Abstract
Fanconi anemia (FA) is an autosomal recessive disease characterized by chromosomal instability, bone marrow failure, and a high risk of developing malignancies. Although the disorder is genetically heterogeneous, all FA cells are defined by their sensitivity to the apoptosis-inducing effect of cross-linking agents, such as mitomycin C (MMC). The cloned FA disease genes, FAC and FAA, encode proteins with no homology to each other or to any known protein. We generated a highly specific antibody against FAA and found the protein in both the cytoplasm and nucleus of mammalian cells. By subcellular fractionation, FAA is also associated with intracellular membranes. To identify the subcellular compartment that is relevant for FAA activity, we appended nuclear export and nuclear localization signals to the carboxy terminus of FAA and enriched its localization in either the cytoplasm or the nucleus. Nuclear localization of FAA was both necessary and sufficient to correct MMC sensitivity in FA-A cells. In addition, we found no evidence for an interaction between FAA and FAC either in vivo or in vitro. Together with a previous finding that FAC is active in the cytoplasm but not in the nucleus, our results indicate that FAA and FAC function in separate subcellular compartments. Thus, FAA and FAC, if functionally linked, are more likely to be in a linear pathway rather than form a macromolecular complex to protect against cross-linker cytotoxicity.
FANCONI ANEMIA (FA) is an autosomal recessive disease that frequently leads to bone marrow failure and myeloid leukemias at childhood.1-3 Cells from FA patients are characterized by hypersensitivity to cross-linking agents, such as mitomycin C (MMC) and diepoxybutane, and a propensity to apoptosis.4-7 By somatic cell fusion studies, eight different complementation groups of FA have been identified, named A to H,8-10 suggesting that the inactivation of at least eight distinct genes could give rise to the clinical and cellular phenotype recognized as FA. One of the central questions in FA research is to elucidate the relationship, if any, of these genes and their products to each other. To date two FA genes, FAC and FAA, have been cloned.11-13 The proteins encoded by these genes are unique and show no homology to each other.
There is conflicting evidence about the possible involvement of FA proteins in DNA repair.3 The FAC protein has been shown to localize predominantly to the cytoplasmic compartment14,15and function in a prerepair pathway.16 In this capacity, cytoplasmic localization of FAC is essential for cross-linker complementation of FA group C (FA-C) cells, while forced nuclear localization completely abolishes this activity.16 In FAA, the presence of a putative nuclear localization signal (NLS) in the amino-terminal region suggests a nuclear function for this protein.12,13 However, in preliminary studies, a chimeric molecule consisting of full-length FAA tagged with the green fluorescent protein (GFP) showed a predominantly cytoplasmic localization in human 293 cells, and inactivation of the NLS motif by site-directed mutagenesis did not abort the complementation function of FAA.17 Although this result suggested that FAA also has a cytoplasmic role, the inclusion of GFP and potential effects on the localization of the protein precluded any definitive conclusions about the localization and function of wild-type FAA.
The participation of two or more FA proteins in a single macromolecular complex that regulates genomic stability is a plausible mechanism that could account for the phenotypic similarity of different FA complementation groups. Previous studies using extracts derived from human lymphoblasts and the human megakaryocytic cell line Dami showed that FAC binds to at least three ubiquitous cytoplasmic proteins.18 However, the expression and size of these FAC-binding proteins in lymphoblasts from FA complementation group A (FA-A) were normal, and there was no interacting protein that, in retrospect, matched the molecular size of FAA. Another possibility is that FA proteins are linked in a linear pathway, and constituents of the pathway may be localized in distinct cellular compartments. The identification of a second FA protein now enables us to test these possibilities.
In this study, we report the generation of an antibody against FAA that is able to detect FAA by various biochemical assays. By immunofluorescence studies, FAA localizes to both the cytoplasmic and nuclear compartment of human and other mammalian cells and shows a clearly different intracellular distribution compared with FAC. We found no evidence for an interaction between the two FA proteins both in vitro and in vivo by coprecipitation assays that have been successfully applied to demonstrate other protein interactions with FAC.18 19 Finally, we provide strong evidence that FAA must reside in the nucleus to protect against cross-linker cytotoxicity. Our data support a model in which FAC and FAA function in different cellular compartments to defend the genome against cross-linking agents.
MATERIALS AND METHODS
Cell culture, transfection, and MMC sensitivity assay.
Lymphoblasts were maintained in RPMI 1640 medium containing 10% fetal calf serum (FCS). Stably transfected lymphoblasts were grown in the same medium supplemented with 200 μg/mL hygromycin. HeLa, COS-1, and 293 cells were grown in Dulbecco's Minimal Essential Medium (DMEM; GIBCO-BRL, Grand Island, NY) with 10% FCS. COS-1 cells were transfected using DEAE-dextran as described previously18and HeLa and 293 cells were transfected using Superfect (QIAGEN, Valencia, CA) according to the manufacturers' instructions. Stably transfected lymphoblasts were generated by electroporation as described previously.16 The MMC growth inhibition assay was performed by exposing parallel cultures (7.5 × 104 cells per mL) to various concentrations of MMC. Cell numbers were determined using a Coulter counter after the untreated control cells had undergone three or more cell divisions.
Constructs.
For the generation of an affinity-purified antibody against FAA, His- and GST-tagged FAA were made as follows: The amino terminal portion of FAA encompassing amino acids 2 to 321 was amplified from pREP4-FAA using the primers FA105 (5′-CGGGATCCGACTCGTGGGTCC-3′) and FA106 (5′-CGGAATTCGTC GACTGAAGAACCTCTTCA-3′). The resulting fragment of approximately 1 kb was digested with BamHI andSal I and subcloned into the corresponding sites of pQE30 (QIAGEN, Santa Clarita, CA) to generate an open reading frame (ORF) for an approximately 37-kD His-FAA fusion protein. The same polymerase chain reaction (PCR) fragment cut withBamHI and EcoRI and subcloned into pGEX2TK yielded pGST-FAA1 encoding an approximately 62-kD fusion protein. Sequences 3′ of the stop codon of FAA were deleted by PCR using the primers FA-3X (5′-TCAGTCTAGATTATTCAGAAGAGATGAGGCTCCTGGGACAGGT-3′) and FA-101 (5′-CTTAATTTTGACCTCTGCTCTGGTGTG-3′), and the resulting fragment was subcloned into pcDNA3 (Invitrogen, San Diego, CA) or pDR2 (CLONTECH, Palo Alto, CA) to generate pcDNAFAA and pDRFAA, respectively. A wild-type or a mutant nuclear export signal (NES) was appended to FAA in several stages. A 2.9-kb KpnI-HindIII fragment of FAA derived from the earlier described FAA-GFP construct17 was subcloned in pBluescript KS. After digestion with HindIII the annealed oligonucleotides NES1 (5′-AGCTTAATGAATTAGCCTTGAAATTAGCAGGTCTTGATATCAACGT-3′) and NES2 (5′-CTGACGTTGATATCAAGACCTGCTAATTTCAAGGCTAATTCATTA-3′) were inserted. After additional subcloning steps, the FAA ORF was reconstructed in pcDNA3 and pDR2 to yield pcDNAFAA-NES and pDRFAA-NES, respectively, which encode FAA fused to wild-type NES. In a similar way, pcDNA- and pDRFAA-P12 were constructed by using primers similar to the NES primers described above in which the underlined nucleotides in NES1 and NES2 were substituted for GC to generate FAA fused to mutant NES. Nuclear-targeted FAA expression plasmids were made following a similar strategy as described above for NES-fusion constructs using the oligonucleotides NLS1 (5′-AGCTTCCGAAGAAAAAGAGAAAGGTGGT-3′), NLS2 (5′-CTAGACC ACCTTTCTCTTTTTCTTCGGA-3′), mutNLS1 (5′-AGCTTCCGAAAGACAAAGAG AAAGGTGGT-3′), and mutNLS2 (5′-CTAGACCACCTTTCTCTTTGTCTTCGGA-3′). The pED6FACγ1 construct has been described.18
Generation of affinity-purified FAA antibody.
Rabbits were immunized against the His-FAA1 protein that was purified from Escherichia coli strain M15 using nickel (Ni)-agarose beads (QIAGEN) according to standard procedures. Affinity-purification of the FAA antibody was performed by passage over a column containing GST-FAA1 bound to glutathionine-Sepharose 4B beads (Pharmacia Biotech, Piscataway, NJ) as described.18 20
Immunofluorescence microscopy and subcellular fractionation.
For colocalization experiments, HeLa and 293 cells were transiently transfected with pcDNAFAA and pED6FACγ1, and replated on glass coverslips after 24 hours. After another 24 hours, cells were processed for immunofluorescence microscopy as described before.15Conjugated antibodies used were fluorescein-conjugated goat–anti-rabbit and rhodamine-conjugated goat–anti-human IgG (Cappel Laboratories, West Chester, PA). To visualize nuclei, cells were stained with bisbenzimide (Sigma Chemical Co, St Louis, MO). Subcellular fractionation steps were performed at 4°C, and all buffers were supplemented with the protease inhibitor phenylmethylsulphonyl-fluoride (PMSF; final concentration 0.5 mM). Transiently transfected or nontransfected cells cultured in 150-mm dishes were rinsed with phosphate-buffered saline and lysed in 1 mL lysis buffer, containing 20 mmol/L Tris-HCl, pH 8.0, 50 mmol/L NaCl, 1 mmol/L EDTA, and 0.5% Nonidet P-40 (NP-40). After 15 minutes, nuclei were pelleted by centrifugation for 5 minutes at 600g and the supernatant was taken as the crude cytoplasmic fraction. After centrifugation of this fraction for 45 minutes at 100,000g, the supernatant (cytosolic fraction) was removed and the pellet consisting of intracellular membranes was resuspended in 250 μL lysis buffer, briefly sonicated, and centrifuged to remove insoluble debris. Nuclei were washed twice in lysis buffer and resuspended in 500 μL lysis buffer supplemented with NaCl to a final concentration of 250 mmol/L, disrupted by shearing with a syringe using a 21-gauge needle, and briefly sonicated. Debris was pelleted and the supernatant was designated as the nuclear extract.
Western blotting, metabolic labeling, and immunoprecipitation.
Western blotting, 35S-methionine labeling of cells, and immunoprecipitation were performed as described earlier.15,18 Cells were lysed in low salt buffer (20 mmol/L Tris-HCl, pH 8.0, 50 mmol/L NaCl, 2 mmol/L EDTA, 0.1% NP-40), in medium salt buffer (20 mmol/L Tris-HCl, pH 8, 100 mmol/L NaCl, 2 mmol/L EDTA, 0.5% NP-40), or in RIPA buffer (50 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 1 % NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]), which were supplemented with the protease inhibitor PMSF and the phosphatase inhibitor sodium orthovanadate at final concentrations of 0.5 mmol/L and 1 mmol/L, respectively. For Western blotting, lysates in low salt buffer representing the equivalent of 2 × 105 cells per lane were subjected to electrophoresis on an 8% polyacrylamide gel (SDS-PAGE) and transferred to Polyscreen membrane (NEN Life Science Products, Boston, MA). Blots were incubated with affinity-purified FAA antibody, anti–β-tubulin antibody (Boehringer Mannheim Biochemicals, Indianapolis, IN) or anti-human topoisomerase II antibody (GenoSys, Houston, TX) in TBST (10 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 0.05% Tween 20) supplemented with 4% nonfat dry milk and 1% bovine serum albumin, followed by incubation in the same buffer with either peroxidase-conjugated goat–anti-rabbit IgG or goat–anti-mouse IgG (GIBCO-BRL) and developed by using enhanced chemiluminescence (Amersham Life Sciences, Arlington Heights, IL). For immunoprecipitation, 1.5 μg purified anti-FAA or anti-FAC antibody was added per sample of cell lysates in either medium salt buffer or RIPA buffer. After incubation for 1 hour at 4°C with rotation, protein A slurry (Bio-Rad Laboratories, Richmond, CA) or protein A/G agarose (Santa Cruz Biotechnology Inc, Santa Cruz, CA) was added, followed by another hour of incubation. Immunocomplexes were washed twice in low salt buffer and once with NET-gel buffer (50 mmol/L Tris-HCl, pH 7.5, 50 mmol/L NaCl, 0.1% NP-40, 0.2% gelatin) for 30 minutes at 4°C. Subsequently, samples were prepared for SDS-PAGE and Western blotting. For immunoprecipitation experiments using lysates derived from metabolically labeled cells, gels were treated with glacial acetic acid and 20% 2,5-diphenyloxazole (Sigma) followed by drying and autoradiography. For coprecipitation experiments, in vitro translation yielding 35S-methionine–labeled FAA and FAC was performed using the TNT T7 coupled reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer's instructions. GST-FAC bound beads used in pull-down experiments have been described.18 Potential complexes were allowed to form in low salt lysis buffer under the same conditions as described above.
RESULTS
Generation and characterization of anti-FAA antibody.
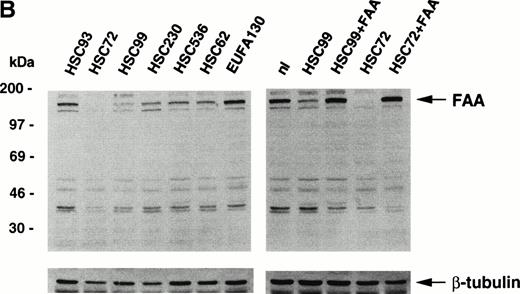
We have generated a rabbit polyclonal antibody directed against the amino-terminal portion of FAA expressed in bacteria. After affinity-purification, Western blotting experiments show that the antibody detects the purified antigen His-FAA1 specifically (Fig 1A). Western blots of extracts derived from lymphoblastoid cell lines show that the antibody can also detect the 163-kD endogenous FAA protein (Fig 1B). Slightly varying levels of FAA are found in cells derived from normal controls or from patients representing FA complementation groups B to E. By contrast, little if any FAA is detected in the FA-A cell line HSC72, which was used previously for expression cloning of the FAA cDNA and has been shown to lack the FAA mRNA.12After transduction of HSC72 cells with a retroviral vector encoding the FAA cDNA, FAA expression is restored (Fig 1B). In another FA-A cell line, HSC99, FAA expression is normal, suggesting that as yet unknown mutations in the two alleles of the FAA gene lead to the expression of defective proteins. After stable transfection with an episomal expression vector containing the FAA cDNA, higher levels of FAA are detected (Fig 1B). As expected, restoration of wild-type FAA expression in both cell lines corrects their sensitivity to MMC (data not shown). FAA can also be immunoprecipitated from extracts of metabolically labeled COS-1 cells that are transiently transfected with a vector encoding FAA cDNA (Fig 1C). These findings show that the anti-FAA antibody specifically detects FAA in different biochemical assays.
Generation of an affinity-purified rabbit polyclonal antibody against FAA that specifically detects FAA in Western blotting and immunoprecipitation experiments. (A) Western blot showing the approximately 37-kD His-FAA1 fusion protein in bacterial extracts and after purification using the affinity-purified antibody. The His-tagged fusion protein encompasses the first 321 amino-terminal residues of FAA and was used to raise the antibody. Lysates derived from noninduced bacteria or isopropyl-β-thiogalactopyranoside (IPTG)-induced bacteria are indicated with (−) or (+), respectively, and the purified protein with (P). (B) Western blot using extracts derived from normal cells or cells representing five different FA complementation groups. The FAA-specific band of approximately 163 kD is indicated. Blots were reprobed with an anti–β-tubulin antibody as a control for the amount of protein loaded per lane. Lymphoblastoid cell lines used represent the following FA subtypes: HSC93 and nl, normal cells; HSC72 and HSC99, FA-A; HSC230, FA-B; HSC536, FA-C; HSC62, FA-D; EUFA130, FA-E. HSC72+FAA indicates HSC72 cells stably transduced with a retroviral vector expressing FAA. HSC99+FAA indicate cells that were stably transfected with an episomal expression vector containing the FAA cDNA. (C) Immunoprecipitation of FAA from whole-cell extracts, generated in RIPA buffer, from metabolically labeled COS1 cells either mock-transfected (−) or transiently transfected with pcDNA-FAA5.5 (+).
Generation of an affinity-purified rabbit polyclonal antibody against FAA that specifically detects FAA in Western blotting and immunoprecipitation experiments. (A) Western blot showing the approximately 37-kD His-FAA1 fusion protein in bacterial extracts and after purification using the affinity-purified antibody. The His-tagged fusion protein encompasses the first 321 amino-terminal residues of FAA and was used to raise the antibody. Lysates derived from noninduced bacteria or isopropyl-β-thiogalactopyranoside (IPTG)-induced bacteria are indicated with (−) or (+), respectively, and the purified protein with (P). (B) Western blot using extracts derived from normal cells or cells representing five different FA complementation groups. The FAA-specific band of approximately 163 kD is indicated. Blots were reprobed with an anti–β-tubulin antibody as a control for the amount of protein loaded per lane. Lymphoblastoid cell lines used represent the following FA subtypes: HSC93 and nl, normal cells; HSC72 and HSC99, FA-A; HSC230, FA-B; HSC536, FA-C; HSC62, FA-D; EUFA130, FA-E. HSC72+FAA indicates HSC72 cells stably transduced with a retroviral vector expressing FAA. HSC99+FAA indicate cells that were stably transfected with an episomal expression vector containing the FAA cDNA. (C) Immunoprecipitation of FAA from whole-cell extracts, generated in RIPA buffer, from metabolically labeled COS1 cells either mock-transfected (−) or transiently transfected with pcDNA-FAA5.5 (+).
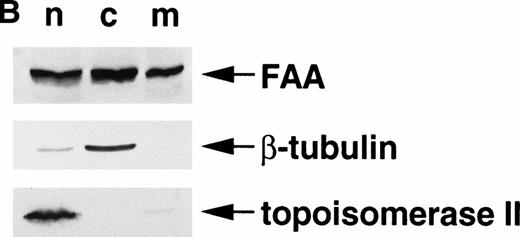
FAA is present in both the nuclear and the cytoplasmic compartment. (A) (see page 2232) Costaining of FAA and IgG heavy chain-tagged FAC in transiently transfected Hela cells by immunofluorescence. (B) Western blotting on subcellular fractions derived from 293 cells transiently expressing FAA. n, nuclear fraction; c, cytosollic fraction; m, membrane fraction.
FAA is present in both the nuclear and the cytoplasmic compartment. (A) (see page 2232) Costaining of FAA and IgG heavy chain-tagged FAC in transiently transfected Hela cells by immunofluorescence. (B) Western blotting on subcellular fractions derived from 293 cells transiently expressing FAA. n, nuclear fraction; c, cytosollic fraction; m, membrane fraction.
Subcellular localization of FAA.
Next, we examined the subcellular localization of FAA in human cells by immunofluorescence microscopy. Simultaneously, we also determined the localization of FAC using a functionally active chimera, FACγ1, derived by fusion of FAC to the IgG1 heavy chain.18 In HeLa cells transiently expressing FAA and FACγ1, the large majority of cells (approximately 90%) show FAA to be present in both the nuclear and cytoplasmic compartment in varying ratios. In about 5% of FAA-positive cells, FAA is detected predominantly in the nucleus and in 5% in the cytoplasm. Nuclear-localized FAA shows punctate forms in occassional cells. Examples of different FAA staining patterns are shown in Fig 2A. No staining is seen without permeabilization, and FAA localization is similar in the absence or presence of overexpressed FACγ1 (data not shown). Consistent with previous studies14,15 and similar to the expression of nontagged FAC, FACγ1 is predominantly localized to the cytoplasmic compartment. Western blotting experiments using subcellular fractions derived from 293 cells transiently expressing FAA show that an estimated 20% of FAA is associated with intracellular membranes (Fig2B), similar to that reported previously for FAC.15 Thus, the distribution of FAA is variable, which suggests that FAA shuttles between the cytoplasm and nucleus.
No evidence for an interaction between FAA and FAC.
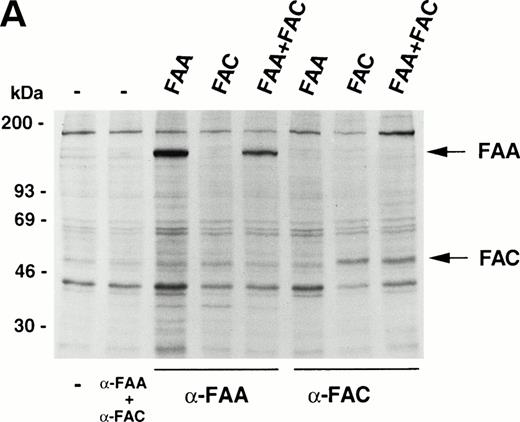
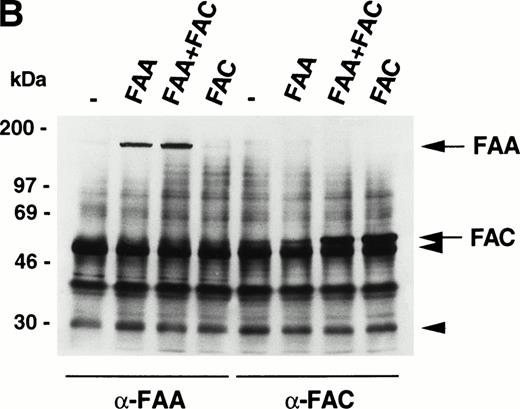
The similarity in the clinical and cellular phenotypes of FA belonging to different complementation groups has led to the suggestion that FA proteins may participate in protein-protein interactions with each other in macromolecular complexes.3,18 Here we have used several strategies to show possible physical interactions between FAA and FAC. First, we do not observe any interactions in coprecipitation experiments using extracts from metabolically labeled 293 cells transiently expressing FAA, FAC, or both (Fig 3A). It should be noted that, under similar conditions, FAC binds to the molecular chaperone GRP9419 and to NADPH cytochrome P450 reductase (Kruyt FAE, Hoshino T, Liu JM, Joseph P, Jaiswal AK, Youssoufian H, manuscript submitted). Second, no FAC-FAA complexes are observed in 293 cells (Fig 3B) or in HeLa and COS-1 cells (data not shown) by sequential immunoprecipitation and Western blotting. Third, mixing35S-labeled FAA and FAC followed by immunoprecipitation with anti-FAA or anti-FAC antibody in vitro (Fig 3C), or by pull-down of 35S-labeled FAA with immobilized GST-FAC (data not shown), reveal no interactions. These findings do not support the existence of a macromolecular complex in which FAA and FAC interact with each other, either directly or indirectly.
FAA and FAC do not interact. (A) Immunoprecipitation experiments using extracts derived from metabolically labeled 293 cells that were transiently transfected with the indicated vectors expressing FAA or FAC. Extracts were prepared in RIPA buffer. (B) Immunoprecipitation of FAA and FAC using extracts derived from 293 cells transiently expressing FAA, FAC, or FAA and FAC as indicated. Extracts were prepared in medium salt buffer. Arrowheads indicate the heavy and light chains of the antibodies. (C) Coprecipitation experiments with in vitro–translated35S-methionine–labeled FAA and FAC under low salt conditions. Total reticulocyte lysate was loaded in lanes marked FAA-lysate and FAC-lysate; control indicates total lysate without template DNA. Full-length FAA and FAC are indicated.
FAA and FAC do not interact. (A) Immunoprecipitation experiments using extracts derived from metabolically labeled 293 cells that were transiently transfected with the indicated vectors expressing FAA or FAC. Extracts were prepared in RIPA buffer. (B) Immunoprecipitation of FAA and FAC using extracts derived from 293 cells transiently expressing FAA, FAC, or FAA and FAC as indicated. Extracts were prepared in medium salt buffer. Arrowheads indicate the heavy and light chains of the antibodies. (C) Coprecipitation experiments with in vitro–translated35S-methionine–labeled FAA and FAC under low salt conditions. Total reticulocyte lysate was loaded in lanes marked FAA-lysate and FAC-lysate; control indicates total lysate without template DNA. Full-length FAA and FAC are indicated.
Nuclear localization of FAA is necessary and sufficient for complementing MMC sensitivity.
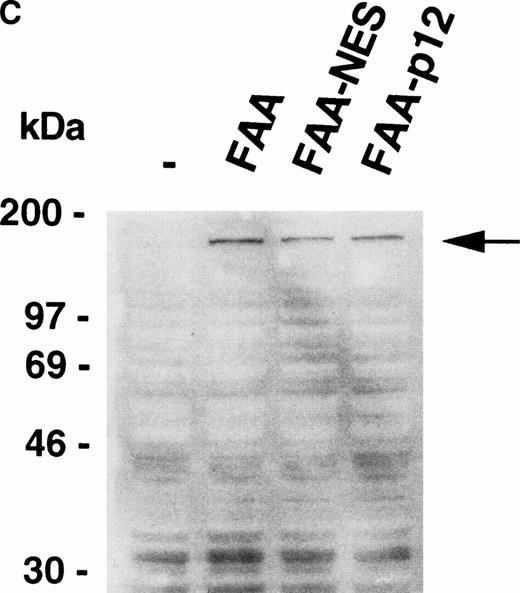
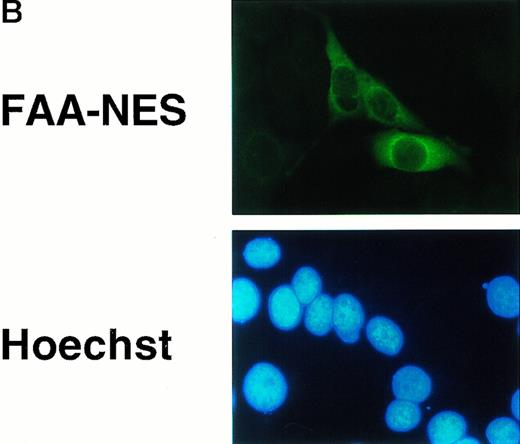
Because FAA is present in both the cytoplasmic and nuclear compartments, we asked whether a particular pool is necessary for protection against cross-linking agents. A previous attempt to address this question by forced nuclear targeting of an FAA-GFP chimera was inconclusive because of a lack of nuclear accumulation of the chimera.17 In a reciprocal approach, we set out to generate a FAA-NES fusion protein to promote the export of nuclear FAA to the cytoplasm. NES motifs have been shown to bind to Exportin1/CRM1 and form complexes with the GTP-bound form of Ran.21,22Subsequently, these complexes are transported through the nuclear pore and dissociate rapidly upon arrival in the cytoplasm, accompanied by the hydolysis of RanGTP to RanGDP.23 We fused the well-characterized NES found in the inhibitor of cAMP-dependent protein kinase24 to the carboxy terminus of FAA to generate FAA-NES. As a control, we introduced an amino acid substitution that renders the NES motif inactive24 to derive FAA-P12 (see Fig 4A). Transient expression of FAA-NES in HeLa cells and subsequent immunofluorescence microscopy shows an apparent exclusive cytoplasmic localization (Fig 4B, see page 2232), whereas FAA-P12 transfected cells show both nuclear and cytoplasmic expression patterns similar to that of nontagged FAA (not shown). In HSC72 cells stably expressing these constructs (Fig 4C), FAA-NES fails to complement, while FAA-P12 is fully capable in restoring MMC resistance (Fig 4D).
Depletion of FAA from the nucleus abolishes its ability to complement MMC hypersensitivity in FA-A lymphoblasts. (A) Schematic representation of FAA-NES and FAA-P12. The double-headed arrow indicates the amino acid substitution leading to inactivation of the NES. (B) (see page 2232) Immunofluorescence microscopy using pcDNAFAA-NES in transiently transfected Hela cells. (C) Western blot showing the stable expression of FAA, FAA-NES, and FAA-P12 in HSC72 cells. (D) MMC sensitivity assays of the HSC72-derived stable transfectants. (□), Control; (▪), FAA; (○), FAA-P12; (•), FAA-NES. A representative experiment is shown.
Depletion of FAA from the nucleus abolishes its ability to complement MMC hypersensitivity in FA-A lymphoblasts. (A) Schematic representation of FAA-NES and FAA-P12. The double-headed arrow indicates the amino acid substitution leading to inactivation of the NES. (B) (see page 2232) Immunofluorescence microscopy using pcDNAFAA-NES in transiently transfected Hela cells. (C) Western blot showing the stable expression of FAA, FAA-NES, and FAA-P12 in HSC72 cells. (D) MMC sensitivity assays of the HSC72-derived stable transfectants. (□), Control; (▪), FAA; (○), FAA-P12; (•), FAA-NES. A representative experiment is shown.
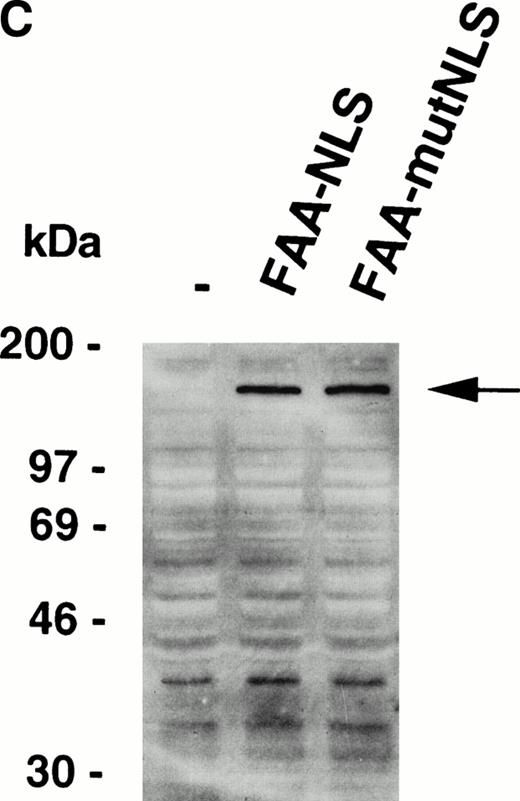
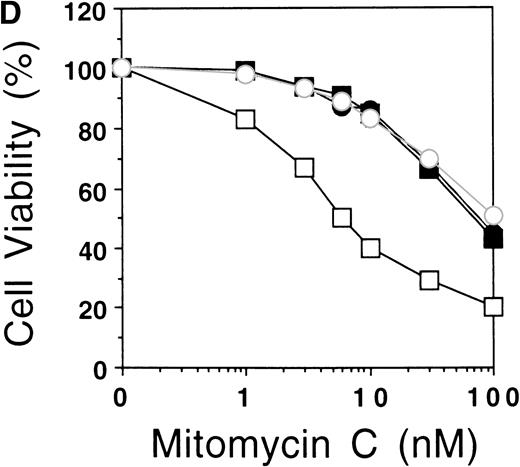
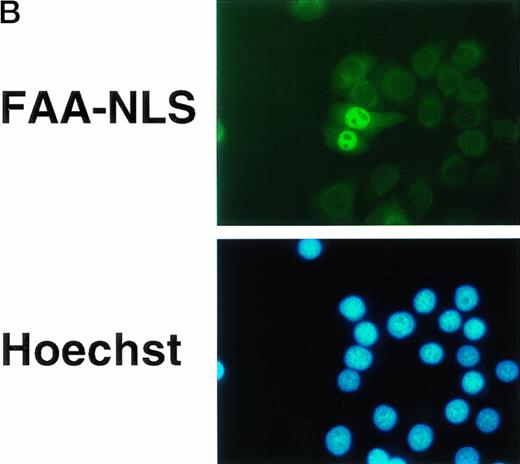
The succesful use of the NES motif led us to reevaluate the effect of a heterologous NLS motif on FAA localization and function. In a previous study, the SV40 large T NLS motif was fused to the amino terminus of FAA-GFP,17 but no change in the localization of FAA was appreciated. In this study, the NLS motif was added to the carboxy terminus of FAA (Fig 5A). As a control, FAA was also fused to a previously described inactive NLS motif.25 Upon expression in HeLa cells, strong nuclear accumulation of FAA-NLS was seen readily by immunofluorescence, but no staining in the cytoplasm could be detected (Fig 5B, see page 2232). FAA-mutNLS localization was similar to the nontagged FAA (not shown). After stable expression in HSC72 (Fig 5C), FAA-NLS was fully able to complement the hypersensitivity of these cells to MMC (Fig 5D), as was FAA-mutNLS. These data show that expression of FAA in the nucleus is essential for its cross-linker complementing function, while depletion of FAA from the nucleus abolishes this activity completely. Thus, the presence of FAA in the nucleus is both necessary and sufficient (ie, coexpression in the cytoplasm is not required) for counteracting the cytotoxic effects of MMC.
Nuclear-targeted FAA is fully functional in complementing MMC sensitivity. (A) Schematic representation of FAA-NLS and FAA-mutNLS. (B) (see page 2232) Visualization of FAA-NLS transiently expressed in HeLa cells by immunofluorescence microscopy. (C) Western blot showing the stable expression of FAA-NLS and FAAmutNLS in HSC72 cells. (D) MMC sensitivity assays of the HSC72-derived stable transfectants. (□), Control; (▪), FAA; (•), FAA-NLS; (○), FAA-mutNLS. A representative experiment is shown.
Nuclear-targeted FAA is fully functional in complementing MMC sensitivity. (A) Schematic representation of FAA-NLS and FAA-mutNLS. (B) (see page 2232) Visualization of FAA-NLS transiently expressed in HeLa cells by immunofluorescence microscopy. (C) Western blot showing the stable expression of FAA-NLS and FAAmutNLS in HSC72 cells. (D) MMC sensitivity assays of the HSC72-derived stable transfectants. (□), Control; (▪), FAA; (•), FAA-NLS; (○), FAA-mutNLS. A representative experiment is shown.
DISCUSSION
In this study we present biochemical, cellular, and genetic data to show that the two currently known FA proteins function in separate subcellular compartments to protect cells against cytotoxicity by cross-linking agents. To validate our conclusions, we have characterized our affinity-purified polyclonal antibody using a number of different strategies and addressed the issue of localization using physical as well as functional endpoints. Overall, we believe that this antibody is monospecific, as indicated by the single FAA-specific band that is immunoprecipitated from COS-1 (Fig 1C) or 293 (Fig 3A and B) cell extracts. However, although overexpressed FAA is readily detectable by a variety of assays, immunodetection of endogenous FAA is more challenging, presumably because of low expression levels. For example, whereas endogenous FAA is clearly detectable by Western analysis, detection by immunofluorescence has not been possible. Thus, we have analyzed endogenous FAA by a more limited repertoire of techniques. Nevertheless, comparative cell fractionation studies show that there are no major differences between the localization of endogenous and transfected FAA (not shown).
Our results show that FAC and FAA differ from each other by several important criteria. For FAC, there is general agreement on the fact that the protein resides predominantly in the cytoplasm.14,15,26 Although Hoatlin et al26recently reported that a small fraction (approximately 10%) of FAC in transfected cells is nuclear, the physiological significance of this observation remains unclear because cytoplasmic localization is required for at least one major function of FAC: Fusion of a foreign NLS to FAC directs the protein to the nucleus and abolishes its complementation function in FA-C cells.16 In addition, the mechanism by which FAC acts to protect against cross-linkers has been at least partly elucidated by our recent finding that FAC binds to the integral microsomal enzyme NADPH cytochrome-P450 reductase. This binding attenuates the activity of the reductase and inhibits conversion of the inert MMC to metabolically active forms (Kruyt FAE, Hoshino T, Liu JM, Joseph P, Jaiswal AK, Youssoufian H, manuscript submitted). FAC may function as a sensor in this major cellular detoxification pathway.
For FAA, there is greater controversy with regard to its subcellular localization. Kruyt et al17 showed that an FAA-GFP chimera is localized primarily to the cytoplasm, and that mutation of the putative NLS does not disturb its complementation function. It is possible that fusion of the GFP epitope to the carboxy terminus interferes with FAA import into the nucleus, and a smaller, yet active, nuclear fraction remains undetectable by this method. Similarly, the lack of nuclear accumulation found with an NLS/FAA-GFP chimera may be due to the GFP epitope. Alternatively, the NLS motif fused to the amino terminus of FAA may be masked. Subsequently, Kupfer at al27showed by using single method, ie, cell fractionation and immunoprecipitation, that FAA is localized both in the nucleus and cytoplasm. In addition, they also demonstrate that FAC is localized in the nucleus with an estimated cytoplasmic/nuclear ratio of 3.2 in extracts prepared from a population of cells, in contrast to their previous reports showing FAC only in the cytoplasm.14 28 To the extent that FAA is found in both the nucleus and cytoplasm, our present results are in agreement with their data. By immunofluorescence studies, we further show that the cytoplasmic/nuclear ratio of FAA can differ in individual cells. This observation suggests that FAA shuttles between the nucleus and cytoplasm. By using foreign nuclear export or nuclear import signals, we have been able to dissect the contributions of each pool to the complementation function of FAA. Our results clearly show that localization of FAA to the nucleus is both necessary and sufficient for its ability to correct the cross-linker sensitivity of FA-A cells. A direct role for FAA in the nucleus may possibly involve DNA repair or recombination.
Unlike Kupfer et al,27 we find no complex formation between FAA and FAC either in vivo or in vitro, and under varying experimental conditions and in different cell lines. It should be noted that these assays have previously allowed us to identify a number of FAC interacting proteins, and we have no obvious explanation for these discrepancies. Although it is possible that the amino-terminal anti-FAA antibody used in our study may block the domain(s) involved in protein interactions and thereby prevent FAA-FAC complex formation, it seems unlikely for several reasons. In contrast to monoclonal antibodies, it would be highly unusual for polyclonal antibodies to function as blocking antibodies. These antibodies recognize multiple epitopes, and it would be reasonable to consider that at least some of the epitopes are physically distinct from sites of protein-protein interaction. Similar to Kupfer et al,27 we also used the reciprocal approach of FAC immunoprecipitation followed by FAA immunoblotting in the hope of further reducing the possibility of epitope-shielding, but again we did not detect any interactions. Certainly, in our experience this approach has not presented any particular difficulties with the demonstration of interactions between FAC and GRP9419 and between FAC and NADPH cytochrome p450 reductase (Kruyt FAE, Hoshino T, Liu JM, Joseph P, Jaiswal AK, Youssoufian H: manuscript submitted). In addition, we eliminated the antibody approach and instead used a functional, epitope-tagged form of FAC, FACγ1,18 but to no avail (data not shown). Finally, we repeated the immunoprecipitation step using either a polyclonal antibody against FAC or against the carboxy terminus of FAA (both antibodies provided by Dr Alan D'Andrea, Dana-Farber Cancer Institute, Boston, MA)27followed by immunoblotting from control and FAA-complemented HSC72 lysates. We found no interaction between FAA and FAC (data not shown). These results force us to conclude that there is no interaction between FAC and FAA.
Although attractive as a hypothesis to explain the similarity in FA pathogenesis, it is by no means obvious that FA proteins must collaborate at some level to guard against cross-linker cytotoxicity. If, indeed, such collaboration exists, our results would be consistent with a model in which the two known FA proteins function in a linear pathway, wherein FAC acts at a proximal step in the cytoplasm and FAA at a more distal step in the nucleus to protect against cross-linker cytotoxicity and chromosomal instability.
ACKNOWLEDGMENT
We thank Dr Hans Joenje (Free University, Amsterdam, The Netherlands) for kindly providing the FAA cDNA and the lymphoblastoid cell lines representing the different FA complementation groups, Dr Christopher Walsh (University of North Carolina, Chapel Hill) for providing the retroviral vector-transduced HSC72 cell line, and Drs Phil Hastings and Huda Zoghbi (Baylor College of Medicine, Houston, TX) for critical comments on the manuscript and use of the fluorescence microscope, respectively.
Supported by grants from the Fanconi Anemia Research Fund and the National Institutes of Health (HL52138).
Address reprint requests to Hagop Youssoufian, MD, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, S840, Houston, TX 77030; e-mail: hagopy@bcm.tmc.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.








This feature is available to Subscribers Only
Sign In or Create an Account Close Modal