Abstract
There is a wide normal range of coagulation factor XIII activity that has never been adequately explained. A polymorphism substituting leucine for valine at position 34 in the activation peptide of the A subunit of factor XIII has recently been discovered in nondeficient individuals, and the present studies indicate that the leucine substitution results in a significant increase in transglutaminase activity. The frequency of the Leu34 allele in the Australian Caucasian population is 0.27, which is high enough to suggest that the inheritance of either the Val34 or Leu34 alleles may contribute to the wide normal range of activity. Although there has been structural evidence indicating that the activation peptide does not dissociate from the enzyme after thrombin cleavage, the discovery of elevated activity resulting from the Leu34 substitution is the first direct evidence that the activation peptide plays a continuing role in the function of factor XIII.
© 1998 by The American Society of Hematology.
COAGULATION FACTOR XIII has a transglutaminase activity that forms γ glutamyl ε lysine isopeptide bonds between fibrin monomers.1 This cross-linking confers added strength to the clot and increases its resistance to degradation by plasmin. Inherited deficiencies of factor XIII lead to delayed bleeding after trauma and poor wound healing. In the absence of replacement therapy, intracranial bleeding can result in severe neurological impairment or death.2
Factor XIII in plasma is a heterotetramer composed of two A subunits arranged as a dimer in association with two B subunits.2,3The A subunits are responsible for the transglutaminase activity after the thrombolytic cleavage of an N-terminal activation peptide.4 The B subunit is a glycoprotein and has no known enzymatic activity. It is thought that the B subunit forms a complex with the A subunit dimer and protects it from elimination from the circulation.5-8 This view is supported by the observation that genetic deficiency of B subunits leads to a secondary deficiency of A subunits in the plasma.8
Previous studies have shown that the A subunit is genetically heterogeneous and a number of polymorphisms have been identified in the protein sequence.9-12 Several studies have shown that there is a large range of plasma A subunit transglutaminase activity in the normal population, and it is possible that the different levels of activity are related to the inheritance of different allelic variants.13-17 Little is known about the effects of most polymorphisms on the enzymatic activity of the A subunit. To our knowledge, this has only been investigated in one study and there was no apparent difference in the transglutaminase activity of FXIIIA*1 and FXIIIA*2, two previously studied electrophoretically detectable variants.18
The factor XIII A subunit is a zymogen that is activated by the cleavage of a 37 residue N-terminal peptide by thrombin. In this study, we have shown that a polymorphic Val34Leu substitution in the activation peptide has a significant effect on activity, demonstrating that the activation peptide contributes to catalytic activity after its cleavage. Furthermore, this polymorphism contributes to the wide range of activity found in the normal population. This variation in activity may also contribute to an explanation of the recently reported association between the Val34Leu polymorphism and myocardial infarction.19
MATERIALS AND METHODS
DNA and plasma samples.
The blood samples were collected from Australian Caucasian blood donors (N = 150) through the Canberra Red Cross Blood Transfusion Service (Canberrra, Australia). The donors were composed of 53% males and 47% females, with an average age of 39 years (range, 16 to 69 years). Plasma was collected after centrifugation of a whole blood sample at 3,000 rpm for 5 minutes and kept at −20°C in small aliquots until used. DNA was isolated from the buffy coat by phenol/chloroform extraction.
DNA amplification and genotyping.
The Val34Leu polymorphism was detected by a previously undescribed method that used a modified primer to introduce a restriction site into one allele during amplification of the DNA. The polymerase chain reaction (PCR) was used to amplify a 192-bp fragment from the factor XIII A subunit gene extending from intron A to exon II and encompassing codon 34. A pair of specific primers (forward primer, 5′ CATGCCTTTTCTGTTGTCTTC 3′; reverse primer, 5′TACCTTGCAGGTTGACGCCCCGGGGCACTA 3′) were used in a 100 μL reaction mix containing 100 to 200 ng genomic DNA, 50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 9.0, 0.1% Triton X-100, 1.5 mmol/L MgCl2, 50 μmol/L dNTPs, 20 pmol of each primer, and 2 U Taq DNA polymerase. Amplification was performed for 1 cycle at 94°C for 3 minutes, followed by 35 cycles of denaturation at 94°C for 1 minute, annealing for 1 minute at 56°C, and extension at 72°C for 1 minute. A final extension at 72°C was performed for 5 minutes. In the case of the reverse primer, a modification (C to T, underlined) was introduced to add a Dde I restriction enzyme site in the Leu34 variant amplified DNA. This Dde I cleavage site is created by an alteration of the Leu34 sequence (CTTGG) to the modified sequence (CTTAG) and allows the PCR-restriction fragment length polymorphism (RFLP) genotyping of a G to T transversion (shown in italics) that causes the Val34Leu polymorphism. Modification of the Val34 sequence (CGTGG to CGTAG) does not create the site for Dde I. The PCR product was digested with Dde I and the fragments were separated by 8% polyacrylamide gel electrophoresis and stained with ethidium bromide. To confirm the accuracy of this method, exon II was amplified and sequenced from several samples shown to be either homozygous for Val34, heterozygous, or homozygous for Leu34. No discordant samples were identified.
Plasma factor XIII activity assay.
Plasma samples were assayed for their factor XIII activity by the method of Dvilansky et al.13 In brief, 100 μL of plasma sample was incubated at 56°C for 3 minutes to remove fibrinogen and centrifuged at 10,000 rpm for 5 minutes. Twenty-five microliters of the defibrinated plasma was then added to an assay reaction containing 10 mmol/L CaCl2, 20 mmol/L dithiothreitol, 5 U/mL human thrombin, 3 mg/mL α casein, and 1 mmol/L putrescine (containing 0.625 μCi [1,4-14C] putrescine dihydrochloride; Amersham, Arlington Heights, IL) in 0.3 mol/L Tris, pH 7.5. Transglutaminase activity was detected between 60 and 90 minutes after the addition of the defibrinated plasma by spotting an aliquot of the reaction mixture onto a 3MM filter paper disc followed by TCA precipitation.20 Transglutaminase activity was expressed as nanomoles of putrescine incorporated into α casein by 1 mg of total plasma protein in 1 hour. The activity of the purified enzyme was determined in a similar manner, with the omission of the heat denaturation. Protein concentration was determined using bovine serum albumin as a standard.21
Expression and purification of recombinant factor XIII A subunit.
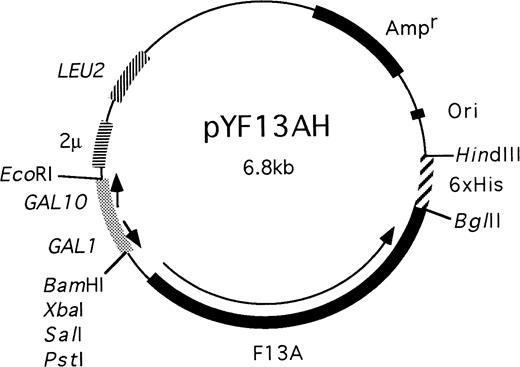
To express and purify recombinant factor XIII A subunit, we generated a yeast plasmid pYF13AH that expressed A subunit with 6 histidine residues at the C-terminal (Fig 1). In brief, the plasmid included a 2.3-kb Pst I cDNA fragment encoding the factor XIII A subunit from pKKF13A. The C-terminal 6 × His sequence was originally derived from pQE-70 (QIAGEN, Clifton Hill, Victoria, Australia). The expression was regulated by a Gal I promoter. The expression plasmid pYF13AH was transfected into the Saccharomyces cerevisiae strain AH22 and recombinants were selected on synthetic media without leucine.
A diagram showing the structure of the plasmid used to express recombinant factor XIII with an extension of 6 histidine residues at the C-terminal.
A diagram showing the structure of the plasmid used to express recombinant factor XIII with an extension of 6 histidine residues at the C-terminal.
For factor XIII expression, cultures were grown in YPD22broth overnight and then transferred to SD/galactose-Leu22broth for 24 hours at 30°C. The yeast pellet from 1 L of culture was washed twice with sterile water and then resuspended in 50 mL yeast lysis buffer (50 mmol/L HEPES, pH 7.5, 150 mmol/L NaCl, 5 mmol/L EDTA, 1% Triton X-100, 1 mmol/L phenylmethylsulphonyl fluoride) and lysed by passage through a Ribi cell disrupter (Sorval). The supernatant (≈50 mL) was collected by centrifugation at 10,000g for 10 minutes at 4°C and was mixed with 4 mL of a 50% suspension of nickel nitrilotriacetic acid agarose beads (Ni-NTA agarose; QIAGEN) at 4°C for 2 to 3 hours. The beads were then washed three times in 4 vol of 50 mmol/L Na-phosphate buffer, pH 7.5, 300 mmol/L NaCl by centrifugation. The beads were subsequently packed in a small column and then eluted with 20 mL steps of 50, 100, and 500 mmol/L imidazole in the washing buffer. Fractions containing transglutaminase activity were checked for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie Blue staining and pooled.
Mutagenesis.
A 2.3-kb Xba I-HindIII fragment containing the A subunit cDNA was transferred from pYF13AH and cloned into M13 mp18. The single-stranded DNA derived from the M13 clone was used as a template to introduce a Leu codon at position 34 by the use of a Sculptor mutagenesis kit (Amersham) and the oligonucleotide primer F13V34L 5′-CCGGGGCACCAAGCCCTGAAG-3′. Mutant clones were identified by sequencing and the modified 2.3-kb Xba I-HindIII cDNA fragment was recloned into pYF13AH to become pYF13Leu34.
RESULTS
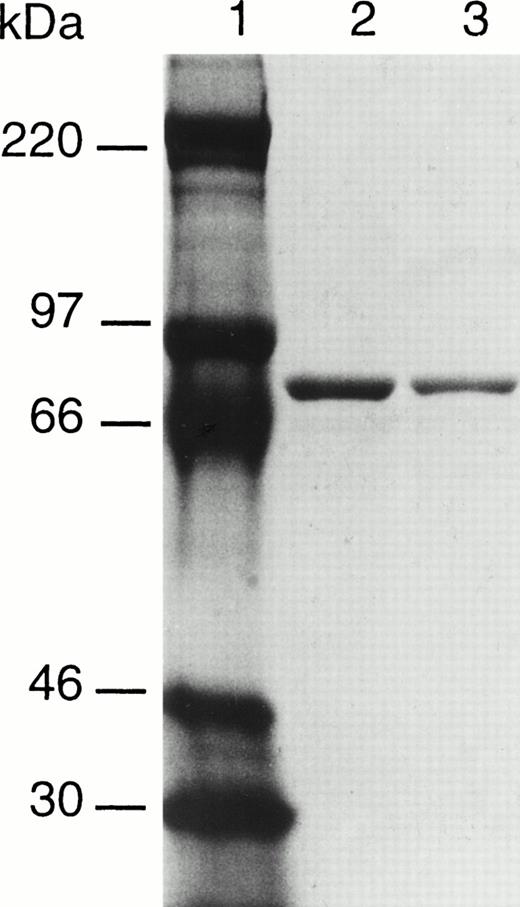
To determine if the Val34Leu polymorphism in factor XIII A subunit affected activity, recombinant factor XIII A subunits with either a valine or a leucine residue in position 34 have been expressed in yeast. The incorporation of an extension of 6 histidine residues at the C-terminal has permitted the purification of the two forms of factor XIII by affinity chromatography on Ni-NTA agarose. The resultant proteins appear to be highly purified and largely free of contaminating yeast proteins as judged by Coomassie Blue staining after SDS-PAGE (Fig 2). Both recombinant enzymes were found to have transglutaminase activity; however, the Leu variant had a significantly higher specific activity than the Val form (Table 1).
SDS-PAGE analysis of purified recombinant factor XIII A subunits. Lane 1, molecular weight makers; lane 2, factor XIII 34Val; lane 3, factor XIII 34Leu.
SDS-PAGE analysis of purified recombinant factor XIII A subunits. Lane 1, molecular weight makers; lane 2, factor XIII 34Val; lane 3, factor XIII 34Leu.
The Specific Activity of Purified Factor XIII Val34 and Factor XIII Leu34
| Purified Enzyme . | Specific Activity (nmol/h/mg) . |
|---|---|
| Factor XIII Val34 | 565 ± 27 |
| Factor XIII Leu34 | 949 ± 70 |
| Purified Enzyme . | Specific Activity (nmol/h/mg) . |
|---|---|
| Factor XIII Val34 | 565 ± 27 |
| Factor XIII Leu34 | 949 ± 70 |
Values are the mean ± SD of three experiments.
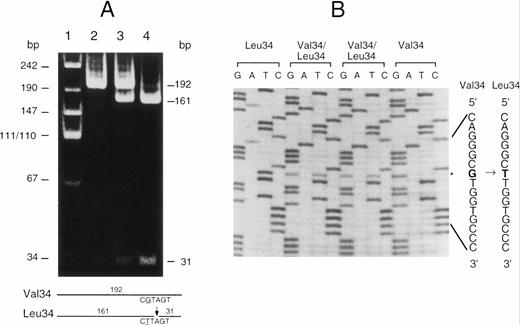
Although both forms of A subunit are found in clinically normal individuals, little is known about the frequency of this polymorphism. To resolve this question, a modified PCR-RFLP procedure was developed to enable the rapid genotyping of individuals. Because there were no restriction enzyme sites associated with this polymorphism, a primer was designed containing a G to A substitution that created aDde I site in the Leu allele but not in the Val allele. After amplification of exon II DNA with the modified primer, the genotype could be readily determined by Dde I digestion and acrylamide gel electrophoresis (Fig 3). The technique was checked by the random sequencing of the region from several individuals and the results were always in complete agreement. This technique was used to genotype a sample of 150 Caucasian blood donors (Table 2). Clearly the Val34 allele is more frequent than the Leu34 allele in this population. The distribution of genotypes is not significantly different from that expected in a Hardy-Weinberg equilibrium. The frequency of these two alleles in other populations has not been published; however, we have calculated these values for several other population samples from previously published haplotype data12 (Table 3). This analysis shows that the Val34 allele is the most frequent in all of the racial groups studied so far. The Leu allele is rare in Japanese, suggesting that the gene frequency may vary significantly between major racial groups.
(A) PCR-RFLP analysis of the Val34Leu polymorphism in exon II of the factor XIII A subunit gene. The fragment amplified from exon II was digested with Dde I. This fragment is 192 bp in length and does not normally contain a Dde I site in either allele. The Dde I site was selectively introduced into the Leu34 allele by the use of a modified primer (see Materials and Methods). Lane 1, molecular size markers; lane 2, DNA from a Val34 homozygote; lane 3, DNA from a Val34/Leu34 heterozygote; lane 4, DNA from a Leu34 homozygote. The expected sizes of the DNA fragments after digestion are shown on the right and schematically at the bottom of the figure. (B) Direct sequencing of amplified exon II from individuals with each genotype. The Val34 and Leu34 sequences are given on the right. The substituted nucleotide G to T causing the Val34Leu substitution is shown in bold type and the position of the base substitution is marked (*).
(A) PCR-RFLP analysis of the Val34Leu polymorphism in exon II of the factor XIII A subunit gene. The fragment amplified from exon II was digested with Dde I. This fragment is 192 bp in length and does not normally contain a Dde I site in either allele. The Dde I site was selectively introduced into the Leu34 allele by the use of a modified primer (see Materials and Methods). Lane 1, molecular size markers; lane 2, DNA from a Val34 homozygote; lane 3, DNA from a Val34/Leu34 heterozygote; lane 4, DNA from a Leu34 homozygote. The expected sizes of the DNA fragments after digestion are shown on the right and schematically at the bottom of the figure. (B) Direct sequencing of amplified exon II from individuals with each genotype. The Val34 and Leu34 sequences are given on the right. The substituted nucleotide G to T causing the Val34Leu substitution is shown in bold type and the position of the base substitution is marked (*).
Gene Frequency of the Factor XIII A Subunit Val34Leu Polymorphism in the Australian Caucasian Population (n = 150)
| . | Genotype . | Gene Frequency . | |||
|---|---|---|---|---|---|
| Val34/Val34 . | Val34/Leu34 . | Leu34/Leu34 . | Val34 . | Leu34 . | |
| Observed | 77 | 64 | 9 | 0.73 | 0.27 |
| Expected | 79.84 | 59.33 | 10.84 | ||
| . | Genotype . | Gene Frequency . | |||
|---|---|---|---|---|---|
| Val34/Val34 . | Val34/Leu34 . | Leu34/Leu34 . | Val34 . | Leu34 . | |
| Observed | 77 | 64 | 9 | 0.73 | 0.27 |
| Expected | 79.84 | 59.33 | 10.84 | ||
χ2 = .78; not significant.
The Gene Frequency of the Val34Leu Polymorphism in Several Populations
| Population . | N* . | Gene Frequency . | |
|---|---|---|---|
| Val34 . | Leu34 . | ||
| Caucasian | |||
| Australian | 300 | 0.73 | 0.27 |
| Finnish† | 58 | 0.86 | 0.14 |
| Russian† | 42 | 0.76 | 0.24 |
| German† | 28 | 0.89 | 0.11 |
| Japanese† | 92 | 0.99 | 0.01 |
| Population . | N* . | Gene Frequency . | |
|---|---|---|---|
| Val34 . | Leu34 . | ||
| Caucasian | |||
| Australian | 300 | 0.73 | 0.27 |
| Finnish† | 58 | 0.86 | 0.14 |
| Russian† | 42 | 0.76 | 0.24 |
| German† | 28 | 0.89 | 0.11 |
| Japanese† | 92 | 0.99 | 0.01 |
*Number of chromosomes tested.
Calculated from previously reported haplotype data.12
It was of interest to determine if the difference in activity of the recombinant proteins was also found in human plasma factor XIII. Comparison of the factor XIII transglutaminase activity in the plasma from individuals with the different genotypes indicated that those that are homozygous for the Leu34 allele have significantly higher activity (P = .016) than those that are homozygous for the Val34 allele, and heterozygotes have an intermediate value (Table 4). This difference in activity is in agreement with the difference found between the purified recombinant Leu34 and Val34 enzymes.
Plasma Factor XIII Activity in Individuals With Different Val34Leu Genotypes
| Genotype . | N . | Enzyme Activity (nmol/h/mg total protein) . |
|---|---|---|
| Val34/Val34 | 10 | 0.92 ± 0.269 |
| Val34/Leu34 | 10 | 1.04 ± 0.231 |
| Leu34/Leu34 | 9 | 1.34 ± 0.405 |
| Genotype . | N . | Enzyme Activity (nmol/h/mg total protein) . |
|---|---|---|
| Val34/Val34 | 10 | 0.92 ± 0.269 |
| Val34/Leu34 | 10 | 1.04 ± 0.231 |
| Leu34/Leu34 | 9 | 1.34 ± 0.405 |
Enzyme activities are expressed as the mean ± SD.
DISCUSSION
Although a number of previous studies have noted a wide range of factor XIII activity in the normal population,13-17 there have been few attempts to explain this phenomena. Because of the wide normal range, it has not been possible to use activity measurements to reliably identify heterozygotes in families in which factor XIII deficiency is segregating. This has presented problems for genetic counselling in affected families. Several genetic polymorphisms in the A subunit of factor XIII have been described, and it is possible that some amino acid substitutions may influence activity.9-12In a previous study, the electrophoretically detectable variants FXIIIA*1 and FXIIIA*2 were purified and characterized.18However, there was no difference in the specific activity of these two isoenzymes. To further investigate the origin of the wide range of normal activity, we have studied the influence of the Val34Leu substitution.11 The specific activity of the purified recombinant Leu34 enzyme was notably higher than that of the Val34 form. To confirm this difference, the investigation was extended to the evaluation of factor XIII activity in plasma from normal blood donors with different genotypes. The blood donors were genotyped by a modified PCR-RFLP method that generated a Dde I site in the Leu34 allele. The activity of Leu34 homozygotes was significantly higher than the activity of the Val34 homozygotes, confirming the difference found between the purified recombinant proteins. Because the site of this polymorphism in the activation peptide is only 4 residues from the thrombin cleavage site, it is possible that the particular residue at this position may influence the rate of activation and this could account for the differences in activity. However, in a previously published study13 and our own unpublished work, it has been shown that, under the conditions used, plasma A subunit is fully activated after 30 minutes. In the present study, the activity was determined between 60 and 90 minutes after thrombin activation. This delay therefore provided adequate time for complete activation.
The frequency of the two alleles in the Australian population is sufficiently high that the difference in activity between their products may contribute substantially to the wide range of factor XIII activity observed in the normal population. However, because a number of polymorphisms have been detected in the A subunit gene, including a tetranucleotide short tandem repeat in the 5′ flanking region,23 the possibility that other polymorphisms in the A subunit gene may also contribute to the wide normal range cannot be ruled out.
Because a polymorphism in the prothrombin gene was found to be associated with elevated plasma prothrombin levels and an increase in venous thrombosis,24 it is possible that the Val34Leu polymorphism in the factor XIII A subunit gene may be another risk factor for venous thrombosis and cardiovascular disease. The recent study of Kohler et al19 suggests that the Leu34 allele protects against myocardial infarction. The mechanism of this protection is not immediately obvious, because the higher level of activity of the Leu34 enzyme might be expected to create a greater risk by increasing resistance of fibrin clots to plasmin degradation. Thus, further study in patients with thrombosis and other forms of cardiovascular disease is required.
It was originally considered that, after thrombin cleavage, the activation peptide played no further part in the activity of the A subunit. However, there are now several lines of evidence to suggest that the activation peptide of factor XIII A might play a continuing role in enzymatic catalysis. First, there is crystallographic evidence that, following thrombin cleavage after Arg37, the activation peptide fragment still remains associated with the A subunit molecule.25 Second, in the A subunit dimer, Arg11 in the activation peptide of one subunit forms a salt-bridge with Asp343 of the other subunit; this interaction may contribute to the proposed role of Asp343 in guiding the lysine substrate to the active site.26 The third line of evidence is the effect described in this study of the amino acid substitution Val34Leu on the transglutaminase activity of the A subunit. This is the first direct evidence indicating the involvement of the activation peptide in the transglutaminase reaction of factor XIII.
The thrombin cleavage site and the Val/Leu34 residue are positioned in a loop that is not clearly defined in the available crystal structures. Thus, it is difficult to even speculate on the mechanism by which this substitution influences transglutaminase activity. However, it is clear that, in the available crystal structures, the active site is buried and inaccessible.25-27 It therefore seems likely that there is a structural change to accommodate the entry of fibrin into the active site and this could involve the activation peptide. The resolution of this question may depend on the solution of the crystal structure with fibrin in the active site.
ACKNOWLEDGMENT
The authors are grateful to the Canberra Red Cross Blood Transfusion Service for providing the blood samples used in this study and to Dr G. Chelvanayagam for advice on the structural implications of this polymorphism.
Address reprint requests to P.G. Board, PhD, Molecular Genetics Group, John Curtin School of Medical Research, GPO Box 334, Canberra ACT 2601, Australia.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal