Abstract
In multiple myeloma (MM), the VDJ rearrangement of the immunoglobulin heavy chain expressed by MM plasma cells provides a unique clonotypic marker. Although clonotypic MM cells have been found in the circulation, their number has been controversial. Our objective was to provide direct evidence, using single-cell assays, for the frequency of clonotypic cells in blood of 18 MM patients, and to confirm their identity as B cells. The clonotypic Ig heavy-chain (IgH) VDJ was determined from single plasma cells using consensus reverse transcriptase-polymerase chain reaction (RT-PCR), subcloning, and sequencing. For all patients, using patient-specific primers, clonotypic transcripts were amplified from 10 or more individual plasma cells. Using in situ RT-PCR, for all patients greater than 80% of plasma cells were found to be clonotypic. Three separate methods, RT-PCR, single-cell RT-PCR, and in situ RT-PCR, were used to analyze clonotypic cells in peripheral blood mononuclear cells (PBMC) from MM patients. Sequencing of the IgH transcripts expressed by individual cells obtained by limiting dilution of freshly isolated PBMC from a MM patient showed that all B cells expressed an identical CDR3. This intraclonal homogeneity indicates an escape from antigenic-selection, characteristic of malignant B cells. For this patient, the frequency of clonotypic PBMC, about 25%, was comparable to the number of PBMC B cells (34%). Because the PBMC included less than 1% plasma cells, virtually all clonotypic PBMC must be B cells. Using single-cell RT-PCR, clonotypic IgH transcripts were identified in individual sorted B cells from blood. To accurately quantify the number of clonotypic B cells, sorted B cells derived from 18 MM patients (36 samples) and 18 healthy donors (53 samples) were analyzed using in situ RT-PCR with patient-specific primers. Clonotypic transcripts were not detectable among normal B cells. For the 18 MM patients, a mean of 66% ± 4% (SE) of blood B cells were clonotypic (range, 9% to 95%), with mean absolute number of 0.15 ± .02 × 109/L blood. Over time in individual patients, conventional chemotherapy transiently decreased circulating clonotypic B cells. Their numbers were increased in granulocyte colony-stimulating factor (G-CSF)– mobilized blood of one patient. However, clonotypic B cells of a one patient became undetectable after allogeneic transplant, correlating with complete remission. Although contributions to MM spread and progression is likely, their malignant status and impact has yet to be clarified. Their high frequency in the blood, and their resistence to conventional chemotherapy suggests that the number of circulating clonotypic cells should be clinically monitored, and that therapeutic targeting of these B cells may benefit myeloma patients.
© 1998 by The American Society of Hematology.
MULTIPLE MYELOMA (MM), a B-cell neoplasia, is characterized by the presence of monoclonal immunoglobulin in the blood, lytic bone lesions, and often large numbers of monoclonal plasma cells in the bone marrow (BM). Although many patients respond initially, nearly all relapse and become refractory to treatment.1,2 While it is clear that monoclonal plasma cells located in the BM directly or indirectly mediate most symptoms of myeloma, these cells do not appear to have the qualities of growth and spread required of a malignant progenitor cell. A number of observations have led to the view that the generative compartment in myeloma includes B-lineage cells found in the BM, the blood, or both, at a stage of differentiation preceding that of plasma cells.3-15
For B-lineage malignancies, unique sequences within the Ig genes, termed complementarity determining regions (CDR1, CDR2, CDR3), provide a consistent molecular marker for unequivocal identification of clonal B-lineage cells. Many studies have shown cells in the blood of myeloma patients with an Ig heavy-chain (IgH) rearrangement identical to that of autologous BM plasma cells.4,8,9,16-23 Although the differentiation stage of these blood cells and their number is controversial, the sharing of IgH rearrangements with the malignant plasma cells is widely accepted as indicative of a clonal relationship, termed clonotypic. A first step towards evaluating the extent to which peripheral blood mononuclear cells (PBMC) include myeloma-associated or precursor cells is to quantitate the number of clonotypic B cells in the circulation. Previous work showing the presence in PBMC of cells with clonotypic rearrangements have provided a wide range of numbers.4,16,17,23,24 Most of these numbers are estimates based on the frequency of a given sequence in nucleic acids purified from heterogeneous cell populations. Circulating plasma cells cannot account for the sometimes large numbers of clonotypic blood cells in myeloma.17 To assess the number of individual cells with clonotypic transcripts in myeloma PBMC and BM cells (BMC), single-cell assays are required. Analysis of myeloma PBMC, using marker sequences confirmed as clonal, indicate a large subset of circulating clonotypic CD19+ cells.24,25 These B cells are morphologically and functionally distinct from plasma cells.4,5,24,25,27 They are CD19+, CD11b+ CD34+ cells4,24,25 that express messenger RNA (mRNA) encoding IgH, CD19, and CD34.24,26 This clonotypic B cell subset has properties consistent with malignant status.24,25,28-31CD34+ CD11b+ B cells are the predominant clonotypic subset in blood, with an average of 81% to 87% clonotypic cells.24,25 The drug resistance of these cells,25,28,32 and their persistence after chemotherapy4 28 implicates them in the events underlying relapse. The frequency of myeloma B cells is most accurately determined using methods that analyze individual B-lineage cells. In this report, using single-cell reverse transcriptase-polymerase chain reaction (RT-PCR) assays and in situ RT-PCR with patient-specific primers, we show that a large proportion of individual myeloma B cells are clonotypic.
MATERIALS AND METHODS
Patients and samples.
Blood and BM were obtained after informed consent from 19 patients with MM, at diagnosis, during intermittent chemotherapy and after treatment (Table 1). Blood was also obtained from 18 healthy donors, and BM from 5 healthy donors. BM aspirates used for deriving the clonotypic IgH VDJ sequence had 11% to 80% plasma cells as identified morphologically and phenotypically. BMC were purified using Ficoll Paque (Pharmacia, Dorval, Quebec, Canada). Peripheral blood was drawn into heparinized tubes and purified over Ficoll Paque (Pharmacia) to give PBMC. Because no EDTA was added, cell-free DNA or RNA from serum would be immediately degraded33 and thus could not be a source of contamination in the analysis described below. All samples were purified immediately after being drawn. For RT-PCR, PBMC were lysed in Trizol (GIBCO-BRL, Burlington, Ontario, Canada) immediately after purification. For in situ RT-PCR, cells were antibody labeled and stored for 18 hours in fixative before sorting. For single-cell RT-PCR, cells were either used immediately in the limiting dilution assay or were antibody labeled, sorted, and processed within 3 to 4 hours after collection of the sample. For limiting dilution analysis, PBMC were diluted to an appropriate concentration as indicated in Results, and immediately deposited into PCR tubes containing lysis buffer (see below). This study analyzed all myeloma patients presenting at our institution for whom a BM sample had been obtained during the course of the study.
Characteristics of Clonotypic IgH VDJ Sequences
| Patient (status-150) . | Stage of Disease . | IgH . | Lt . | % PC-153 . | Vh Family . | Jh Family . | CDR3 Length (NT) . |
|---|---|---|---|---|---|---|---|
| 1. (Unt) | 3A | IgG | K | 23 | Vh2 (S12-12) | Jh4 (b) | 42 |
| 2. (Unt) | 3A | IgG | K | 31 | Vh5 (DP73) | Jh3 (a) | 30 |
| 3. (Tr) | Prog. | IgA | K | 33 | Vh3 (DP77) | Jh2 | 51 |
| 4. (Unt) | 3 | IgA | K | 30 | Vh4 (DP65) | nd | 36 |
| 5. (Unt) | 2A | IgG | L | 23 | VH3 (DP49) | Jh6 (b) | 48 |
| 6. (Unt) | 3 | IgG | K | 30 | Vh4 (DP71) | Jh4 (a) | 48 |
| 7. (Unt) | 3A | IgG | K | 25 | VH3 (DP77) | Jh2 | 48 |
| 8. (Off) | Relapse | IgA | L | 57 | Vh3 (DP46) | Jh3 (b) | 42 |
| 9. (Unt) | 3 | Nda | L | 80 | Vh3-21 (DP77) | Jh6 (c) | 51 |
| 10. (Unt) | Prog. | IgG | 90 | Vh2 (S12-12) | Jh4 (a) | 42 | |
| 11. (Ind) | 1A | IgA | K | 51 | Vh3-30 (DP49) | Jh4 (a) | 33 |
| 12. (Tr)-151 | Stable Resp. | IgG | L | 11 | Vh3-15 (DP389) | Jh4 (a) | 42 |
| 13. (Unt) | 3 | IgA | K | 70 | Vh3-8 (DP58) | Jh4 (a) | 39 |
| 14. (Off)-152 | Relapse | IgG | K | 67 | Vh3-30 (DP49) | Jh4 (b) | 39 |
| 15. (Unt) | 3 | IgG | K | 75 | Vh4-31 (DP78) | Jh4 (b) | 24 |
| 16. (Unt) | 3 | IgG | K | 22 | Vh1 (DP88) | Jh4 (b) | 51 |
| 17. (Unt) | 3A | IgG | K | 50 | Vh3-49 (DP57) | Jh4 (c) | 84 |
| 18. (Tr) | 3 | IgG | K | 41 | Vh5-51 (DP73) | Jh4 (b) | 27 |
| 19. (Unt) | 3 | IgG | K | 55 | Vh-3 (DP47) | Jh4 (b) | 45 |
| Patient (status-150) . | Stage of Disease . | IgH . | Lt . | % PC-153 . | Vh Family . | Jh Family . | CDR3 Length (NT) . |
|---|---|---|---|---|---|---|---|
| 1. (Unt) | 3A | IgG | K | 23 | Vh2 (S12-12) | Jh4 (b) | 42 |
| 2. (Unt) | 3A | IgG | K | 31 | Vh5 (DP73) | Jh3 (a) | 30 |
| 3. (Tr) | Prog. | IgA | K | 33 | Vh3 (DP77) | Jh2 | 51 |
| 4. (Unt) | 3 | IgA | K | 30 | Vh4 (DP65) | nd | 36 |
| 5. (Unt) | 2A | IgG | L | 23 | VH3 (DP49) | Jh6 (b) | 48 |
| 6. (Unt) | 3 | IgG | K | 30 | Vh4 (DP71) | Jh4 (a) | 48 |
| 7. (Unt) | 3A | IgG | K | 25 | VH3 (DP77) | Jh2 | 48 |
| 8. (Off) | Relapse | IgA | L | 57 | Vh3 (DP46) | Jh3 (b) | 42 |
| 9. (Unt) | 3 | Nda | L | 80 | Vh3-21 (DP77) | Jh6 (c) | 51 |
| 10. (Unt) | Prog. | IgG | 90 | Vh2 (S12-12) | Jh4 (a) | 42 | |
| 11. (Ind) | 1A | IgA | K | 51 | Vh3-30 (DP49) | Jh4 (a) | 33 |
| 12. (Tr)-151 | Stable Resp. | IgG | L | 11 | Vh3-15 (DP389) | Jh4 (a) | 42 |
| 13. (Unt) | 3 | IgA | K | 70 | Vh3-8 (DP58) | Jh4 (a) | 39 |
| 14. (Off)-152 | Relapse | IgG | K | 67 | Vh3-30 (DP49) | Jh4 (b) | 39 |
| 15. (Unt) | 3 | IgG | K | 75 | Vh4-31 (DP78) | Jh4 (b) | 24 |
| 16. (Unt) | 3 | IgG | K | 22 | Vh1 (DP88) | Jh4 (b) | 51 |
| 17. (Unt) | 3A | IgG | K | 50 | Vh3-49 (DP57) | Jh4 (c) | 84 |
| 18. (Tr) | 3 | IgG | K | 41 | Vh5-51 (DP73) | Jh4 (b) | 27 |
| 19. (Unt) | 3 | IgG | K | 55 | Vh-3 (DP47) | Jh4 (b) | 45 |
Abbreviations: ND, not detectable by routine clinical methods; Lt, light chain; K, kappa; L, lambda; Unt, untreated; Tr, treated; Off, Off therapy; Ind, indolent myeloma; Prog, progressive disease; Resp, responder.
These patients were all diagnosed with myeloma; patients 8 and 14 were in relapse at the time their BM was obtained. Patient 10 died 1 month after diagnosis. Their treatment status at the time the bone marrow sample was obtained is indicated.
This sample was taken immediately before autologous hematopoietic transplantation.
This patient was first diagnosed in 1988 and is thus a long-term survivor. In 1993/94, she had clonotypic DNA sequences detectable in her blood (4, Patient 3 in that study). The sequence of IgH VDJ transcripts in her plasma cells was determined at relapse for this study (in 1997) using single-sorted BM plasma cells. The CDR3 sequence obtained was identical to that determined previously.4 All of her relapse plasma cells express this sequence.
%PC = percent of plasma cells in the bone marrow sample used to derive the clonotypic sequence.
Antibodies and reagents.
Immunofluorescence (IF) and sorting.
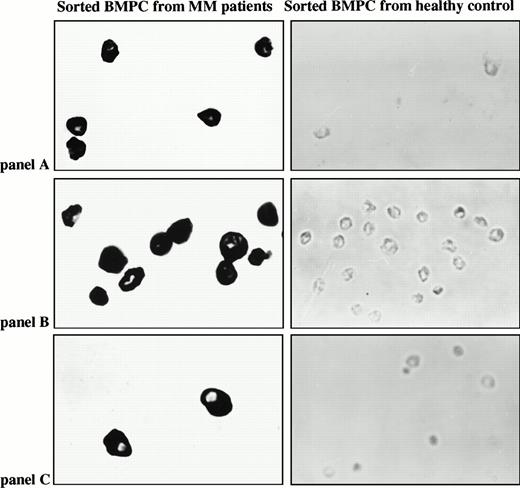
Staining for surface phenotype used one- or two-color IF with CD19-FITC or CD38-PE/antihuman Ig-FITC, as well as all relevant isotype controls, as described previously.4,24,28 BMC were stained with CD38-PE and antihuman Ig-FITC followed by sorting of CD38hiIg+ BMC with high forward and side scatter. PBMC were sorted for CD19+ cells with no gates set on scatter beyond those used to exclude red and dead cells. The CD19+ B cells sorted here were detected by their staining with either FMC63 or B4 (Coulter, Oakville, Ontario, Canada) but were not detected by Leu12 (Becton Dickinson)1A,5,36 (International Myeloma Workshop, Boston, 1997), or by monoclonal antibody (MoAb) to CD19 from Sigma (Oakville, Ontario, Canada), Dako (Detroit, MI), or Immunotech/Coulter (Burlington, Ontario, Canada). Neuraminidase treatment shows Leu12 epitopes indicating they are present but cryptic on these B cells.36 For single-cell experiments, individual CD19+ PBMC or CD38hiIg+ large BMC were sorted directly into 0.2 mL thin-walled PCR tubes or onto slides. On reanalysis, sorted populations had a purity of 96% or greater for the defining phenotype. Less than 1% of morphologically identifiable plasma cells were observed in cytospins of sorted B cells (performed for six patients), in cytospins of PBMC (for four patients), or in smears of patient blood (all patients).
Identification of patient-specific (clonotypic) IgH sequences and primer selection.
The clonotypic sequence was determined from mRNA of 1 to 1000 BM plasma cells using consensus RT-PCR, followed by sequencing of the amplified product from at least 3 individual plasma cells or from 6 subclones of the consensus product. Primers to histone, a housekeeping mRNA, were used as a positive control to ensure mRNA integrity. IgH VDJ region primers to FR2, J region, or to Vh3 family4 37 were as previously described. CDR2 and CDR3 patient-specific primers are given in Results. Histone primers were as follows: 5’ CCACTGAACTTCTGATTCGC, 3’ GCGTGCTAGCTGGATGTCTT. The V, D, and J families of clonotypic IgH sequences were analyzed, and patient-specific primers annealing to the CDR2 and CDR3 portions of the identified IgH sequence were chosen using the V-base sequence directory (http://www.mrc-cpe.cam.ac.uk/imt-doc/vbase-home-page.html), DNAPLOT (http://www.mrc-cpe.cam.ac. uk/imt-doc/DNAsearch.html), and Primer3 (Picks PCR primers from nucleotide sequence) (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi). Primers were synthesized on ABI PCR mate 391 (Perkin Elmer, Applied Biosystems, Mississauga, Ontario, Canada). To confirm the specificity of the chosen primers, they were used in PCR to identify a product of the expected size from complementary DNA (cDNA) of individual autologous BM plasma cells, but not from cDNA of plasma cells or PBMC of unrelated myeloma patients.
Patient-specific amplification (PSA).
For amplification of PS sequences, primers to the CDR2 and the CDR3 regions of the rearranged IgH VDJ from individual BM plasma cells were designed and used for in situ RT-PCR. The size of PCR product varied in individual patients within the range of 120 to 180 bp, and gave a discrete single band when mRNA was analyzed. For all patients, the specificity of the PSA was confirmed by testing the primers using RNA isolated from PBMC B cells of healthy donors, CD38hicIg+ BM plasma cells of healthy donors, and unrelated myeloma B and plasma cells as negative controls.
RT-PCR: (consensus and specific).
Using Trizol according to manufacturers directions (GIBCO-BRL, Burlington, Ontario, Canada), RNA was prepared from 0.1 to 10 × 106 unfractionated BMC, PBMC or sorted populations of B cells from myeloma patients and normal donors. After purification, 1 μg of RNA was reverse transcribed using SuperScript reverse transcriptase (GIBCO-BRL) and universal primer oligo dT15, according to the manufacturers instructions. PCR was performed under standard conditions. Briefly, 2 μL of cDNA was added to 48 μL of PCR buffer (GIBCO-BRL), 2 mmol/L MgCl2, mixed with 0.2 μmol/L consensus primers FR2 and Jh, and 1 U/reaction tube of TAQ polymerase: 25 cycles of 30 seconds at 94°C, 30 seconds at 50°C, and 45 seconds at 72°C were performed on the PCR Thermal Cycler Perkin Elmer 9600 (Perkin Elmer). For consensus RT-PCR, a second round of amplification used FR2 and a nested Jh consensus primer. For PSA, as described above, the second round of amplification used patient-specific primers (CDR2 and CDR3) for 25 cycles at an annealing temperature of 60°C. The PCR product was analyzed on a 2% agarose gel in tris-borate/EDTA (TBE) buffer, soaked in ethidium bromide, and visualized under ultraviolet light.
Single-cell RT-PCR.
Using the ELITE flow cytometer with an Autoclone Cell Deposition Unit (Coulter, Hiale, FL), single cells were sorted into 0.2 mL PCR tubes containing 8 μL of RT-Lysis solution (SuperScript first-strand buffer, 0.5% NP-40 (vol/vol), 0.01 mol/L dithiothreitol (DTT), 0.25 mmol/L dNTPs, 0.006 mmol/L dT16, 200 U of RNAse inhibitor (GIBCO-BRL). Immediately after the sort, tubes were centrifuged for 1 minute and then frozen at −80°C. After thawing, samples were heated to 70°C for 10 minutes, placed on ice, and 2 μL (100 U) of reverse transcriptase SuperScript (GIBCO-BRL) was added into each tube and incubated at 42°C for 60 minutes. The reaction was stopped by heating at 99°C for 3 minutes. Alternatively, immediately after purification, within 1 to 2 hours after the sample was drawn, PBMC were diluted to an appropriate concentration as indicated in results and 1 μL of this suspension was deposited into 9 μL of lysis buffer containing the reverse transcriptase, in PCR tubes. This step was followed by an immediate incubation at 42°C for 60 minutes and next at 99°C for 3 minutes. For either method, all of the obtained single-cell cDNA was then used in a two-step nested PCR. Briefly, PCR mix (0.2 mmol/L dNTPs, 10 mmol/L Tris-HCL pH 8.3, 0.2 μmol/L each of sense and antisense primers, 2 mmol/L MgCl2, and 2 U of TAQ polymerase) in a final volume of 50 μL was added to tubes, followed by 30 cycles of 30 seconds at 94°C, 30 seconds at 52°C and 1 minute at 72°C. Four percent (vol/vol) of this PCR amplified mixture was transferred into a secondary PCR mix with 2 mmol MgCl2, FR2, and JH2 primers and cycled as previously for 35 cycles. Finally, 20% of the product was analyzed on 2% agarose gels (GIBCO/BRL). For all samples, amplified products were subcloned into a TA cloning vector (Invitrogen, Mississauga, Ontario, Canada) and cycle sequenced with universal M13 forward and reverse primers. For the PSA, 25 cycles were performed using CDR2 and CDR3 primers and an annealing temperature of 60°C. The final product was analyzed on 2% agarose gel in 0.5 × TBE buffer.
For the limiting dilution analysis of single PBMC from patient 19, for the first 30 cycles of PCR the Vh-3 (family specific) primer was used in combination with the Jh consensus primer. For the next 35 cycles of nested PCR, patient-specific primers for CDR2 and CDR3 were used (expected product size = 176 bp). Samples were analyzed by electrophoresis on 2% agarose gels and the product visualized by ethidium bromide staining. To permit sequencing of the CDR3 region, the PCR product from the first-stage PCR reaction (Vh3 and Jh primers) was reamplified with a second set of primers, using a patient-specific primer to CDR2 and a consensus Jh2 primer. For analysis of clonotypic sequences from individual B cells, the product of the nested PCR was subcloned into TA cloning vector. Plasmids were isolated from three to five randomly chosen colonies from each transformation and sequenced using dRhodamine terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Mississauga, Ontario, Canada). The sequencing products were analyzed on a capillary sequencer ABI 310 (PE Applied Biosystems).
In situ RT-PCR.
RESULTS
Generation of patient-specific IgH VDJ sequences using single plasma cell RT-PCR.
To identify clonotypic IgH VDJ sequences for individual myeloma patients, single BM plasma cells were sorted directly into PCR tubes containing RT-lysis buffer, followed by reverse transcription and amplification of cDNA with consensus primers to FR2 and Jh of IgH.4,24 25 For all patients analyzed, a PCR product was obtained from the majority of these sorted plasma cells (Fig 1A).
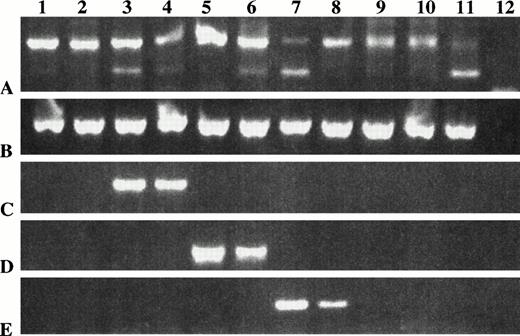
RT-PCR amplification of IgH VDJ using consensus primers from single BM plasma cells of a myeloma patient. (A) Individual BM plasma cells from patient 2 were purified, stained, and sorted into PCR tubes containing lysis buffer, followed by reverse transcription and seminested amplification of IgH VDJ mRNA using consensus primers to FR2 and Jh as indicated in methods. Product was amplified from 10 of 14 plasma cells (lines 1-14) (M: molecular weight marker − 100 bp ladder). (B) Individual BM plasma cells from patient 2 were purified, stained, and sorted into PCR tubes containing lysis buffer, followed by reverse transcription and nested amplification of IgH VDJ mRNA first using consensus primers to FR2 and Jh and next using specific primers to CDR2 and CDR3 regions of clonotypic IgH for patient 2. Product was amplified from 12 of 14 plasma cells (lines 1-14) (M: molecular weight marker − 100 bp ladder).
RT-PCR amplification of IgH VDJ using consensus primers from single BM plasma cells of a myeloma patient. (A) Individual BM plasma cells from patient 2 were purified, stained, and sorted into PCR tubes containing lysis buffer, followed by reverse transcription and seminested amplification of IgH VDJ mRNA using consensus primers to FR2 and Jh as indicated in methods. Product was amplified from 10 of 14 plasma cells (lines 1-14) (M: molecular weight marker − 100 bp ladder). (B) Individual BM plasma cells from patient 2 were purified, stained, and sorted into PCR tubes containing lysis buffer, followed by reverse transcription and nested amplification of IgH VDJ mRNA first using consensus primers to FR2 and Jh and next using specific primers to CDR2 and CDR3 regions of clonotypic IgH for patient 2. Product was amplified from 12 of 14 plasma cells (lines 1-14) (M: molecular weight marker − 100 bp ladder).
The amplified products from these individual plasma cells were pooled, subcloned, and sequenced. The predominant sequence thus obtained was tentatively selected as the clonotypic sequence pending further testing. In all patients analyzed there were at least two other species of IgH transcript detected among the amplified sequences, suggestive of nonmalignant clones. Patient-specific primers were designed and tested for their ability to amplify IgH mRNA in RT-PCR from individual sorted plasma cells. For most patients the sequence initially selected was detectable in nearly all of the sorted autologous plasma cells analyzed (Fig 1B). However for 3 of 19 patients, the sequence initially identified was present in only 10% or fewer of BM plasma cells and was completely different from the actual MM clonal marker sequence. For these patients, additional cloning and sequencing reactions were necessary to identify the clonotypic myeloma sequence. For all patients, the expression of an IgH VDJ sequence defined as clonotypic was validated using single-cell RT-PCR on sorted BM plasma cells (Fig1B), analysis of RT-PCR product from 1000 plasma cells, and amplification of a clonotypic product from purified RNA of BMC for the appropriate patient. Controls confirmed that the clonotypic MM sequences were not detected in plasma cells of unrelated patients or normal donors (see below).
To rigorously confirm the specificity of patient-specific priming and the unique expression of sequences identified as clonotypic, total RNA prepared from patient and normal donor BMC was analyzed by RT-PCR using PSA. For all 19 patients, we detected a clonotypic transcript only when patient-specific primers were used in combination with RNA isolated from the appropriate patient. The clonal product was not detected in total RNA from BMC from unrelated patients or normal donors (see below). Next, in situ RT-PCR using patient-specific primers was performed on sorted plasma cells from myeloma patients and from normal donors. Patient-specific primers were used for further analysis only if they amplified a product from the majority (>80%) of autologous plasma cells, but did not amplify a product from BM plasma cells of an unrelated myeloma patient or of healthy donors (Fig 2).
In situ RT-PCR on BM plasma cells with patient-specific primers. (A) Plasma cells sorted from a BM of MM patient 10 and a healthy donor underwent in situ amplification with patient 10 CDR2/CDR3-IgH–specific primers. (B) Plasma cells sorted from a BM of MM patient 2 and a healthy donor underwent in situ amplification with patient 2 CDR2/CDR3-IgH–specific primers. (C) Plasma cells sorted from a BM of MM patient 11 and a healthy donor underwent in situ amplification with patient 11 CDR2/CDR3-IgH–specific primers.
In situ RT-PCR on BM plasma cells with patient-specific primers. (A) Plasma cells sorted from a BM of MM patient 10 and a healthy donor underwent in situ amplification with patient 10 CDR2/CDR3-IgH–specific primers. (B) Plasma cells sorted from a BM of MM patient 2 and a healthy donor underwent in situ amplification with patient 2 CDR2/CDR3-IgH–specific primers. (C) Plasma cells sorted from a BM of MM patient 11 and a healthy donor underwent in situ amplification with patient 11 CDR2/CDR3-IgH–specific primers.
The characteristics of the clonotypic sequences derived from BM plasma cells of 19 myeloma patients are presented in Tables 1 and 2. The patients have the Ig isotype distribution typical of myeloma and a normal Vh and Jh usage. The full sequences of the CDR3 portion of IgH, and the portion of the CDR2 sequences chosen as primers, are presented in Table 2.
Clonotypic IgH VDJ sequences are detectable in the blood of patients with MM.
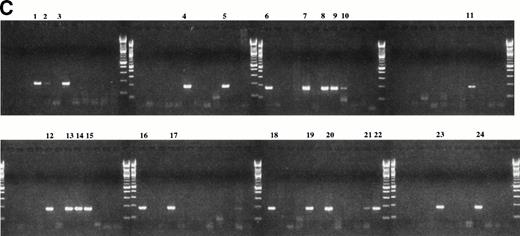
To determine if clonotypic sequences were detectable in PBMC, total RNA was purified from PBMC of myeloma patients, as well as from normal donors, and subjected to PSA. All PBMC and BMC tested expressed mRNA encoding IgH and histone. Figure 3 presents data for three representative myeloma patients, and shows that patient-specific clonotypic IgH transcripts were present in PBMC and BMC of the relevant patient, but not in PBMC of unrelated myeloma patients or normal-donor PBMC. The presence of clonotypic IgH transcripts in RNA from myeloma PBMC was confirmed for all 19 patients.
RT-PCR with patient-specific primers detects clonotypic transcripts from both blood and BM of the relevant patient but not from unrelated patients or healthy donors. One microgram of total RNA isolated from myeloma and control BM and PBMC was reverse transcribed and amplified with patient-specific primers. A clonal product was detected only in BM and PBMC of the patient for whom the primers were generated. The following primer pairs were used in PCR: (A) consensus IgH VDJ, (B) histone (housekeeping gene), (C) primers for CDR2/CDR3 of patient 10, (D) primers for CDR2/CDR3 of patient 11, (E) primers for CDR2/CDR3 of patient 12. Lanes 1 and 2, unrelated myeloma patient; lane 3 patient 10 BM; lane 4, patient 10 PBMC; lane 5, patient 11 BM; lane 6, patient 11 PBMC; lane 7, patient #12 BM; lane 8, patient 12 PBMC; lanes 9 and 10 healthy control BM; lane 11 healthy control PBMC; lane 12 water control.
RT-PCR with patient-specific primers detects clonotypic transcripts from both blood and BM of the relevant patient but not from unrelated patients or healthy donors. One microgram of total RNA isolated from myeloma and control BM and PBMC was reverse transcribed and amplified with patient-specific primers. A clonal product was detected only in BM and PBMC of the patient for whom the primers were generated. The following primer pairs were used in PCR: (A) consensus IgH VDJ, (B) histone (housekeeping gene), (C) primers for CDR2/CDR3 of patient 10, (D) primers for CDR2/CDR3 of patient 11, (E) primers for CDR2/CDR3 of patient 12. Lanes 1 and 2, unrelated myeloma patient; lane 3 patient 10 BM; lane 4, patient 10 PBMC; lane 5, patient 11 BM; lane 6, patient 11 PBMC; lane 7, patient #12 BM; lane 8, patient 12 PBMC; lanes 9 and 10 healthy control BM; lane 11 healthy control PBMC; lane 12 water control.
Clonotypic cells are frequent in freshly isolated myeloma PBMC: PBMC B cells express clonotypic transcripts, as confirmed by sequencing of the amplified product.
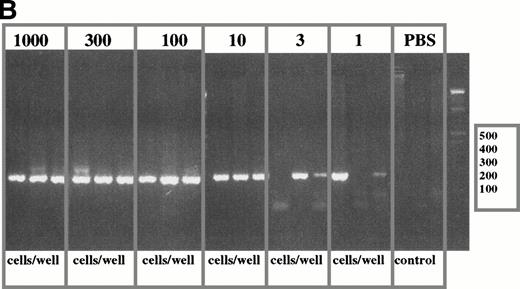
To determine the frequency of clonotypic cells, a limiting dilution analysis was performed using defined numbers of PBMC in threefold dilutions to give from 1000 to 1 cells per tube. PBMC were diluted immediately after purification. Two representative patients are shown in Fig 4A and 4B. For freshly diluted tubes containing 1000, 100, 10, or 3 PBMC per tube, all replicates were positive indicating that every aliquot of 3 to 1000 cells included at least 1 clonotypic cell. At 1 cell per tube, 1/3 (Fig4A and 4B) or 24 of 96 tubes (Fig 4C) were positive. The frequency of clonal cells was in the range of 25% to 33% of PBMC for these two patients, comparable with that obtained for the same samples using in situ RT-PCR (see below). A clonotypic RT-PCR product was not detected in PBS control tubes. This type of experiment has been done for PBMC samples from six other myeloma patients, and the frequency of clonotypic PBMC was within the range of that calculated from analysis by in situ RT-PCR.
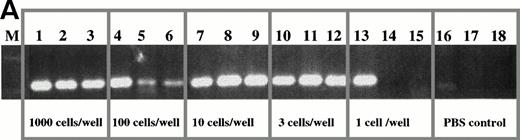
Limiting dilution analysis of freshly isolated PBMC from patient 1 and 19 using PSA. (A) Limiting dilution assay of PBMC from patient 1. The number of cells indicated in the figure were added to PCR tubes, reverse transcribed, and amplified in PSA with patient-specific primers. The transcripts in each tube were amplified in nested PCR using consensus primers to FR2 and Jh1 followed by a second amplification using patient-specific CDR2 and CDR3 primers. Of three tubes containing one cell, one was positive and two were negative suggesting an approximate concentration of the clonotypic cells in the blood of about 33%. The number of B cells as measured by flow cytometry for an aliquot of the same PBMC sample was 35%. This PBMC sample is the same as that described in Table 3, line 2, where clonotypic cells represented 18% of PBMC as measured by in situ RT-PCR on sorted B cells. (B) Limiting dilution of fresh PBMC from patient 19. IgH transcripts were amplified using VH3 family specific,40and Jh consensus primers, followed by patient-specific CDR2 and CDR3 primers in nested PCR. For this patient, 2 of 3 tubes were positive at the concentration of three cells per well (with 37% negative tubes at this concentration, the approximate frequency of clonotypic cells is one in every three PBMC, based on Poisson statistics), and 2 of 3 positive at one cell per well. By flow cytometry, patient 19 had 34% B cells in the PBMC samples used for the limiting dilution assay. (C) Clonotypic IgH transcripts at one cell per tube from patient 19. To more accurately determine the number of clonotypic cells, 96 tubes of one cell/tube were analyzed and 24 were positive (25%), as shown in the figure. With 75% negative tubes at this concentration, the chances of a given tube receiving more than one clonotypic cell are very low.
Limiting dilution analysis of freshly isolated PBMC from patient 1 and 19 using PSA. (A) Limiting dilution assay of PBMC from patient 1. The number of cells indicated in the figure were added to PCR tubes, reverse transcribed, and amplified in PSA with patient-specific primers. The transcripts in each tube were amplified in nested PCR using consensus primers to FR2 and Jh1 followed by a second amplification using patient-specific CDR2 and CDR3 primers. Of three tubes containing one cell, one was positive and two were negative suggesting an approximate concentration of the clonotypic cells in the blood of about 33%. The number of B cells as measured by flow cytometry for an aliquot of the same PBMC sample was 35%. This PBMC sample is the same as that described in Table 3, line 2, where clonotypic cells represented 18% of PBMC as measured by in situ RT-PCR on sorted B cells. (B) Limiting dilution of fresh PBMC from patient 19. IgH transcripts were amplified using VH3 family specific,40and Jh consensus primers, followed by patient-specific CDR2 and CDR3 primers in nested PCR. For this patient, 2 of 3 tubes were positive at the concentration of three cells per well (with 37% negative tubes at this concentration, the approximate frequency of clonotypic cells is one in every three PBMC, based on Poisson statistics), and 2 of 3 positive at one cell per well. By flow cytometry, patient 19 had 34% B cells in the PBMC samples used for the limiting dilution assay. (C) Clonotypic IgH transcripts at one cell per tube from patient 19. To more accurately determine the number of clonotypic cells, 96 tubes of one cell/tube were analyzed and 24 were positive (25%), as shown in the figure. With 75% negative tubes at this concentration, the chances of a given tube receiving more than one clonotypic cell are very low.
At the time of the limiting dilution analysis, patient 19 had stable disease with only 5% to 6% BM plasma cells, mIg of 5 gm/L, and no detectable circulating plasma cells in blood smears. Clonotypic PBMC are unlikely to be circulating plasma cells based on the low frequency of such cells in blood of patients throughout most of their disease.38,39 To confirm the relative absence of plasma cells from myeloma PBMC, cytospins of PBMC from patient 19 were Giemsa stained and analyzed by an experienced hematologist. Less than 1% of PBMC (14/1576 or 0.89%) were morphologically identifiable as plasma cells. However, at one cell per tube, 25% of tubes were positive for this patient, indicating that, at most, one tube might contain a plasma cell. This analysis shows that circulating plasma cells cannot possibly account for all the clonotypic cells detected in myeloma PBMC. Thus, clonotypic PBMC are B cells, as previously shown.24 25
To confirm that patient-specific RT-PCR was in fact detecting clonotypic sequences, we sequenced the amplified product from 18 of the positive PBMC B cells of patient 19 shown in Fig 4C. The first-stage reaction mixtures were reamplified using a patient-specific CDR2 primer and the Jh consensus primer. For each positive tube, the amplified product was subcloned and sequenced, after randomly picking several colonies with an Ig insert from each transformation (three to five transformants for each individual B cell, for a total of 74 inserts). All IgH subclones analyzed had CDR3 sequences completely identical to those of autologous plasma cells (Table 2). This confirms the specificity of the patient-specific RT-PCR, and indicates the population of circulating B cells lacks intraclonal heterogeneity. Because VH3 is the most frequently used VH gene family,40-42 residual polyclonal B cells likely exist,1A,36 but they were excluded from analysis by our use of the patient-specific CDR2 primer in the second stage of the nested PCR reaction. The analysis of Fig 4 shows that in fresh PBMC, clonotypic B cells are frequent.
Clonotypic IgH CDR3 Sequences and Primers Used for PSA
| ....CDR2 PRIMER .................FR3 | CDR3............................. JH | ||
| 01. | CACCAAGGACTCCTCCAAAA//TGTGCTCAC | ... | AAACTTATCACTGGTTGGGACGGTAGTAGTTACTTTGACCAG.TGGGGC |
| 02. | TGGGAGTCATCTATCCTGGTG//TGTGCGACA | ... | CAACACTACTATGATAGTTATATTGACTTC.TGGGGC |
| 03. | AGACTCAATGAAGGGCCGAT//TGTGCGAGA | ... | GATACCTATTATTATGGTTCAGGGAGTTATTCATACTGGTACTTCGATCTC.TGGGGC |
| 04. | ACTTCTACGACAATGGCGAAAC//TGTGCGGGT | ... | GGCACCACGTCCTCCCAGGGTCAGAGGTTGGAACTC.TGGGGC |
| 05. | CACCATTCAGATTCCGTGAAG//TGTGCGAAG | ... | CTCGTGGTTGTGGCGGTGGAAGCTCTAACCCAT GATAATCTTGATATT.TGGGGC |
| 06. | ACCAACATACAATCCCTCCCTC//TGTGCGAGG | ... | GTCCCCATGAACTATGCTATAAGGGGAAACTTAGGT TCCATTGACTAC.TGGGGC |
| 07. | AGTGGGGACAGCAGTGAAAT//TGTGCGAGA | ... | GAATGGTCGTACTTCTATGAAAGTTATTGGTTA TTACCCTTTGACTTC.TGGGGC |
| 08. | CTTATGTCATATGATGAAGGC//TGTGCGAGA | ... | GACGGAAGCAGAGATGGCTACAACTCG GGTGTTTTTGATATC.TGGGGC |
| 09. | ACCATCTCCAGAGACAACGC//TGTGCGAGA | ... | GGGGATGGTTCGGGAGAGATCTTT CCTTACTACTACTATCACATGGACGTC.TGGGGC |
| 10. | ATGAGCGCTACAACCCATCT//TGTTCACAC | ... | ACGCGTTTCATGCCTGCGGATGTGAACAACTTCTTTGACTAC.TGGGGC |
| 11. | TGGCACTTATTTCAGATGATGG//TGTGCGCCA | ... | GTTCTTGCCAACTGGTTTCGCCCCTTTGACCAC.TGGGGC |
| 12. | GTGGTGGGACAACAGACTCC//TGTACCACA | ... | GCGTTCAGTGAGCCCTCCAGCGACTACTACACGATGGACTTC.TGGGGC |
| 13. | AGTACTGGCGCTGTCACAAA//TGTGCGACA | ... | GATCAAGATGACTACGGTGACTACGGGACCTTTAACTCC.TGGGGC |
| 14. | TGGAGTGGGTGGCTTCTTAT//TGTACGAGA | ... | GTAAATCCTTTCTATGAAGGTAGTCGTTATCCCATA TACTACTTTGGCTAC.TGGGGC |
| 15. | TCCTCCAGTGGAAGCACCTA//TGCGCCAGA | ... | GATCCCTCTGACTACTTTGACCTC.TGGGGC |
| 16. | TCATCCCTATCTTTGGCACA//TGTGCGACA | ... | GTAAATCCTTTCTATGAAGGTAGTCGTTATCCCATA TACTACTTTGGCTAC.TGGGGC |
| 17. | GCTGGAGTGGGTAGGTTTCA//TGTACTAGA | ... | GATAGGGAGGATACTGTAGTAGGAACAGTTACTATGGGCCGAATACCCACGGTT AAATACTACTACTCCACCACATGGACGTC.TGGGGC |
| 18. | TGGGTGGGAATCATCTATCC//TGTGCGAGA | ... | CATTATCACGGTTACCGATCGGACGTC.TGGGGC |
| 19. | CCATCAACTACATTGCTGACA//TGTGCGAAG | ... | GGCCATCGCCATTGTAATGGAGGTGGCTGCGCCGTCCTGGACCTC.TGGGGC |
| ....CDR2 PRIMER .................FR3 | CDR3............................. JH | ||
| 01. | CACCAAGGACTCCTCCAAAA//TGTGCTCAC | ... | AAACTTATCACTGGTTGGGACGGTAGTAGTTACTTTGACCAG.TGGGGC |
| 02. | TGGGAGTCATCTATCCTGGTG//TGTGCGACA | ... | CAACACTACTATGATAGTTATATTGACTTC.TGGGGC |
| 03. | AGACTCAATGAAGGGCCGAT//TGTGCGAGA | ... | GATACCTATTATTATGGTTCAGGGAGTTATTCATACTGGTACTTCGATCTC.TGGGGC |
| 04. | ACTTCTACGACAATGGCGAAAC//TGTGCGGGT | ... | GGCACCACGTCCTCCCAGGGTCAGAGGTTGGAACTC.TGGGGC |
| 05. | CACCATTCAGATTCCGTGAAG//TGTGCGAAG | ... | CTCGTGGTTGTGGCGGTGGAAGCTCTAACCCAT GATAATCTTGATATT.TGGGGC |
| 06. | ACCAACATACAATCCCTCCCTC//TGTGCGAGG | ... | GTCCCCATGAACTATGCTATAAGGGGAAACTTAGGT TCCATTGACTAC.TGGGGC |
| 07. | AGTGGGGACAGCAGTGAAAT//TGTGCGAGA | ... | GAATGGTCGTACTTCTATGAAAGTTATTGGTTA TTACCCTTTGACTTC.TGGGGC |
| 08. | CTTATGTCATATGATGAAGGC//TGTGCGAGA | ... | GACGGAAGCAGAGATGGCTACAACTCG GGTGTTTTTGATATC.TGGGGC |
| 09. | ACCATCTCCAGAGACAACGC//TGTGCGAGA | ... | GGGGATGGTTCGGGAGAGATCTTT CCTTACTACTACTATCACATGGACGTC.TGGGGC |
| 10. | ATGAGCGCTACAACCCATCT//TGTTCACAC | ... | ACGCGTTTCATGCCTGCGGATGTGAACAACTTCTTTGACTAC.TGGGGC |
| 11. | TGGCACTTATTTCAGATGATGG//TGTGCGCCA | ... | GTTCTTGCCAACTGGTTTCGCCCCTTTGACCAC.TGGGGC |
| 12. | GTGGTGGGACAACAGACTCC//TGTACCACA | ... | GCGTTCAGTGAGCCCTCCAGCGACTACTACACGATGGACTTC.TGGGGC |
| 13. | AGTACTGGCGCTGTCACAAA//TGTGCGACA | ... | GATCAAGATGACTACGGTGACTACGGGACCTTTAACTCC.TGGGGC |
| 14. | TGGAGTGGGTGGCTTCTTAT//TGTACGAGA | ... | GTAAATCCTTTCTATGAAGGTAGTCGTTATCCCATA TACTACTTTGGCTAC.TGGGGC |
| 15. | TCCTCCAGTGGAAGCACCTA//TGCGCCAGA | ... | GATCCCTCTGACTACTTTGACCTC.TGGGGC |
| 16. | TCATCCCTATCTTTGGCACA//TGTGCGACA | ... | GTAAATCCTTTCTATGAAGGTAGTCGTTATCCCATA TACTACTTTGGCTAC.TGGGGC |
| 17. | GCTGGAGTGGGTAGGTTTCA//TGTACTAGA | ... | GATAGGGAGGATACTGTAGTAGGAACAGTTACTATGGGCCGAATACCCACGGTT AAATACTACTACTCCACCACATGGACGTC.TGGGGC |
| 18. | TGGGTGGGAATCATCTATCC//TGTGCGAGA | ... | CATTATCACGGTTACCGATCGGACGTC.TGGGGC |
| 19. | CCATCAACTACATTGCTGACA//TGTGCGAAG | ... | GGCCATCGCCATTGTAATGGAGGTGGCTGCGCCGTCCTGGACCTC.TGGGGC |
The sequence of CDR2 chosen for PSA priming is indicated, followed by a break in the continuous VDJ sequence and then by the complete CDR3 sequence. Regions used as patient-specific CDR3 primers are highlighted in bold italics.
Clonotypic PBMC are CD19+ B cells.
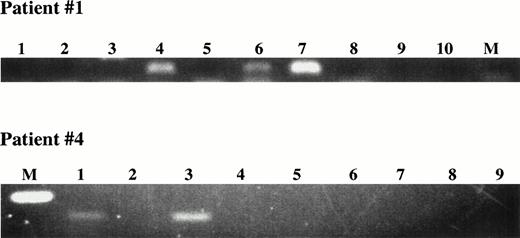
To confirm that B cells expressed the clonotypic sequence, single CD19+ B cells were sorted into PCR tubes for direct lysis followed by amplification of IgH transcripts using liquid phase patient-specific RT-PCR. Results for two representative patients are shown in Fig 5A. Clonotypic IgH transcripts were amplified from three of eight B cells from patient 1, and for two of eight B cells from patient 4.
Detection of circulating clonotypic cells among sorted PBMC B cells using single-cell in situ RT-PCR with patient-specific primers. (A) Single-cell RT-PCR using sorted B cells. Individual FMC63+ B cells were sorted directly into PCR tubes. Nested PCR first with consensus, and second with patient-specific primers was performed. The products had the expected size of 111 bp (Patient 1) or 160 bp (patient 4). M = molecular weight markers; Lanes 1 to 8 = sorted B cells; lanes 9,10 water controls. (B) In situ RT-PCR using sorted B cells. Row 1, CD19+ cells sorted from a peripheral blood of MM patient 11 and from a healthy donor underwent in situ RT-PCR amplification with patient 11 CDR2/CDR3-IgH–specific primers. Row 2, CD19+cells sorted from a peripheral blood of MM patient 12 and from a healthy donor underwent in situ amplification with CDR2/CDR3-IgH–specific primers for patient #12. Row 3, CD19+ cells sorted from a peripheral blood of MM patient 1 and from a healthy donor underwent in situ amplification with CDR2/CDR3-IgH–specific primers for patient 1. Row 4, CD19+ cells sorted from a peripheral blood of MM patient 5 and from a healthy donor underwent in situ amplification with CDR2/CDR3-IgH–specific primers for patient 5.
Detection of circulating clonotypic cells among sorted PBMC B cells using single-cell in situ RT-PCR with patient-specific primers. (A) Single-cell RT-PCR using sorted B cells. Individual FMC63+ B cells were sorted directly into PCR tubes. Nested PCR first with consensus, and second with patient-specific primers was performed. The products had the expected size of 111 bp (Patient 1) or 160 bp (patient 4). M = molecular weight markers; Lanes 1 to 8 = sorted B cells; lanes 9,10 water controls. (B) In situ RT-PCR using sorted B cells. Row 1, CD19+ cells sorted from a peripheral blood of MM patient 11 and from a healthy donor underwent in situ RT-PCR amplification with patient 11 CDR2/CDR3-IgH–specific primers. Row 2, CD19+cells sorted from a peripheral blood of MM patient 12 and from a healthy donor underwent in situ amplification with CDR2/CDR3-IgH–specific primers for patient #12. Row 3, CD19+ cells sorted from a peripheral blood of MM patient 1 and from a healthy donor underwent in situ amplification with CDR2/CDR3-IgH–specific primers for patient 1. Row 4, CD19+ cells sorted from a peripheral blood of MM patient 5 and from a healthy donor underwent in situ amplification with CDR2/CDR3-IgH–specific primers for patient 5.
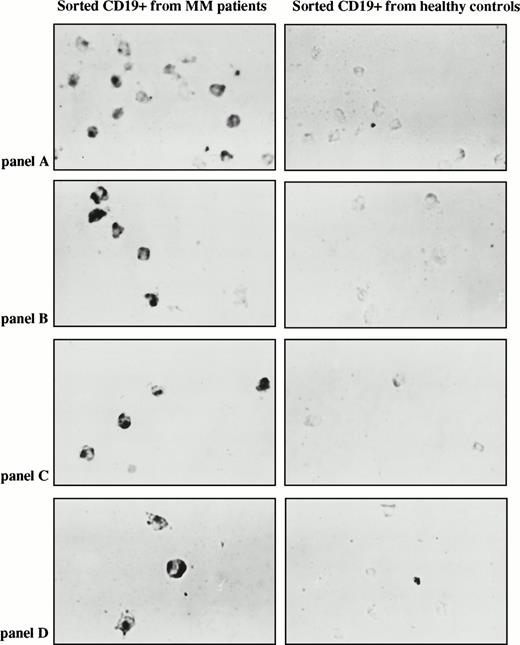
To further confirm identification and more precisely quantify the clonotypic B cells in myeloma PBMC, we used in situ RT-PCR to analyze sorted CD19+ B cells for clonotypic sequences. CD19+ B cells were rapidly stained and fixed, followed by sorting onto slides for in situ RT-PCR using patient-specific primers. For all patient-specific primer pairs, in situ RT-PCR PSA was performed on sorted B cells from PBMC of at least two normal donors and sorted plasma cells from BMC of at least one normal donor, with consistently negative results. Representative PSA in situ RT-PCR of sorted B cells from four myeloma patients and four normal donors is shown in Fig 5B.
On average, 66% of B cells in myeloma PBMC are clonotypic as detected by in situ RT-PCR.
Table 3 records the frequency of clonotypic B cells for each myeloma patient, as measured by in situ RT-PCR, for one or more blood samples taken at regular clinical visits. In blood, the proportion of sorted B cells expressing clonotypic IgH mRNA ranged from 9% to 95% with a mean of 66% ± 4% standard error (SE). These values were used to calculate that 14% ± 2% of PBMC were clonotypic cells (range, 0.9% to 50% of PBMC). The proportion of clonotypic cells among total white blood cells was calculated as 3.5% ± 1% (range, 1% to 9%) (not shown). The absolute number of circulating clonotypic B cells was 0.15 ± 0.02 × 109/L of blood (range, 0.01 to 0.61 × 109/L).
Clonotypic B Cells Are Frequent in the Blood of Myeloma Patients
| Patient Status at End of Study . | Sequential Time Points (#) . | Cells That Are Clonotypic (%) . | ||
|---|---|---|---|---|
| B Cells . | PBMC . | Absolute # ×109/L Blood . | ||
| 1. Tr | 5 | 69, 95, 93 | 24, 3, 14 | 0.1, 0.35, 0.09 |
| 95, 64 | 10, 18 | 0.05, 0.11 | ||
| 2. Deceased | 2 | 74, 74 | 19, 31 | 0.17, 0.15 |
| 3. Tr | 2 | 71, 54 | 14, 9 | 0.22, 0.12 |
| 4. Allo transplant | 6 | 45, 46, 46, 62 | 9, 9, 10, 19 | 0.12, 0.08, 0.06, 0.11 |
| 2, <0.2 | 0.6, <0.1 | 0.004, <0.000004 | ||
| 5. Off | 3 | 91, 40, 73 | 18, 8, 16 | 0.24, 0.1, 0.21 |
| 6. Tr | 3 | 60, 77, 90 | 10, 10, 50 | 0.08, 0.03, 0.1 |
| 7. Deceased | 1 | 93 | 12 | 0.06 |
| 8. Deceased | 1 | 9 | 0.9 | 0.01 |
| 9. Tr | 1 | 52 | 7 | 0.09 |
| 11. Unt | 2 | 65, 64 | 10, 6 | 0.08, 0.06 |
| 12. Auto transplant | 2 | 46, 64 | 15, 7 | 0.16, 0.13 |
| 13. Tr | 1 | 73 | 12 | 0.18 |
| 14. Tr | 2 | 28, 93 | 8, 21 | 0.03, 0.08 |
| 15. Tr | 2 | 66, 95 | 11, 13 | 0.17, 0.13 |
| 16. Tr | 1 | 56 | 13 | 0.17 |
| 17. Tr | 1 | 53 | 20 | 0.42 |
| 18. Tr | 2 | 68, 49 | 20, 18 | 0.61, 0.39 |
| 19. Tr | 1 | 50 | 21 | 0.27 |
| Mean ± SE | 66 ± 4 | 14 ± 2 | 0.15 ± .02 | |
| Normal PBMC (18 donors) | ||||
| CD19+ PBMC (55 slides) | <0.3 | |||
| Normal plasma cells (BM, 5 donors) | ||||
| CD38hiIg+ BMC (20 slides) | <0.3 | |||
| Patient Status at End of Study . | Sequential Time Points (#) . | Cells That Are Clonotypic (%) . | ||
|---|---|---|---|---|
| B Cells . | PBMC . | Absolute # ×109/L Blood . | ||
| 1. Tr | 5 | 69, 95, 93 | 24, 3, 14 | 0.1, 0.35, 0.09 |
| 95, 64 | 10, 18 | 0.05, 0.11 | ||
| 2. Deceased | 2 | 74, 74 | 19, 31 | 0.17, 0.15 |
| 3. Tr | 2 | 71, 54 | 14, 9 | 0.22, 0.12 |
| 4. Allo transplant | 6 | 45, 46, 46, 62 | 9, 9, 10, 19 | 0.12, 0.08, 0.06, 0.11 |
| 2, <0.2 | 0.6, <0.1 | 0.004, <0.000004 | ||
| 5. Off | 3 | 91, 40, 73 | 18, 8, 16 | 0.24, 0.1, 0.21 |
| 6. Tr | 3 | 60, 77, 90 | 10, 10, 50 | 0.08, 0.03, 0.1 |
| 7. Deceased | 1 | 93 | 12 | 0.06 |
| 8. Deceased | 1 | 9 | 0.9 | 0.01 |
| 9. Tr | 1 | 52 | 7 | 0.09 |
| 11. Unt | 2 | 65, 64 | 10, 6 | 0.08, 0.06 |
| 12. Auto transplant | 2 | 46, 64 | 15, 7 | 0.16, 0.13 |
| 13. Tr | 1 | 73 | 12 | 0.18 |
| 14. Tr | 2 | 28, 93 | 8, 21 | 0.03, 0.08 |
| 15. Tr | 2 | 66, 95 | 11, 13 | 0.17, 0.13 |
| 16. Tr | 1 | 56 | 13 | 0.17 |
| 17. Tr | 1 | 53 | 20 | 0.42 |
| 18. Tr | 2 | 68, 49 | 20, 18 | 0.61, 0.39 |
| 19. Tr | 1 | 50 | 21 | 0.27 |
| Mean ± SE | 66 ± 4 | 14 ± 2 | 0.15 ± .02 | |
| Normal PBMC (18 donors) | ||||
| CD19+ PBMC (55 slides) | <0.3 | |||
| Normal plasma cells (BM, 5 donors) | ||||
| CD38hiIg+ BMC (20 slides) | <0.3 | |||
Analysis was by in situ RT-PCR. Individual time points are listed sequentially in the table. The percent of PBMC and the absolute number in WBC was calculated from the percent of clonotypic B cells detected after sorting, using the percent of total PBMC that are B cells (determined by staining with anti-CD19 and FCA), and the number of mononuclear cells in blood. The mean ± SE values were calculated from the aggregate of individual time points for all patients using SigmaStat. Time points at which analysis was done are shown in Fig 6for patients 1-6, 11, 12, 15, and 18. Patient 7 was analyzed at 3 months after diagnosis, 8 was analyzed 5 years/5 months and 5 years/6 months after diagnosis while on treatment, 9 at diagnosis, 13 at diagnosis, 14 at 9 years/1 month and 9 years/3 months while on treatment, 16, 17, and 19 at diagnosis. For all samples, a minimum of 300 cells and frequently 500 to 1000 cells were viewed.
Abbreviations: Tr, treated; Off, off treatment; Unt, untreated; Allo, allogeneic; Auto, autologous.
The number of clonotypic B cells in blood is reduced after chemotherapy but most are drug-resistant survivors.
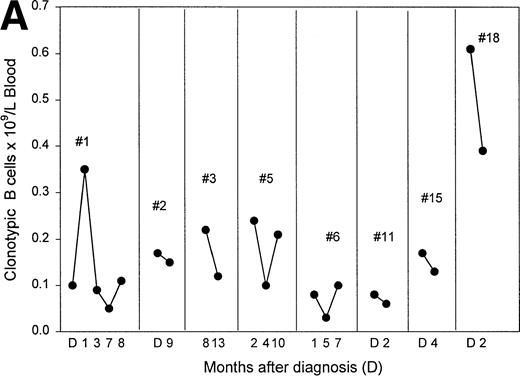
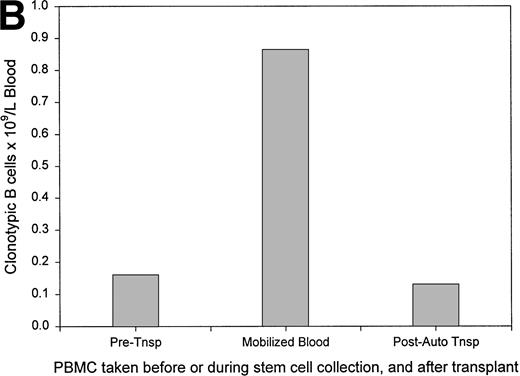
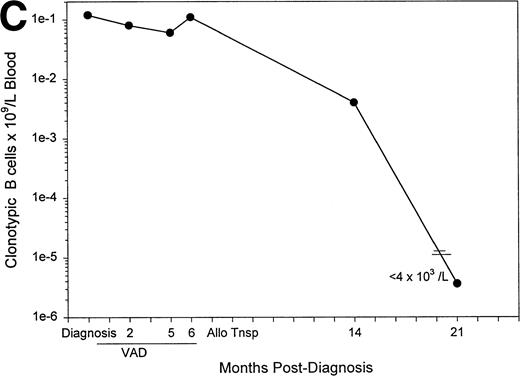
Figure 6A plots the absolute numbers of circulating clonotypic B cells in individual myeloma patients as a function of time and exposure to chemotherapy. Although the absolute numbers decreased slightly in most of the patients who had been evaluated at multiple time points, clonotypic cells persist in blood during and after therapy. For one patient (#12) analyzed before and after autologous stem-cell transplantation, a similar frequency of clonotypic B cells was detectable before and after the transplantation, and a high frequency was detected in the G-CSF–mobilized blood used for transplant (Fig 6B). For this patient, circulating clonotypic cells appear to be mobilized by G-CSF and escape cytoreduction. A sequential time course was performed for a second patient who subsequently underwent allogeneic stem-cell transplantation. Figure 6C shows that for patient 4, whose clonotypic B cells were only mildly depleted by vincristine/adriamycin/dexamethasone (VAD) therapy, the number of circulating clonotypic B cells was reduced 28-fold at 7 months, and became undetectable at 14 months after transplant.
The frequency of clonotypic B cells is slightly reduced by conventional chemotherapy in most patients, and is maintained after autologous transplantation (auto-tnsp), but clonotypic B cells are depleted after cytoreduction therapy and allogeneic transplantion. (A) The number of circulating clonotypic B cells was monitored at the time points after diagnosis as indicated. Data points are from Table 3. Patient ID numbers are given in the figure. For patients 1, 2, 7, 15, and 18 chemotherapy was initiated immediately after diagnosis and continued at monthly intervals until month 7. With the exception of the 10- and 13-month time points, patients 3, 5, and 6 were on chemotherapy for the time points recorded here. Patient 11 remained untreated for both samples shown here. (B) This patient was analyzed for clonotypic B cells a few days before stem-cell mobilization with G-CSF (mIg = 16 gm/L and 11% plasma cells in BM, a 75% decrement in response to treatment). The frequency of clonotypic B cells was analyzed in the G-CSF–mobilized blood cells, and in a blood sample taken at 1 month after cytoreduction and autologous transplantation, at which time the patient had a further 20% reduction in mIg for a total decrement of 95%. This patient has since remained in partial remission. (C) This patient was treated with VAD for 6 cycles followed by high-dose cytoxan and radiation, followed by allogeneic hematopoietic transplantation (Allo Tsnp) and immunosuppression with cyclosporin A. This patient remains in complete remission.
The frequency of clonotypic B cells is slightly reduced by conventional chemotherapy in most patients, and is maintained after autologous transplantation (auto-tnsp), but clonotypic B cells are depleted after cytoreduction therapy and allogeneic transplantion. (A) The number of circulating clonotypic B cells was monitored at the time points after diagnosis as indicated. Data points are from Table 3. Patient ID numbers are given in the figure. For patients 1, 2, 7, 15, and 18 chemotherapy was initiated immediately after diagnosis and continued at monthly intervals until month 7. With the exception of the 10- and 13-month time points, patients 3, 5, and 6 were on chemotherapy for the time points recorded here. Patient 11 remained untreated for both samples shown here. (B) This patient was analyzed for clonotypic B cells a few days before stem-cell mobilization with G-CSF (mIg = 16 gm/L and 11% plasma cells in BM, a 75% decrement in response to treatment). The frequency of clonotypic B cells was analyzed in the G-CSF–mobilized blood cells, and in a blood sample taken at 1 month after cytoreduction and autologous transplantation, at which time the patient had a further 20% reduction in mIg for a total decrement of 95%. This patient has since remained in partial remission. (C) This patient was treated with VAD for 6 cycles followed by high-dose cytoxan and radiation, followed by allogeneic hematopoietic transplantation (Allo Tsnp) and immunosuppression with cyclosporin A. This patient remains in complete remission.
DISCUSSION
The work presented here shows that 66% ± 4% of total circulating blood B cells in 18 myeloma patients, or on average 14% of PBMC, have an IgH rearrangement identical to that of autologous BM plasma cells. The absolute number ranges from 0.01 to 0.61 × 109clonotypic B cells/L of blood (mean, 0.15 ± .02 × 109/L). Using single-cell patient-specific RT-PCR of freshly isolated myeloma PBMC, the amplified CDR3 transcripts were sequenced and found to be identical for 18 individual B cells, confirming the high frequency of clonotypic B cells, the specificity of the patient-specific RT-PCR, and indicating that circulating myeloma B cells exhibit intraclonal homogeneity. This implies that expansion of B cells within the myeloma clone is independent of antigen-mediated selective pressure, a characteristic of malignant lymphocytes. Confirming the identity of clontypic PBMC, clonal IgH transcripts were identified among 3 of 8 and 2 of 8 sorted B cells, from 2 different patients, in single-cell RT-PCR. The clonotypic cells in the blood express CD19, as well as CD34 and CD11b as shown previously.24,25 The clonotypic PBMC detected cannot be accounted for by circulating plasma cells, which are infrequent38,39 and lack CD3.24,43,44 Thus, based on their high frequency in blood of myeloma patients, their phenotypic profile,4,24 and their morphology,27,29 these clonotypic cells are B cells. Consistent with our identification of the clonotypic B cell set as adherent cells,6 Berenson et al14 have shown by Southern blotting that rearranged myeloma IgH in blood derives from adherent cells. To accurately quantify the number of clonotypic B cells, patient-specific in situ RT-PCR was performed on sorted B cells. The number of clonotypic B cells as measured by in situ RT-PCR correlated with the number of clonotypic PBMC as measured by an RT-PCR limiting dilution analysis of PBMC having patient-specific IgH transcripts.
Only an IgH CDR3 sequence that identifies most BM plasma cells would be expected to have a high frequency in the blood. Thus, it was essential to confirm that the IgH sequence identified as clonotypic was expressed by the majority of individual BM plasma cells in a patient. Most studies do not control for the possible presence of frequent but nonmalignant plasma cell clones in the BM. The number of residual polyclonal B cells in myeloma patients is low,45,46 with an abnormally restricted specificity repertoire,47-50 further supporting the idea that expanded but nonmalignant B-cell clones, unrelated to the myeloma clone, might be detected among BMC of some myeloma patients. To address this problem, clonotypic transcripts were measured using single-cell and in situ RT-PCR. For all patients, greater than 80% of BM plasma cells expressed the sequence identified as clonotypic. Although fully expected, this work represents the first formal proof that nearly all individual plasma cells from myeloma BM express the same IgH VDJ sequence.
Based on previous work analyzing nearly 500 patients, the absolute numbers of circulating B cells represent about 3% to 10% of total white blood cells, or about 0.4 × 109 B cells/L of blood.4 On average for the smaller patient cohort analyzed here, the absolute number of clonotypic B cells was 0.15 × 109/L. Based on mean values from this previous group,4 clonotypic B cells are expected to represent about 38% of the total circulating B cells in blood. This is within the actual range (mean, 66%) reported here for the 18 individual myeloma patients for whom clonotypic sequences are available. Our evidence shows that for myeloma patients assayed at more than one time point, the absolute number of circulating clonotypic B cells decreased only partially after chemotherapy. For one patient who had a 75% decrement in mIg at the time of transplant, a high frequency of clonotypic B cells was found in the G-CSF–mobilized blood used for autologous transplant. At 1 month after transplant the patient had a 95% total decrement in mIg, although clonotypic B cells persisted in blood. For a second patient, clonotypic B cells became undetectable after allogeneic transplantation, when the patient was clinically in complete remission.
The high proportion of monoclonal B cells in myeloma patients contrasts with the extensive diversity of the B-cell population from normal donors. Little or no overlap was detected among randomly picked VDJ rearrangements from normal PBMC51 or from IgH sequences of 70 individual B cells from a normal donor.41 Thus at a molecular level, this work confirms the restricted diversity/specificity repertoire among the B cells from each myeloma patient.24,27,49,52 The lack of diversity is also indicated by the monotypic expression of light-chain Ig4 and of light chain mRNA.53 In situ RT-PCR confirms that circulating B cells have monotypic expression of light-chain mRNA (data not shown). The relative absence of polyclonal B cells in many of the myeloma patients analyzed here confirms our previous reports that the number of normal polyclonal B cells is reduced in myeloma patients.45 46
The number of clonotypic B cells detected here range from 0.9% to 50% of total PBMC, within the range of values obtained by Billadeau et al,16,17 although we found much higher mean values. Billadeau et al16,17 report 7 of 39 patients who had 1% or more clonal PBMC. These values did not correlate with the number of circulating plasma cells measured at the same time points in the patients.17 This implies that the clonal cells detected by Billadeau et al16,17 were B cells. The differences between the analysis done here and the Billadeau study may reflect their use of cryopreserved cells. We find that clonotypic B and plasma cells are often lost on thawing of cryopreserved PBMC or BMC (unpublished data). Brown et al54 have also found frequent clonal cells, with 1% to 4% clonal PBMC in two myeloma patients. However, our results are in contrast to those reported by Chen and Epstein23 who detected infrequent clonotypic cells and low numbers of total circulating B cells as compared to ourselves and others,4 55 suggesting that their sort strategy may have inadvertently selected a polyclonal subset of B cells.
Expression of clonotypic sequences by B cells in myeloma indicates a direct relationship with the malignant clone of plasma cells, but not necessarily the same malignant status.5 The extent to which these clonotypic B cells have undergone the progressive steps required for full fledged malignancy is unknown. Broadly speaking, there are three possible stages en route to malignancy and myeloma in which clonotypic B cells might be expected. Firstly, B cells having the clonotypic IgH rearrangement may have progressed through all stages required for malignant growth. Secondly, they may have undergone only some of the steps necessary for full malignancy. Third, they may be nonmalignant remnants of the original B cell clone that gave rise to the myeloma. This third possibility assumes that the monoclonal gammopathy of uncertain significance from which myeloma may originate, persists after evolution of frank myeloma. The population of clonotypic B cells described here is heterogeneous by many measures and may include representatives of all these stages, as a mixture of coexisting malignant, partially malignant, and nonmalignant clonal relatives of the BM plasma cells. However, a variety of evidence including oncogene expression,29,56 DNA aneuploidy,27,28functional multidrug resistance,25,28,32 stem-cell–like characteristic5,24 27 and the clonal homogeneity shown here, support options one or two, suggesting that the majority of these B cells are malignant and have escaped the need for antigen-driven clonal expansion.
In summary, this work was designed to resolve the controversy surrounding the frequency of clonotypic cells in blood. We find that substantial numbers of clonotypic B cells (0.01 to 0.61 × 109/L of blood) colonize the blood of myeloma patients. Clonotypic B cells are found in indolent myeloma, during stable phases of disease, and after autologous stem-cell transplantation in one patient. For a second transplant patient, clonotypic B cells became undetectable after allogeneic transplantation. The frequent relapse rate for myeloma indicates that treatment does not eliminate the generative compartment of myeloma and that the malignant clone may include less differentiated B lineage cells that are drug resistant. Circulating clonotypic B cells, as measured using in situ RT-PCR with patient-specific primers, provide a marker of blood involvement in myeloma that complements measures of plasma cell kill. It seems likely that at least some of these clonotypic B cells are malignant.5 If so, treatments to eradicate or reduce their numbers might confer enhanced survival. The relatively large numbers of circulating clonotypic B cells, shown here and elsewhere24 25 to survive chemotherapy, suggest that targeting clonotypic B cells should be a therapeutic priority.
ACKNOWLEDGMENT
Dr Tony Fields, Director of the Cross Cancer Institute, has been and continues to be a supportive facilitator for this work. The Edmonton Blood Transfusion Service provided blood samples from healthy normal individuals. The Department of Surgery and the Hematology laboratory of the University of Alberta Hospitals provided normal bone marrow. We thank the myeloma patients at the Cross Cancer Institute for their generous donations of tissues and their willingness to participate in this study.
Funded by a grant from the National Cancer Institute of Canada with funds from the Canadian Cancer Society, and by the Alberta Cancer Board Research Initiatives Program. A.J.S. was supported by a 75th Anniversary Award from the Faculty of Medicine and currently holds an AHFMR studentship award. The Faculty of Medicine Flow Cytometry Facility was supported by grants from the Medical Research Council of Canada and the Alberta Cancer Board Research Initiatives Program.
Address correspondence to Linda M. Pilarski, Department of Oncology, University of Alberta, Edmonton, Alberta, T6G1Z2, Canada; e-mail:lpilarsk@gpu.srv.ualberta.ca.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


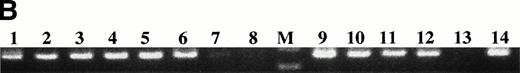
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal