in the inoculum. Very faint bands could be seen from thymocytes on overexposure.
Absence of replication-competent virus in reconstituted recipients.
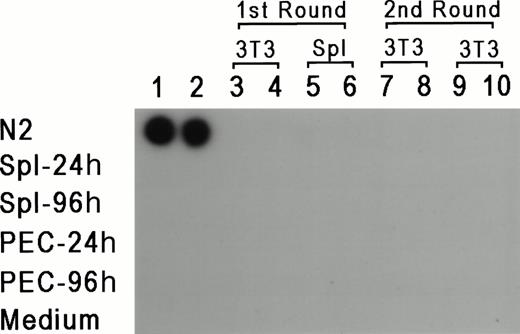
To rule out the possibility that the results reflect a lateral spread of vector virus due to activation of an endogenous retrovirus, assays were performed to determine whether replication-competent virus could be detected in PEC or LPS-stimulated spleen cells isolated from reconstituted mice 24 or 96 hours after thioglycollate injection. For 7 days, 1 × 106 PEC or spleen cells were cocultivated with either 2 × 105 NIH 3T3 cells or 1 × 106 primary syngeneic spleen cells. Supernatant was obtained from these cultures every 24 hours and tested for reverse transcriptase (RT) activity according to standard procedures.28 If replication-competent virus was present, it should have been propagated in fresh cells, and supernatant from these cultures should be positive for RT activity. In parallel, each supernatant sample was added to fresh cultures of NIH 3T3 cells for a second round of expansion for an additional 7 days, and RT activity was also checked every 24 hours. Figure 4represents the results from supernatants obtained from day 3 of the primary and secondary cultures, and as can be seen, all of the samples were negative except for the positive control. RT activity was negative for all time points tested. Moreover, the NIH 3T3 cultures from both rounds of amplification were also scored for transfer of theneo-containing N2 vector by selection with G418 (350 μg/mL). If replication-competent virus was present, it should provide viral proteins for passage of N2 vector virus. No G418-resistant colonies were detected (data not shown), providing additional support that replication-competent virus was not present in vector positive animals, and ruling out horizontal spread of the vector virus due to activation of a latent endogenous retrovirus.
RT assay for the detection of replication competent virus. PEC and spleen cells were obtained 24 and 48 hours after thioglycollate injection and cocultured with either 3T3 cells or LPS-stimulated spleen cells. Supernatants were obtained every 24 hours for 7 days and added to fresh 3T3 cells. Two samples of supernatants harvested at day 3 of the first and second cultures were checked for the presence of RT (columns 3 through 10). Columns 1 and 2 represent control supernatants taken from cultures of N2 producer line, spleen cells, PEC, and media control, as marked in the figure.
RT assay for the detection of replication competent virus. PEC and spleen cells were obtained 24 and 48 hours after thioglycollate injection and cocultured with either 3T3 cells or LPS-stimulated spleen cells. Supernatants were obtained every 24 hours for 7 days and added to fresh 3T3 cells. Two samples of supernatants harvested at day 3 of the first and second cultures were checked for the presence of RT (columns 3 through 10). Columns 1 and 2 represent control supernatants taken from cultures of N2 producer line, spleen cells, PEC, and media control, as marked in the figure.
Lack of expression of a transduced human ADA gene in granulocytes and macrophages.
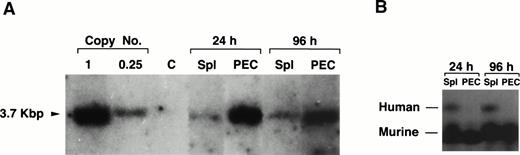
We have previously shown that the human ADA gene can be efficiently introduced into, and expressed in mature B and T cells using the AsADA retroviral vector (Fig 1).22,26However, in the experiments mentioned earlier in which we first used T-cell–depleted spleen as target cells, although the gene could be detected in tissues other than lymphoid organs, expression of the ADA protein could not be detected in these tissues. One explanation for this could be that the relative number of transduced cells in nonlymphoid organs is too low to allow protein detection. Because the ADA assay is quite sensitive, we preferred an alternative explanation, which is that the ADA gene when introduced into early progenitor cells is shut off during the differentiation to mature granulocytes and macrophages. To test this, the same viral vector used in previous studies (Fig 1)22,26,27 was used to infect T-cell–depleted spleen cells. As shown in Fig 5A, gene transfer into the myeloid lineage, as assessed by Southern blotting, was as efficient for the human ADA gene as it was for the neo gene. The transferred ADA gene could be detected in spleen and PEC 6 months after adoptive transfer of infected target cells (1 copy/cell for PEC and 0.2 copies/cell for spleen cells, respectively). The humanADA gene allows for a simple enzymatic measurement of protein expression and it can be distinguished from the endogenous murine ADA because of its different electrophoretic mobility.22,26 As shown in Fig 5B, we could not detect any human ADA expression in PEC using an ADA-specific enzymatic assay. On the other hand, as expected, human protein expression was detected in spleen cells because the spleen contains large numbers of B cells which are also transduced in this protocol. The lack of expression in myeloid cells is not surprising. It is not uncommon that xenogeneic tissue-specific promoters are turned off during differentiation.29 Also, the human ADA gene is driven by a minimal promoter which was shown to be active in mature lymphocytes and in various lymphoid cell lines, but to the best of our knowledge, it was never tested in myeloid cells.30 However, the fact that there is no expression of the ADA gene in PEC provides further support to the notion that the proviral signal observed by Southern blotting in PEC is only due to positive myeloid cells and not to contaminating B cells, which is consistent with the esterase staining results. The levels of expression of the endogenous murine ADA are obviously much higher than the human gene because most of the cells in the spleen express the endogenous murine ADA.
Lack of expression of the exogenous gene in granulocytes and macropahges. (A) Southern blotting of genomic DNA prepared from PEC 24 and 96 hours after thioglycollate treatment and spleen from two representative mice reconstituted 6 months earlier with 20 × 106 cells of T-cell–depleted spleen cells infected with pAsADA. DNA was digested with SacI and probed with aneo-specific probe. SacI-digested pAsADA plasmid DNA equivalent to 0.25 and 1 copies/cell are as indicated. (B) Human and mouse ADA protein assay. Cell lysates prepared from 1 × 106 cells from the same spleen and PEC samples used for Southern blotting were electrophoresed on a cellulose acetate plate to separate human from murine ADA enzymes. ADA activity was detected by the standard colorimetric enzymatic assay. PEC were analyzed 24 hours (granulocytes) and 96 hours (macrophages) after thioglycollate induction, respectively.
Lack of expression of the exogenous gene in granulocytes and macropahges. (A) Southern blotting of genomic DNA prepared from PEC 24 and 96 hours after thioglycollate treatment and spleen from two representative mice reconstituted 6 months earlier with 20 × 106 cells of T-cell–depleted spleen cells infected with pAsADA. DNA was digested with SacI and probed with aneo-specific probe. SacI-digested pAsADA plasmid DNA equivalent to 0.25 and 1 copies/cell are as indicated. (B) Human and mouse ADA protein assay. Cell lysates prepared from 1 × 106 cells from the same spleen and PEC samples used for Southern blotting were electrophoresed on a cellulose acetate plate to separate human from murine ADA enzymes. ADA activity was detected by the standard colorimetric enzymatic assay. PEC were analyzed 24 hours (granulocytes) and 96 hours (macrophages) after thioglycollate induction, respectively.
In situ PCR of peritoneal exudate cells.
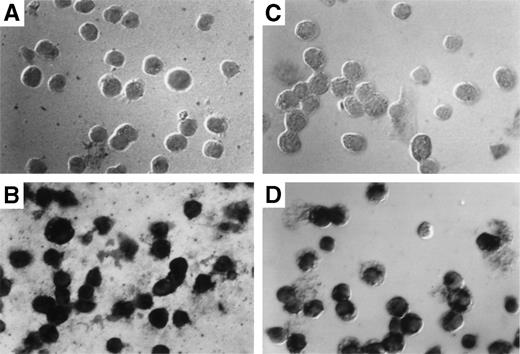
The results obtained by Southern blotting can only indicate the average number of exogenous gene copies per cell. They cannot indicate whether most cells harbor 1 copy or whether a few cells harbor many copies of the gene. To determine the exact percentage of myeloid cells that harbor the transduced gene, in situ PCR was used to amplify the transduced neo gene followed by specific hybridization of biotinylated neo-specific probes. This allows the visualization of positive signals and the morphology of the target cell on a cell per cell basis. At least 80% of both granulocytes (Fig 6C) and macrophages (Fig 6D) from reconstituted animals harbored at least 1 copy of the transducedneo gene. Cells from control animals were totally negative (Fig6A and B).
Detection of provirus sequences by in situ PCR amplification. Peritoneal granulocytes and macrophages were obtained 24 and 96 hours, respectively, after thioglycollate injection from control (A and C) or from mice reconstituted with targeted spleen cells (B and D) 12 months earlier and subjected to in situ PCR hybridization using biotinylated probes. Amplified neo-specific sequences were visualized on a cell per cell basis using an alkaline phosphatase-based colorimetric assay.
Detection of provirus sequences by in situ PCR amplification. Peritoneal granulocytes and macrophages were obtained 24 and 96 hours, respectively, after thioglycollate injection from control (A and C) or from mice reconstituted with targeted spleen cells (B and D) 12 months earlier and subjected to in situ PCR hybridization using biotinylated probes. Amplified neo-specific sequences were visualized on a cell per cell basis using an alkaline phosphatase-based colorimetric assay.
Clonal analysis of the targeted myeloid population.
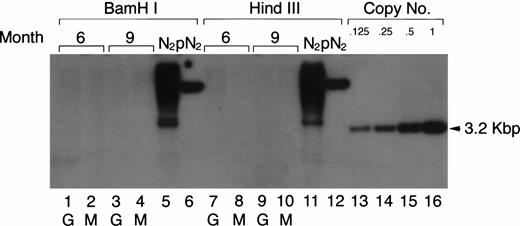
Previous retroviral gene tagging of pluripotential stem cells suggested that at any particular time point, very few clones contribute to the generation of the various hematopoietic lineages.31 32 To test the degree of clonality in the mature myeloid lineage after adoptive transfer of retrovirus-tagged spleen cells, the same DNA used in the Southern blotting analysis for the detection of the proviral DNA in PEC-derived granulocytes and macrophages was digested with eitherBamHI or HindIII. These enzymes only cut once within the retroviral vector. Therefore, if the mature PEC arise from one or few precursor clones, one would expect to see only a few bands in a Southern blot when neo-specific probes are used. The results shown in Fig 7 clearly show that the populations of peritoneal granulocytes and macrophages are not oligoclonal. Discrete bands were not detected in the Southern blot analysis after exposure for 1 to 2 days, even though the blot was sensitive to below 0.125 copies of the exogenous gene/cell. After longer exposure, some faint bands (corresponding to 0.001 copies/cell) could be detected, which might suggest that some infected clones contribute a little more than others to the mature myeloid population.
Clonal analysis for the populations of PEC-derived granulocytes and macrophages. The PEC isolated from the same mice described in Fig 2A were used for clonal analysis. Genomic DNA was prepared from PEC 24 and 96 hours after injection of thioglycollate and digested with the restriction enzyme BamHI or HindIII and probed using a neo-specific probe. Lanes 1, 3, 7, and 9 represent DNA from granulocytes (24 hours) and lanes 2, 4, 8, and 10 represent DNA from macrophages (96 hours) as indicated by the lettering below the lanes. Genomic DNA from the N2 producer cell (lanes 5 and 11) and 1 copy of plasmid DNA, pN2 (lanes 6 and 12), were used for oligoclonal and monoclonal controls, respectively. The last four lanes represent SacI-digested pN2 plasmid DNA equivalent to 0.125, 0.25, 0.5, and 1.0 copies/cell, respectively.
Clonal analysis for the populations of PEC-derived granulocytes and macrophages. The PEC isolated from the same mice described in Fig 2A were used for clonal analysis. Genomic DNA was prepared from PEC 24 and 96 hours after injection of thioglycollate and digested with the restriction enzyme BamHI or HindIII and probed using a neo-specific probe. Lanes 1, 3, 7, and 9 represent DNA from granulocytes (24 hours) and lanes 2, 4, 8, and 10 represent DNA from macrophages (96 hours) as indicated by the lettering below the lanes. Genomic DNA from the N2 producer cell (lanes 5 and 11) and 1 copy of plasmid DNA, pN2 (lanes 6 and 12), were used for oligoclonal and monoclonal controls, respectively. The last four lanes represent SacI-digested pN2 plasmid DNA equivalent to 0.125, 0.25, 0.5, and 1.0 copies/cell, respectively.
DISCUSSION
In this study we present evidence for the existence of a long-lived, spleen-residing, myeloid-committed stem cell population. These stem cells can be targeted with a retroviral vector after stimulation of T-cell–depleted spleen cells with LPS. Because current retroviral vectors, including the ones used in this study, require at least one round of cell replication to integrate into the cell’s genome, the targeted stem cells are induced to proliferate either directly or indirectly by LPS. There was no gene transduction in the absence of LPS; however, transduction was independent on the presence of B cells because spleen cells from B-cell–deficient (μ knockout) mice were as efficiently transduced as normal B cells. This suggests that the targeted cells themselves are responsive to LPS. The evidence that this cell population is not pluripotential is three-fold. First, the targeted cells do not home back to the BM and therefore, secondary BM transfers from positive mice into normal irradiated recipients do not transfer the exogenous gene. Second, whereas at least 80% of mature thioglycollate-induced granulocytes and macrophages were positive for the exogenous gene, as determined by in situ PCR, newly generated T and B lymphocytes were not. Third, clonal analysis of the targeted macrophages and granulocytes populations (Fig7) indicated that the cells were not of oligoclonal origin, which should have been the case if pluripotential cells were targeted.14 15 However, it is not clear how homogenous the targeted stem cell population is and it can represent either a pan myeloid stem cell population or a pool of more than one precursor cells such as macrophage- and granulocyte-committed stem cells.
Surprisingly, these myeloid-committed stem cells contributed to the replenishment of the mature myeloid population for at least 9 months, as assessed by Southern blot analysis and for 12 months as assessed by in situ PCR hybridization. Their ability to replenish the mature myeloid cell populations for prolonged periods is seemingly antithetical to the current dogma that cells other than PSC can only contribute to the hematopoietic lineage for a limited period. However, this dogma is based on studies using BM cell transfers and our findings are based on spleen cell transfers. It is therefore possible that the targeted population we describe only resides in the spleen. It is equally plausible that this population is present in BM but copurifies with the enriched BM population that contains pluripotential cells. Only 10% of this population are PSC and the rest of the cells are at present not well characterized.2 6 The myeloid precursor(s) we describe might be part of the uncharacterized fraction of this population. If these myeloid precursor cells also reside in BM, why they were not detected by others is unclear. One possibility could be that they do not respond to any of the stimuli used by others and would have been detected if LPS was used. This possibility is currently under investigation.
Aside from the implication these findings have on our understanding of hematopoiesis, the ability to specifically introduce exogenous genes into the myeloid lineage offers an opportunity to study the consequences of expressing an exogenous gene exclusively in the myeloid lineage using somatic cell gene transfer. So far, retroviral-mediated gene transfer into the myeloid lineage has mainly been achieved by the transduction of BM stem cells and the efficiency was rarely higher than 5%.33 Other disadvantages of using BM cells is that PSC comprise a rare population in BM, they are very hard to purify, they have not been ideal targets for gene transfer, and they are not lineage specific. Even if the efficiency of gene transfer into pluripotential cells improves in the future, transduction of exogenous genes into these cells would probably result in the expression of the gene in all hematopoietic cell lineages, which may not always be desirable. In contrast, transduction of myeloid-committed stem cells allows specific expression in the myeloid compartment. This cell would therefore make a more suitable target for gene therapy–based treatment of genetic disorders inherent to the myeloid lineage such as Gaucher’s disease. Restricted expression in the myeloid lineage might also facilitate selective induction and potentiation of immune responses against tumor or viral antigens.
Supported in part by the National Multiple Sclerosis Society Grant No. 2626A2/1 (Y.R.), Milstein family foundation (Y.R.), and by an Individual National Research Service Award, Grant No. GM19331 (A.R.).
Address reprint requests to Yacov Ron, PhD, Department of Molecular Genetics and Microbiology, University of Medicine and Dentistry of New Jersey, Robert Wood Johnson Medical School, 675 Hoes Lane, Piscataway, NJ 08854; e-mail: yron@umdnj.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal