Abstract
The objective of this study is to investigate the role of DNA-dependent protein kinase (DNA-PK) in the chronic lymphocytic leukemia (CLL) lymphocyte response to nitrogen mustard therapy. DNA-PK is a nuclear serine/threonine kinase that functions in DNA double-strand break repair and in the joining process in recombination mechanisms. In a series of 34 patients with B-CLL, either untreated (n = 16) or resistant to chlorambucil (n = 18), the kinase activity of the complex, as determined by its capacity to phosphorylate a peptide substrate in vitro, is increased in the resistant samples as compared with the untreated ones (24.4 ± 2.6 arbitrary units [a.u.] [range, 12.7 to 55.8 a.u.] versus 8.1 ± 2.8 a.u. [range, 0.9 to 44.5 a.u.], respectively (P < .0001]), independent of other clinical and biological factors. Linear regression analysis shows an excellent correlation between the level of DNA-PK activity and the inherent in vitro sensitivity of CLL lymphocytes to chlorambucil (r = .875, P =.0001). The regulation of DNA-PK activity was associated with increased DNA-binding activity of its regulatory subunit, the Ku heterodimer, in resistant samples. These results suggest that this activity is a determinant in the cellular response to chlorambucil and participates in the development of nitrogen mustard–resistant disease. The increase in DNA-PK activity might contribute to the enhanced cross-link repair that we previously postulated to be a primary mechanism of resistance to nitrogen mustards in CLL.
THE NITROGEN MUSTARDS (NMs), mechlorethamine, melphalan, chlorambucil, and cyclophosphamide, are used in the treatment of a wide variety of cancers. Alkylation of the DNA and, more importantly, the interstrand cross-linking of DNA, is considered to be responsible for the toxicity of NMs.1-3Although these drugs have been in use for over 30 years, factors underlying in vivo–acquired resistance of human malignancies to NMs are still poorly understood. In an effort to develop a clinically applicable model, we have been studying the mechanisms of resistance to NMs in primary human tumor cells by using B-cell chronic lymphocytic leukemia (B-CLL) as a model. CLL is a disease characterized by the proliferation of abnormal, developmentally regulated immature B cells that accumulate in the peripheral blood of affected patients. The nitrogen mustard, chlorambucil (CLB), is commonly used as first-line therapy for CLL, with an initial response rate of 60% to 80%, but the gradual development of resistance eventually renders the drug ineffective.4 We have previously shown that lymphocytes from treated-resistant patients have an enhanced capacity to remove cross-links compared with those from untreated patients.5In addition, we recently observed that the development of CLB resistance in CLL appears to be specifically associated with cross-resistance to other bifunctionnal alkylating agents, which produces interstrand cross-links (ICL) in DNA.6 Thus, enhanced ICL-specific repair appears to be one of the primary mechanisms of NM resistance in CLL. DNA interstrand cross-links and certain double-strand breaks need information supplied by another chromosome or chromatid for error-free repair. Based on the genetic and biochemical evidence from bacterial and yeast systems, it is thought that cross-links are removed from the DNA of mammalian cells by the combined actions of excision repair (NER) and recombination systems.7,8 It has been suggested that there may be at least two recombinational mechanisms by which interstrand DNA cross-links can be removed, which include the following: (1) an intramolecular pathway operating throughout the cell cycle requiring ERCC-1 and ERCC-4 plus possibly other proteins from the nucleotide excision repair pathway; and (2) a second intermolecular pathway operating between sister chromatids after replication.8However, our results suggest that the incision/excision step of ICL repair is not rate-limiting in CLB-resistant lymphocytes6,9,10 and suggest that the development of enhanced repair of DNA cross-links in CLB-resistant CLL is likely to be associated with an enhanced recombination pathway. In a preliminary study, using a small sample of patients, we found an increase in the DNA-dependent protein kinase activity in resistant samples.11
DNA-dependent protein kinase (DNA-PK) is a recently identified nuclear protein serine/threonine kinase comprising a large catalytic subunit of 460 kD, DNA-PKcs, and a DNA binding sub-unit, the Ku autoantigen (a dimer of the Ku 70 and Ku 86 proteins). Ku binds to DNA double-strand ends and other discontinuities in the DNA12-14 and recruits the catalytic subunit of the complex.15 The active DNA-PK complex then acquires the capacity, at least in vitro, to phosphorylate many DNA-bound proteins in the vicinity.16 DNA-PK has been unequivocally identified as an important mammalian DNA repair complex involved in recombination processes.17 Mutations in either DNA-PKcs or in the 86-kD subunit of Ku result in DNA double-strand break (DSB) repair defects that manifest themselves as x-ray sensitivity and impaired V(D)J recombination.18,19 In addition, previous reports showed that mutant cells deficient either in DNA-PKcs or in the Ku DNA-end binding activity also exhibit significant hypersensitivity to NMs (melphalan and mechlorethamine) and cisplatin.20,21Furthermore, the hypersensitivity of the rodent xrs6 cell line (which lacks the 86-kD subunit of Ku) to NMs appears to be related to the DNA-PK defect because enhanced resistance to NMs was regained along with bleomycin resistance and Ku DNA end-binding activity in a revertant cell line, xrs6rev.20 Similar restoration of NMs sensitivity was observed in the xrs6/Ku80 cell line stably transfected with the human Ku 86 gene (C.M., unpublished results). In addition, we have found that cisplatin-resistant L1210 murine cell lines exhibited cross-resistance to ionizing radiation and to NMs, associated with an overexpression of the Ku 86 sub-unit (P. Frit, Y. Canitrot, J.M. Barret, P. Calsou, and B. Salles, submitted for publication). Although its precise mechanism of action remains unknown, these results provide evidence in favor of a role for the DNA-PK complex in regulating cell sensitivity to NMs.
The aim of the present study is to determine whether regulation of DNA-PK activity is involved in the in vivo development of CLB resistance in CLL. To test this hypothesis, we examined the activity of DNA-PK in lymphocytes obtained from 34 CLL patients either untreated or resistant to NM therapy. Our results clearly show that changes in DNA-PK activity correlate with CLB resistance, suggesting that DNA-PK plays an important role in regulating tumor treatment response to cross-linking agents as demonstrated both at the clinical and in vitro levels.
MATERIALS AND METHODS
Patients.
Thirty-four patients with a diagnosis of B-CLL according to cytologic and immunologic criteria who were followed at the Jewish General Hospital of Montreal between October 1993 and May 1997 were enrolled in the study after informed consent.22 Their main clinical features are summarized in Table 1. Patients were classified into two groups as previously described.23 Patients were either untreated (n = 16) or treated and resistant to nitrogen mustards (n = 18). All the patients with resistant CLL had received 4 to 6 mg of CLB daily for 6 months or more. All the patients with resistant disease had previously responded to CLB with the exception of two patients who showed de novo resistance. In some cases, patients also received cyclophosphamide either alone (50 to 100 mg daily for 3 months or more) or in combination (CHOP [cyclophosphamide, doxorubicin, vincristine, prednisone] and CVP [cyclophosphamide, vincristine, prednisone] protocols).
Clinical Characteristics of the Patients
| Patient No. . | Sex/ Age-150 . | Lymphocyte Count/μL . | Stage-151 . | Treatment Received-152 . | Time Since Last Treatment . | DNA-PK Activity-153 . |
|---|---|---|---|---|---|---|
| Untreated patients | ||||||
| U1 | F/51 | 80,700 | B | 6.9 | ||
| U2 | F/59 | 28,800 | B | 5.3 | ||
| U3 | F/84 | 83,100 | B | 9.8 | ||
| U4 | F/64 | 76,200 | B | 3.4 | ||
| U5 | F/65 | 31,000 | C | 2.2 | ||
| U6 | M/81 | 49,900 | A | 2.2 | ||
| U7 | M/59 | 48,300 | A | 1.7 | ||
| U8 | M/85 | 58,500 | B | 1.9 | ||
| U9 | M/50 | 40,200 | A | 22.8 | ||
| U10 | M/92 | 53,500 | A | 14.9 | ||
| U11 | F/71 | 29,800 | A | 44.5 | ||
| U12 | M/66 | 136,400 | B | 0.9 | ||
| U13 | F/69 | 49,900 | A | 3.4 | ||
| U14 | M/74 | 35,600 | A | 2.9 | ||
| U15 | F/52 | 42,000 | B | 5.8 | ||
| U16 | F/52 | 110,400 | B | 1.2 | ||
| Treated patients with resistance to NMs | ||||||
| T1 | M/53 | 57,500 | C | CLB, CYC, CHOP | 12 mo | 19.0 |
| T2 | F/78 | 233,500 | C | Fluda | On treatment | 13.0 |
| T3 | F/59 | 74,800 | C | CLB, CYC, Fluda | 6 mo | 13.4 |
| T4 | F/87 | 77,800 | C | CLB | On treatment | 16.9 |
| T5 | F/82 | 32,700 | C | CLB, CYC | On treatment | 12.7 |
| T6 | F/72 | 128,100 | C | CLB | On treatment | 32.5 |
| T7 | F/56 | 102,100 | B | CLB, CYC | On treatment | 28.3 |
| T8 | F/60 | 158,500 | C | CLB, CYC | On treatment | 26.3 |
| T9 | M/63 | 104,700 | C | CLB, CYC | 3 mo | 16.2 |
| T10 | M/86 | 58,100 | B | CLB | On treatment | 28.8 |
| T11 | F/76 | 127,300 | C | CLB | On treatment | 55.8 |
| T12 | F/70 | 61,900 | B | CLB | 12 mo | 35.6 |
| T13 | M/54 | 127,300 | C | CLB | 1 mo | 32.3 |
| T14 | M/76 | 167,800 | B | CLB, CVP, Fluda | 12 mo | 20.7 |
| T15 | M/69 | 398,500 | C | CLB, CVP, Fluda | 3 mo | 19.4 |
| T16 | M/77 | 88,400 | B | CLB | 5 mo | 25.6 |
| T17 | M/66 | 31,300 | B | CLB, CYC, Fluda | On treatment | 34.9 |
| T18 | M/52 | 56,700 | B | CLB, CHOP CLB | On treatment | 14.9 |
| Patient No. . | Sex/ Age-150 . | Lymphocyte Count/μL . | Stage-151 . | Treatment Received-152 . | Time Since Last Treatment . | DNA-PK Activity-153 . |
|---|---|---|---|---|---|---|
| Untreated patients | ||||||
| U1 | F/51 | 80,700 | B | 6.9 | ||
| U2 | F/59 | 28,800 | B | 5.3 | ||
| U3 | F/84 | 83,100 | B | 9.8 | ||
| U4 | F/64 | 76,200 | B | 3.4 | ||
| U5 | F/65 | 31,000 | C | 2.2 | ||
| U6 | M/81 | 49,900 | A | 2.2 | ||
| U7 | M/59 | 48,300 | A | 1.7 | ||
| U8 | M/85 | 58,500 | B | 1.9 | ||
| U9 | M/50 | 40,200 | A | 22.8 | ||
| U10 | M/92 | 53,500 | A | 14.9 | ||
| U11 | F/71 | 29,800 | A | 44.5 | ||
| U12 | M/66 | 136,400 | B | 0.9 | ||
| U13 | F/69 | 49,900 | A | 3.4 | ||
| U14 | M/74 | 35,600 | A | 2.9 | ||
| U15 | F/52 | 42,000 | B | 5.8 | ||
| U16 | F/52 | 110,400 | B | 1.2 | ||
| Treated patients with resistance to NMs | ||||||
| T1 | M/53 | 57,500 | C | CLB, CYC, CHOP | 12 mo | 19.0 |
| T2 | F/78 | 233,500 | C | Fluda | On treatment | 13.0 |
| T3 | F/59 | 74,800 | C | CLB, CYC, Fluda | 6 mo | 13.4 |
| T4 | F/87 | 77,800 | C | CLB | On treatment | 16.9 |
| T5 | F/82 | 32,700 | C | CLB, CYC | On treatment | 12.7 |
| T6 | F/72 | 128,100 | C | CLB | On treatment | 32.5 |
| T7 | F/56 | 102,100 | B | CLB, CYC | On treatment | 28.3 |
| T8 | F/60 | 158,500 | C | CLB, CYC | On treatment | 26.3 |
| T9 | M/63 | 104,700 | C | CLB, CYC | 3 mo | 16.2 |
| T10 | M/86 | 58,100 | B | CLB | On treatment | 28.8 |
| T11 | F/76 | 127,300 | C | CLB | On treatment | 55.8 |
| T12 | F/70 | 61,900 | B | CLB | 12 mo | 35.6 |
| T13 | M/54 | 127,300 | C | CLB | 1 mo | 32.3 |
| T14 | M/76 | 167,800 | B | CLB, CVP, Fluda | 12 mo | 20.7 |
| T15 | M/69 | 398,500 | C | CLB, CVP, Fluda | 3 mo | 19.4 |
| T16 | M/77 | 88,400 | B | CLB | 5 mo | 25.6 |
| T17 | M/66 | 31,300 | B | CLB, CYC, Fluda | On treatment | 34.9 |
| T18 | M/52 | 56,700 | B | CLB, CHOP CLB | On treatment | 14.9 |
Response to therapy to date for the untreated patients is as follows: U1, U2, U3, U6, U7, U8, U10, U11, U13, and U16 have not received any treatment. U4 was treated for 8+ months and had an excellent response to CLB (normalization of lymphocyte count, lymphadenopathy: pretreatment node size of 3 to 4 cm vposttreatment node size of ≤0.5 cm). U5 and U12 were treated for 8+ and 36 months, respectively, and had a complete response to CLB (normalization of lymphocyte count, disappearance of lymphadenopathy, and no other evidence of disease). U9 had no response to chlorambucil as previously defined24 (lymphocyte count >35,000 and no decrease in lymphadenopathy after 6 months of chlorambucil treatment). U14 was treated with CLB for 3 months. He had an excellent response as evidenced by a normalization of his lymphocyte count, disappearance of lymphadenopathy and splenomegaly, but his platelet count did not return to normal (95,000/μL). U15 did not tolerate CLB treatment because of nausea and vomiting. She took 30% to 40% of the first two courses (CLB 20 mg and 12 mg every day ×5 every 4 weeks). She took only 1 day of the third course of CLB 4 mg daily. Despite this she showed a partial response as evidenced by a decrease in the size of a large cervical lymph node mass (9 × 9 cm to <3 × 3 cm).
Sex: M, male; F, female; age in years.
Staging is according to Binet et al.22
CYC, cyclosphosphamide; Fluda, fludarabine.
DNA-PK activity is expressed in arbitrary units. For each sample, the results represent the mean of at least two experiments with variability of <15%.
Isolation of CLL lymphocytes and cell culture.
Lymphocytes were isolated from the peripheral blood of CLL patients by sedimentation centrifugation on Ficoll Hypaque (Pharmacia, Uppsala, Sweden) as previously described.6 Isolated lymphocytes were washed twice with phosphate-buffered saline, pelleted by centrifugation at 500g, and either resuspended in culture medium for MTT (3-[4,5-dimethylthiazol-2-yl]2,5-diphenyl-tetrazolium bromide) cytotoxic assay (see below) or stored as a pellet in liquid nitrogen before use for whole cell extract preparation (see below). The percentage of contaminating T cells was determined using fluorescence-activated cell sorting analysis with CD3 antibody. The mean T-cell contamination in our population was 4.4% ± 0.9% (SE), with a range of 1% to 8%. The percentage of contaminating T cells was not statistically different between the untreated and resistant samples (4.7 ± 0.9 v 4.2% ± 0.9%, respectively; not significant [NS]).
The MO59J and MO59K cell lines24 were provided by Dr J.A. Turner (Cross Cancer Institute, Edmonton, Canada). Cells were grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO-BRL, Grand Island, NY) supplemented with 10% fetal calf serum, 2 mmol/L glutamine, 125 U/mL penicillin, and 125 μg/mL streptomycin.
MTT cytotoxic assay.
After the purification procedure, the B-CLL lymphocytes were immediately resuspended in culture medium (RPMI 1640, 10% fetal calf serum, 20 mmol/L HEPES, 10 μg/mL gentamycin) and the samples were screened for their sensitivity to CLB using the MTT assay, as previously described.6
Cell extracts.
Whole cell extracts were prepared as previously described with slight modifications.25 Briefly, cell pellets were quickly thawed and resuspended in extraction buffer (1 × 108 cells per 100 μL) containing 50 mmol/L NaF, 20 mmol/L HEPES (pH 7.8), 450 mmol/L NaCl, 25% glycerol, 0.2 mmol/L EDTA, 0.5 mmol/L dithiothreitol in the presence of proteinase inhibitors (0.5 mmol/L phenylmethylsulfonyl fluoride, 0.5 μg/mL apoprotin, 0.5 μg/mL leupeptin and 1.5 μg/mL pepstatin), then frozen on dry ice and thawed at 30°C three times. After centrifugation for 30 minutes at 4°C, supernatants were stored at −70°C. Protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad, Hercules, CA). The protein concentration ranged from 7.4 to 39.0 mg/mL and was not significantly different between the untreated and treated-resistant samples (13.4 ± 2.0 versus 19. 0 ± 2.8 mg/mL, P = .11).
DNA-PK activity.
The DNA-PK “pulldown” kinase assay was performed as previously described.25,26 For each sample, three kinase assays were conducted in parallel: 1 in the absence of peptide, 1 in the presence of a wild-type peptide (EPPLSQEAFADLLKK) that is a good substrate for DNA-PK, and 1 in the presence of a mutated peptide (EPPLSEQAFADLLKK) that is an ineffective DNA-PK substrate. Values plotted are mean values of at least two experiments and were derived as previously described27 by subtracting the value for no peptide from the values for wild-type and mutated peptide and then dividing these two resulting figures by the no peptide value. The results are expressed as arbitrary units (a.u.). Consistent with previous work,24 we found that, whereas the radioresistant human malignant glioma M059K cell line contains DNA-PK activity, the radiosensitive M059J cell line does not (data not shown). In preliminary experiments, we verified that no significant differences in DNA-PK activity were found between extracts from fresh and frozen samples (data not shown).
In addition, we compared the phosphorylation of the wild-type and mutated peptides from extracts that underwent the “pulldown” assay, where protein binds selectively to double-stranded DNA-cellulose beads, to extracts that were not subjected to the “pulldown” assay but that were incubated in the presence of 10 μg/mL of salmon sperm DNA. This was done with three untreated and three resistant samples. The activity of the extracts that were not subjected to the “pulldown” assay was 28.8% ± 2.9% of the activity determined with the pulldown assay. In particular, there was no significant difference in the percentage of activity between the untreated samples (32.4% ± 2.6%) and the resistant samples (25.2% ± 1.3%).
Band shift assay.
The double-stranded 25-mer DNA was prepared as previously described.28 Briefly, radiolabeled DNA (4 ng, 100,000 cpm) was incubated with extracts (0.25 to 0.75 μg) in 20 μL of binding buffer (10 mmol/L Tris-HCl pH 8.0, 1 mmol/L EDTA, 10% glycerol, 50 mmol/L KCl) at room temperature for 10 minutes. The samples were electrophoresed on a 12% polyacrylamide gel at 4°C, for 2 hours at 100 V. The gel was dried on Whatman paper (Whatman, Maidstone, UK) and exposed to x-ray film. Excised bands that corresponded either to free probe or to DNA-protein complexes were quantified by scintillation counting. The Ku DNA-end binding activity was expressed as the percentage of radioactivity that corresponded to the DNA-protein complexes (cpm in DNA − protein complexes/[cpm in DNA − complexes + cpm in free probe] × 100). To ensure quantitative measurements of Ku DEB activity, we assessed in preliminary experiments that the results obtained in these experimental conditions are in the linear range between 0.25 and 0.75 μg of protein extracts for the control cell line MO59K cell line. For the CLL extracts, two different protein concentrations were used (0.25 and 0.5 μg) for each experiment.
Western blot.
Proteins were subjected to electrophoresis on an 8% sodium dodecyl sulfate-polyacrylamide gel under reducing conditions. The proteins were then electroblotted onto a nitrocellullose filter in a Bio-Rad Trans-Blot chamber and detected as previously described.11Antibodies used in Western blotting experiments were as follows. The monoclonal antibodies (MoAbs) directed against human Ku 70 (MoAb N3H10) and human Ku 86 (MoAb S10B1)29 were obtained from Interchim (France), and used at a 1:1,000 dilution (secondary antibody goat anti-mouse at a 1:2,500 dilution). The MoAb directed against the catalytic subunit of DNA-PKcs antibody (MoAb 18-2, a gift from Dr T. Carter, St John's University, New York, NY)30 was used at a 1:2,500 dilution (secondary antibody goat anti-mouse at a 1:2,500 dilution).
Statistical analysis.
The P values for the group comparisons (treated-resistant versus untreated) were obtained using an unpaired t-test analysis. The r values for the correlation curves were obtained using a simple linear regression analysis. All computations were performed using StatView 512 V.12 (Calabasas, CA).
RESULTS
DNA-PK activity correlates with sensitivity of CLL lymphocytes to chlorambucil.
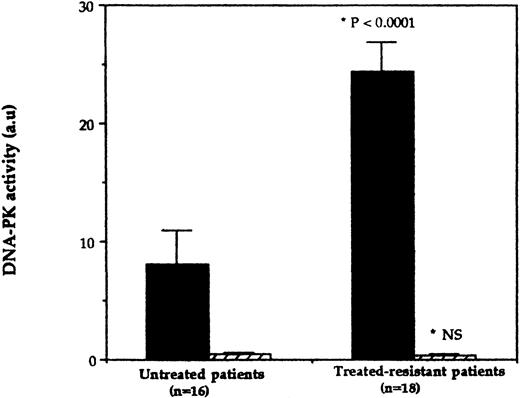
In CLL extracts, the level of DNA-PK activity varied considerably between the different samples analyzed (see Table 1 for individual values). In the samples obtained from resistant patients, the level of DNA-PK activity is similar to that observed in a panel of established cell lines extracts,11 whereas there is less activity in the majority of the samples obtained from untreated patients. The mean DNA-PK activity was found to be significantly higher in the resistant samples compared with untreated ones (24.4 ± 2.6 a.u. v 8.1 ± 2.8 a.u., respectively, P < .0001) (Fig 1). No differences were observed between the two groups regarding the incorporation into the mutated peptide (Fig1).
DNA-PK activity in CLL samples. Whole cell extracts (50 μg) derived from CLL lymphocytes obtained either from untreated or treated-resistant patients were tested for DNA-PK activity using standard DNA-PK microfractionation/peptide assays in the presence of wild-type (▪) or mutated (▨) p53 peptide as indicated in Materials and Methods. The panel represents the mean activity (±SE) for the two groups of samples. *P value as determined by using an unpaired Student's t-test.
DNA-PK activity in CLL samples. Whole cell extracts (50 μg) derived from CLL lymphocytes obtained either from untreated or treated-resistant patients were tested for DNA-PK activity using standard DNA-PK microfractionation/peptide assays in the presence of wild-type (▪) or mutated (▨) p53 peptide as indicated in Materials and Methods. The panel represents the mean activity (±SE) for the two groups of samples. *P value as determined by using an unpaired Student's t-test.
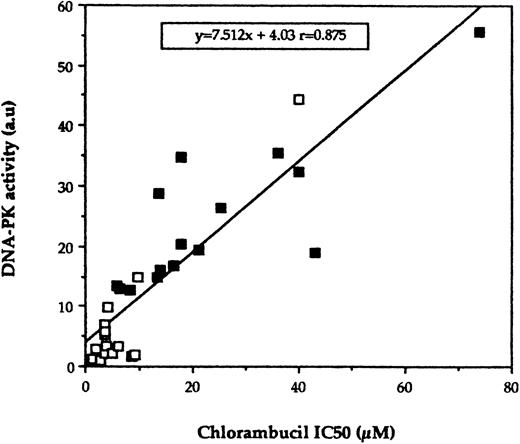
Previous clinical studies showed that 60% to 80% of the patients with CLL will respond to CLB administered as first-line therapy.4 Thus, it is expected that the vast majority of the untreated patients included in our study will respond to this drug if and when they are treated. Our laboratories have shown previously, by using the MTT assay, that lymphocytes from patients with B-CLL display in vitro sensitivity to nitrogen mustards (chlorambucil and melphalan) that correlates with their clinical status.6,11Thus, to analyze the implication of DNA-PK activity in the cellular response of B-CLL lymphocytes to nitrogen mustards, the relationship between this activity and in vitro sensitivity to CLB was examined in 30 of the B-CLL samples included in this study. As expected, the treated-resistant population (n = 15) was found to be 3.3-fold more resistant to CLB than the untreated one (n = 15) with a mean IC50 of 23.5 ± 4.7 μmol/L (range, 5.8 to 74 μmol/L) versus 7.2 ± 2.4 μmol/L (range, 1.5 to 40 μmol/L), respectively, P = .002. However, there are variations in CLB sensitivity within the group of untreated patients because some patients might be de novo resistant to CLB. Linear regression analysis shows a highly significant correlation between the level of DNA-PK activity and in vitro CLB sensitivity (P = .0001; Fig 2). Six of the patients in the untreated group have now been treated with CLB. One patient who exhibited elevated DNA-PK activity (U9) and who was treated with CLB is resistant to this treatment as previously defined,23 while five other untreated patients with low DNA-PK activity responded to chlorambucil treatment (see legend of Table 1). The level of DNA-PK activity did not correlate with the age of the patients (P = .7), the lymphocyte count of the blood samples (P = .4), the percentage of contaminating T cells in purified cell population (P = .98), or the protein concentration of the extracts (P = .68). In addition, disease duration was not a significant variable associated with the level of DNA-PK activity (P = .2). Moreover, when the group of previously treated patients was considered, the level of DNA-PK activity was not different between the patients that were on therapy (n = 10) and those that were off therapy (n = 8) (26.1 ± 4.1 a.u. v 22.2 ± 2.9 a.u., respectively, P =.2). For the patients that were off therapy, the DNA-PK activity did not correlate with the time since last treatment was received (P = .7).
Correlation of DNA-PK activity with CLB sensitivity as determined in vitro. CLL lymphocytes were screened for CLB sensitivity in vitro by using the MTT assay as described in Materials and Methods, and the IC50 for this drug was compared with DNA-PK activity. Each point represents the result from an individual patient's sample. (□), Lymphocytes obtained from untreated patients; (▪), lymphocytes obtained from treated-resistant patients. (Four patients were not included in this analysis bacause in vitro sensitivity to melphalan was done instead of chlorambucil.)
Correlation of DNA-PK activity with CLB sensitivity as determined in vitro. CLL lymphocytes were screened for CLB sensitivity in vitro by using the MTT assay as described in Materials and Methods, and the IC50 for this drug was compared with DNA-PK activity. Each point represents the result from an individual patient's sample. (□), Lymphocytes obtained from untreated patients; (▪), lymphocytes obtained from treated-resistant patients. (Four patients were not included in this analysis bacause in vitro sensitivity to melphalan was done instead of chlorambucil.)
Regulation of DNA-PK activity in CLL extracts.
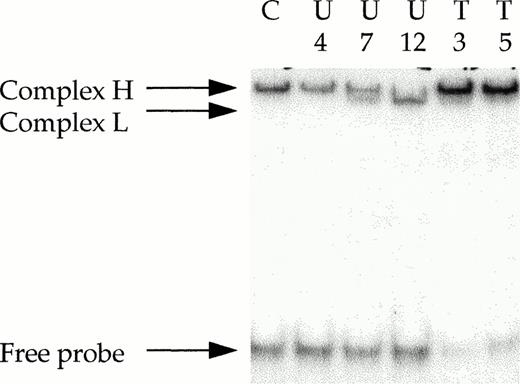
We then examined the mechanism(s) involved in the regulation of DNA-PK activity in the CLL samples. First, we showed that the DNA-PKcs protein was expressed at similar levels between the different samples (Table 2). Second, the DNA-end binding activity was determined by electrophoretic mobility shift assays because the DNA end-binding (DEB) activity of the Ku heterodimer is the early limiting step for the formation of an active DNA-PK complex. In human cell lines, Ku represents the major double-strand (ds) DEB protein and its activity can be easily detected by using ds DNA fragments in an electrophoretic mobility shift assay (EMSA). It has been shown that a 25- to 30-bp dsDNA fragment is the minimum length required for the binding of a single Ku heterodimer.13 As previously described,28 we used a 25-mer ds probe for EMSA analysis. As shown in Fig 3, a unique DNA-protein complex that corresponded to Ku DEB activity (complex H) is detected in the control cell line extracts. In some cases, when the CLL extracts were considered, a second complex with faster electrophoretic mobility is observed (see complex L, lanes 3 and 4). We have previously shown that this complex corresponded to the expression in these cells of a variant form of the Ku 86 protein.11 Although the Ku 70/ Ku 86 variant heterodimer binds to DNA ends, this altered form of the Ku heterodimer has a decreased ability to recruit the catalytic subunit, DNA-PKcs, and contributes to a negative regulation of the kinase activity of the complex.28 31 The expression of this altered form of the Ku heterodimer was detected in 4 of the 16 samples obtained from untreated patients (U5 , U7, U8, and U12), as shown in Fig 3 for samples U7 and U12 (lanes 3 and 4). In addition, we verified by Western blot that the variant form of the Ku 86 protein is expressed in these four samples (data not shown). In these samples, the expression of the altered form of the heterodimer was associated with low DNA-PK activity (mean, 1.67 a.u.; see Table 1 for individual values) and high sensitivity to CLB (mean IC50, 2.4 μmol/L). Interestingly, the altered form of the Ku heterodimer was not detected in samples obtained from treated-resistant patients or in the patients that exhibited de novo high levels of DNA-PK activity.
Western Blot Analysis
| Samples . | DNA-PKcs . | Ku 86 . | Ku 70 . |
|---|---|---|---|
| Untreated | 1.22 ± 0.11 | 1.18 ± 0.16 | 1.12 ± 0.13 |
| Treated-resistant | 1.16 ± 0.11 | 3.19 ± 0.82 | 3.11 ± 0.19 |
| P value determined by t-test | NS | P < .00001 | P < .00001 |
| Samples . | DNA-PKcs . | Ku 86 . | Ku 70 . |
|---|---|---|---|
| Untreated | 1.22 ± 0.11 | 1.18 ± 0.16 | 1.12 ± 0.13 |
| Treated-resistant | 1.16 ± 0.11 | 3.19 ± 0.82 | 3.11 ± 0.19 |
| P value determined by t-test | NS | P < .00001 | P < .00001 |
Ku 86, Ku 70, and DNA-PKcs protein levels were determined in B-CLL lymphocytes obtained from untreated and treated patients with resistance to NMs. Data were normalized to α-tubulin in order to account for loading differences and expressed relative to normalized U1 sample extracts which was assigned the arbitrary value of 1 for the Ku 86, Ku 70, and DNA-PKcs expression.
Ku DEB activity in CLL extracts as determined by EMSA. Protein extracts (0.5 μg) obtained from purified B-CLL cells were incubated with 32P-labeled 25-bp DNA probe in the presence of 1 μg of closed circular plasmid DNA, as a nonspecific competitor. The electrophoretic mobility of the protein-DNA complexes were analyzed in a 12% polyacrylamide gel as described in Materials and Methods. The positions of protein-DNA complexes consisting of DNA-end binding activity are indicated (H, heterodimer that corresponds to full-length Ku 70/full-length Ku 86 subunits; L, heterodimer that corresponds to full-length Ku 70/variant Ku 86 subunits). U, untreated CLL lymphocytes; T, treated-resistant CLL lymphocytes; C, MO59K control cells.
Ku DEB activity in CLL extracts as determined by EMSA. Protein extracts (0.5 μg) obtained from purified B-CLL cells were incubated with 32P-labeled 25-bp DNA probe in the presence of 1 μg of closed circular plasmid DNA, as a nonspecific competitor. The electrophoretic mobility of the protein-DNA complexes were analyzed in a 12% polyacrylamide gel as described in Materials and Methods. The positions of protein-DNA complexes consisting of DNA-end binding activity are indicated (H, heterodimer that corresponds to full-length Ku 70/full-length Ku 86 subunits; L, heterodimer that corresponds to full-length Ku 70/variant Ku 86 subunits). U, untreated CLL lymphocytes; T, treated-resistant CLL lymphocytes; C, MO59K control cells.
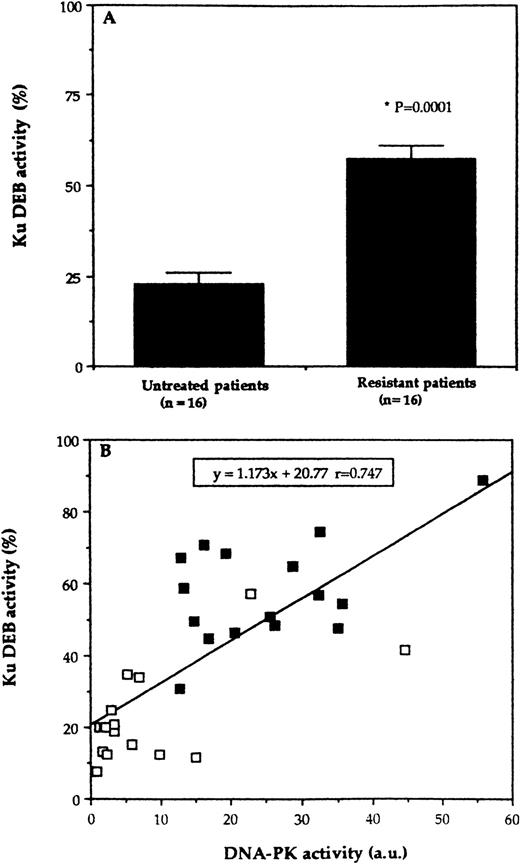
The Ku DEB activity was quantitated in 32 samples of our series (see Materials and Methods). As for DNA-PK activity, Ku DEB activity was significantly higher in the treated-resistant samples as compared to the untreated ones (57.6% ± 3.5% v 22.8% ± 3.3%,P =.0001). These results suggest that an increase in the level of Ku DEB activity contributes to the observed increase in DNA-PK activity. Indeed, as shown in Fig 4, in the 32 samples tested, a linear correlation was observed between the Ku DEB activity and the kinase activity of the complex (P = .0001), supporting the concept that regulation of the activity of the complex occurs primarily at the Ku level. Furthermore, the Western analyses confirms an increase in Ku70/Ku86 protein levels but not in DNA-PKcs (Table 2).
Ku DEB activity according to the two groups of samples. (A) The Ku DEB activity was determined by EMSA experiments and bands that corresponded to the Ku-DNA complexes were excised and counted by scintillation as described in Materials and Methods. For each sample, the results represent the mean of at least two experiments with variability of less than 10%. (B) Correlation of Ku DEB activity with the DNA-PK kinase activity as determined for 32 samples. Each point represents the result from an individual patient's sample. (□), Lymphocytes obtained from untreated patients; (▪), lymphocytes obtained from treated-resistant patients.
Ku DEB activity according to the two groups of samples. (A) The Ku DEB activity was determined by EMSA experiments and bands that corresponded to the Ku-DNA complexes were excised and counted by scintillation as described in Materials and Methods. For each sample, the results represent the mean of at least two experiments with variability of less than 10%. (B) Correlation of Ku DEB activity with the DNA-PK kinase activity as determined for 32 samples. Each point represents the result from an individual patient's sample. (□), Lymphocytes obtained from untreated patients; (▪), lymphocytes obtained from treated-resistant patients.
DISCUSSION
To the best of our knowledge, the implication of DNA-PK in regulating the response of primary tumor specimens to treatment (radiotherapy or chemotherapeutic treatment with NMs) has never been characterized, although this activity is likely to play a role in regulating cell sensitivity to these cytotoxic agents, as shown in mutant cell lines.20,21 Our present data show that, in CLL samples, DNA-PK activity is increased in samples that exhibited a phenotype of resistance to NMs determined either both in vivo and in vitro or in vitro only for those untreated patients that have not started therapy at the time of the analysis. In addition, the excellent linear correlation between DNA-PK activity and in vitro CLB toxicity strongly suggest that DNA-PK is likely to contribute to this phenotype. In fact, the higher DNA-PK activity observed is unlikely to reflect a transient induction by the treatment itself because the increase in DNA-PK activity with a corresponding decrease in CLB sensitivity in vitro was observed even in patients that were not actively receiving therapy at the time of the sampling (8 of the 18 resistant samples analyzed and 3 of the 16 previously untreated patients). Moreover, the level of DNA-PK activity was not different between the resistant patients that were on therapy compared with those who had interrupted treatment. In accordance, no changes in the levels of Ku polypeptides or DNA-PK activity have been detected after treatment of established cell lines with genotoxic compounds.24,32 The increase of DNA-PK activity in CLB-resistant samples is unlikely to be caused by the presence of a subpopulation of actively dividing cells. Previous studies have suggested that Ku expression may be dependent on the proliferative status of the cells.33-35 However, despite dramatic changes in Ku mRNA expression, the total level of Ku has been reported to be only slightly (about twofold) increased in proliferative versus quiescent cells.33,35 In CLL, even in advanced disease, cell-cycle analysis consistently showed that most malignant cells are in G0/G1 phase and very few are in S phase.36 37Our results suggest that a higher DNA-PK activity is a marker for the presence of resistant cells to NMs in the tumor cell population, and this activity appears to play a determining role in the resistance phenotype.
In our study we have shown that DNA-PK activity is positively regulated in cells that exhibited a resistant phenotype to CLB. Our results also provide the basis for a new approach to understanding the mechanisms that contribute to the development of drug resistance in CLL. Indeed, it is noticeable from our results that most leukemic extracts obtained from untreated patients exhibited very low levels of DNA-PK activity (see Fig 1 and Table 1). Thus, resistance in CLL may be simply a state in which tumor cells lose an abnormal sensitivity to alkylating agents, rather than a process in which they gain an abnormal insensitivity. Our results show that in CLL, changes in DNA-PK activity appear to be regulated through a variation in the Ku heterodimer DEB activity, but also, in some samples, potentially through the expression of an altered form of the heterodimer. Interestingly, we recently showed that the normal human peripheral blood CD19+ B lymphocytes express solely this altered form of the Ku heterodimer that is no longer able to recruit the catalytic component of the complex when bound to DNA.31 Thus, our present results extend our previous reports11,31 and show that in B-CLL lymphocytes its expression may be persistent despite the acquisition of a malignant phenotype in some, but not all, samples. Independent of the expression of this altered form of the Ku heterodimer, our results show that the kinase activity of the complex is regulated through the activity of its regulatory sub-unit Ku, because the level of the kinase activity of the complex observed in CLL samples correlated with the level of Ku DEB activity (see Fig 4). In addition, we verified by Western blot that the level of expression of the catalytic subunit of the complex was not increased in the resistant samples with elevated levels of kinase activity. Taken together, these results show that regulation of Ku expression and function plays a pivotal role during the acquisition of resistance to NMs therapy. This regulation contributes to the increased activity of the kinase of the complex as Ku DNA-binding represents the predominant mechanism for DNA-PK activation.15 25
In conclusion, although uncertainties remain concerning its precise mechanism of action, our results strongly suggest that Ku/DNA-PK activity contributes to resistance to NM therapy in B-CLL. Because its appears that Ku/DNA-PK is tightly regulated in normal lymphoid tissues, it would be of interest to determine if this activity is involved in the response of other tumor cell types to this class of drugs or if these results are restricted to tumors of lymphoid origin. Finally, to the extent that enhanced kinase activity does contribute to resistance, it should be possible to improve the efficacy of nitrogen mustards against currently resistant tumors by inhibiting this activity. Accordingly, results obtained in our laboratory show that wortmannin, a nonspecific inhibitor of DNA-PK activity, is able to potentiate CLB toxicity in resistant samples.38 Thus, our findings point to new possibilities to improve the effectiveness of NM therapy.
ACKNOWLEDGMENT
The authors thank Dr P. Calsou for critical reading of the manuscript. We also thank Franka Sicilia for excellent technical support in T-cell typing.
C.M. and G.C. contributed equally to this investigation.
Supported by the Cancer Research Society, Montreal, Quebec, Canada. L.P. is a recipient of the Gertrude and Stanley Vineberg Clinical Scientist Award. C.M. is a recipient of a postdoctoral fellowship from the “Comité leucémie de la Fondation de France.”
Address reprint requests to Lawrence Panasci, MD, Lady Davis Institute for Medical Research, The Sir Mortimer B. Davis-Jewish General Hospital, 3755 Côte Sainte Catherine, Montreal, Quebec, Canada, H3T 1E2; e-mail: gchris1@po-box.mcgill.ca.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal