To the Editor:
The Rh system is genetically controlled by two different but highly homologous genes on chromosome 1p34-36. The RHCE gene encodes a different RhCcEe polypeptide and the RHD gene encodes the D polypeptide, and there are a large number of antigenic polymorphisms between these two peptides. Of these, the RhDelis characterized as RhD− by using a conventional serological test, but it does show absorption and elution of anti-D.1 The molecular basis of RhDel is not known.
The blood of 21 Del (21.6%) of 102 serological RHD− patients was obtained after an absorption and elution test. A modified polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method based on the polymorphisms between RHCE and RHD genes was used to analyze the Del gene structure,2 and the results showed that there was no difference between the RhD and RhDel gene except that at the BspHI site of exon 9, the Del gene lacked theBspHI site that was similar with the RHCE gene. Haplotyping by Sph I bands showed no gross difference between RHD and RHDel genes3 (data not shown).
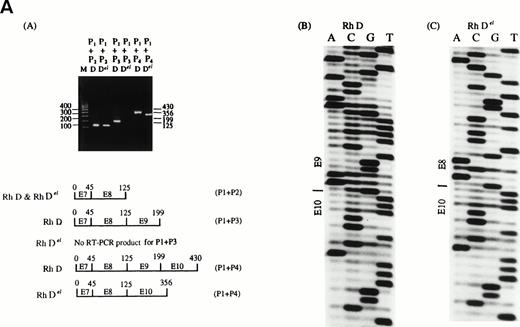
To further characterize the Del gene and its expression, a nested reverse tranascriptase-polymerase chain reaction (RT-PCR) method was used to amplify the different region between RHD and Del genes.4 The RNA was extracted from red blood cells and white blood cells, and the RT-PCR was performed as described.5 The first PCR amplified the RHD or Del gene from the exon 7 to exon 10 region (using upstream primer P0: 5′-TCCCCACAGCTCCATCATGGG-3′ and downstream primer P4: 5′-GTATTCTACAGTGCATAATAAATGGTG-3′, both are RHD gene-specific primers). The PCR products were subjected to nested-PCR: using an RHD specific upstream primer P1: 5′-CATTGTGCTGCTGGTGCTTG-3′, and downstream primer P2: 5′-CTGTCAGGAGACCAGACGTG-3′ to amplify part of exon 7 and exon 8; using P1 and downstream primer P3: 5′-CTTCCAGAAAACTTGGTCATC-3′ to amplify from exon 7 to exon 9; using primer P1 and P4 to amplify from exon 7 to exon 10. The results showed that the D and Del genes were similar at exon 7 and 8, but there was no nested RT-PCR product for Del gene for the primers P1 and P3, and the nested PT-PCR product of Del gene for the primers P1 and P4 was shorter than the product of normal D gene. Direct sequencing of the nested RT-PCR products of D and Del genes showed that there was an exon 9 deletion of Del gene (Fig1A).
(A) By using different pairs of RH D specific primers and nested RT-PCR to amplify the RH Del gene, there was a deletion of exon 9 of RH Del gene (a). The results were further confirmed by direct sequencing of the nested RT-PCR product (b) and (c). The sequences of primers P1, P2, P3 and P4 are shown in the text. (B) The breakpoint region sequence of Del in comparison with D, Ce, and ce alleles.
(A) By using different pairs of RH D specific primers and nested RT-PCR to amplify the RH Del gene, there was a deletion of exon 9 of RH Del gene (a). The results were further confirmed by direct sequencing of the nested RT-PCR product (b) and (c). The sequences of primers P1, P2, P3 and P4 are shown in the text. (B) The breakpoint region sequence of Del in comparison with D, Ce, and ce alleles.
To characterize the breakpoint of Del gene, a nested PCR method was used to amplify the breakpoint region. For the first PCR, an RHD gene-specific downstream primer (primer P4) and a nonspecific upstream primer (5′-GATTGGCTTCCAGGTCCTCC-3′) were used to amplify part of the RhD or RhDel gene from exon 8 to 3′ noncoding region using genomic DNA. For the second PCR, two nonspecific RH gene primers were used (upstream primer 5′-TCAGCATTGGGGAACTCAGC-3′, and downstream primer: 5′-GCCTTGTTTTTCTTGGATG-3′) to amplify from part of exon 8 to exon 10. The PCR products were subjected to direct sequencing or subcloning sequencing analysis. The results showed that the Del had a 1,013-bp deletion between introns 8 and 9, including whole exon 9 (Fig 1B).
The Del gene transcript maintains a normal open reading frame and thus should encode a protein with 463 amino acid residues with a new C-terminal extension from codon 384 as compared with the normal D protein of 417 amino acid residues (Fig2). Although the Del show some D activity after an absorption and elution test, a case of Rh− with Del activity patient was transfused with RHD+ blood did develop anti-D antibody by a traditional serological test several weeks after transfusion. From this point of view, RhDel should be recognized as a type of RhD−, and whether the Del blood transfused to RhD+ or other types of RhD−cases will develop anti-Del antibody needs further study.
Amino acid sequence of RH Del are shown. The amino acids of Rh Del are different from RH D gene after codon 384, and there are 46 amino acids more in Rh Del protein.
Amino acid sequence of RH Del are shown. The amino acids of Rh Del are different from RH D gene after codon 384, and there are 46 amino acids more in Rh Del protein.
ACKNOWLEDGMENT
We thank the Taichung Blood Donation Center and the Kaohsiung Blood Donation Center for kindly providing the blood samples. This work was supported in part by a grant from China Medical College Hospital (DMR-87-007). This work was first submitted to Blood as a regular paper on April 2, 1998, and subsequently condensed as a letter.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal