Abstract
The serine protease cathepsin G is synthesized during the promyelomonocytic stage of neutrophil and monocyte differentiation. After processing, including removal of an amino-terminal propeptide from the catalytically inactive proform, the active protease acquires a mature conformation and is stored in azurophil granules. To investigate the importance of the proform-conformation for targeting to granules, a cDNA encoding a double-mutant form of human preprocathepsin G lacking functional catalytic site and amino-terminal prodipeptide (CatG/Gly201/▵Gly19Glu20) was constructed, because we were not able to stably express a mutant lacking only the propeptide. Transfection of the cDNA to the rat basophilic leukemia RBL-1 and the murine myeloblast-like 32D cl3 cell lines resulted in stable, protein-expressing clones. In contrast to wild-type proenzyme, CatG/Gly201/▵Gly19Glu20 adopted a mature conformation cotranslationally, as judged by the early acquisition of affinity to the serine protease inhibitor aprotinin, appearing before the carboxyl-terminal processing and also in the presence of the Golgi-disrupting agent brefeldin A. The presence of a mature amino-terminus was confirmed by amino-terminal radiosequencing. As with wild-type proenzyme, CatG/Gly201/▵Gly19Glu20 was proteolytically processed carboxyl-terminally and glycosylated with asparagine-linked carbohydrates that were converted into complex forms. Furthermore, it was targeted to granules, as determined by subcellular fractionation. Our results show that the initial proform-conformation is not critical for intracellular sorting of human cathepsin G. Moreover, we demonstrate that double-mutant cathepsin G can achieve a mature conformation before carboxyl-terminal processing of the proform.
© 1998 by The American Society of Hematology.
CATHEPSIN G IS A chymotrypsin-like protease with antimicrobial activity1 and belongs to a superfamily of serine proteases stored in cytoplasmic granules of hematopoietic cells.2 Together with leukocyte elastase, proteinase 3, and the catalytically inactive protease-homologue azurocidin, whose genes are clustered on chromosome 19p13.3,3 cathepsin G (encoded on chromosome 14q11.2)4 is synthesized during the promyelocyte stage of neutrophil differentiation.5,6 These 4 proteins, collectively called serprocidins, are stored in the azurophil (peroxidase-positive; primary) granules.1,2,6 Cathepsin G or a cathepsin G-like enzyme has also been detected in monocytes, a subset of mast cells, and basophils.7-9 In addition to its capability of killing bacteria and fungi (a function that seems to be independent of its catalytic activity), cathepsin G has several possible catalytic functions in normal and pathological events, indicated by in vitro studies; eg, leukocyte migration by acting as a chemotactic agent for neutrophils and monocytes10 and participation in an alternative pathway of leukocyte initiation of coagulation by activating coagulation factor X11 and factor V.12
The hematopoietic serine proteases are synthesized as catalytically inactive proenzymes (zymogens) that, after removal of an amino-terminal propeptide, often a dipeptide, are stored in granules as active enzymes.13-17 Procathepsin G is activated simultaneously with, or directly after, transfer to granules,13,18-20 but a fraction of the zymogen is constitutively secreted. The activation is thought to be catalyzed by dipeptidyl peptidase I,21 a thiol protease that removes the amino-terminal dipeptide of several members of the hematopoietic serine protease family.16,17,22 In addition to the amino-terminal propeptide, procathepsin G has a carboxyl-terminal extension of 11 to 12 amino acids, which is removed during processing of the proform.13,18,19,23 24 The enzyme mediating this removal has not been identified and the role of the carboxyl-terminal extension is unclear.
Several mechanisms have been proposed to be responsible for sorting of proteins destined for storage in granules.2,25 However, the mechanisms for sorting of the serprocidins are unknown. It could be hypothesized that sorting of cathepsin G is restricted to protein in proform-conformation, in analogy to recent reports on pro-opiomelanocortin26 and prorenin,27 in which binding of a processing enzyme is important for sorting to granules.
The main aim of the present study was to investigate whether the proform-conformation of cathepsin G is important for intracellular sorting of the zymogen. To this end, we transfected cDNA encoding mutated forms of human preprocathepsin G lacking the amino-terminal propeptide (expected to adopt a mature conformation cotranslationally) to the rat basophilic/mast cell line RBL-1 and the murine myeloblast like 32D cl3 cell line, cellular models recently used for investigations on the processing of neutrophil serine proteases.19,20,28 29 We demonstrate that the proform-conformation of procathepsin G is not critical for sorting to granules or for constitutive secretion, but rather may serve to protect the cell from the catalytic activity of cathepsin G. In addition, we show that conversion into a mature conformation can occur independently of carboxyl-terminal processing.
MATERIALS AND METHODS
Materials.
The eukaryotic expression vectors pCEP4 and pcDNA3 were from Invitrogen (Leek, The Netherlands). The vectors provide a cytomegalovirus promoter-driven expression of introduced cDNA. The plasmids also confer resistance to hygromycin B and geneticin, respectively, allowing selection of recombinant cells.35S-methionine/35S-cysteine (cell labeling grade) and 3H-isoleucine were from Amersham (Amersham, UK). Before use, 3H-isoleucine was concentrated 10-fold in a vacuum centrifuge. Percoll and Protein A-Sepharose CL4-B were from Pharmacia (Uppsala, Sweden). Aprotinin-agarose was from Sigma (St Louis, MO). N-glycosidase F, endoglycosidase H, and geneticin were from Boehringer Mannheim (Mannheim, Germany). Hygromycin B was from Calbiochem (La Jolla, CA). A polyclonal rabbit antiserum to cathepsin G was obtained by immunization of rabbits.30 Brefeldin A (BFA) was a gift from Sandoz (Basel, Switzerland). The polyvinylidene difluoride (PVDF) membranes were from Millipore (Bedford, MA). The β-galactosidase (β-gal) staining kit and the eukaryotic expression vector pCMVβ, carrying the reporter gene β-gal, were from Invitrogen.
cDNA, mutagenesis, and construction of expression vectors.
Full-length cDNA for human preprocathepsin G (a gift from Dr Guy Salvesen, Duke University, Durham, NC) was cloned into pCEP4, thus creating the expression vector pCEP4/CatG.19 For site-directed mutagenesis, preprocathepsin G cDNA was used as template in a two-step spliced overlap extension polymerase chain reaction (SOE PCR)31 in the following way. In the first reaction, 2 separate amplifications with 100 ng of DNA template in 10-cycle PCRs produced 2 fragments of preprocathepsin G cDNA overlapping each other on the sequence around the amino-terminal dipeptide, Gly19and Glu20 (numeration from the initial ATG translational initiation site). By design of the primers, Gly19 and Glu20 were deleted. The 5′- and 3′-primers for the cDNA sequence used were identical to those earlier used for producing pCEP4/CatG, thus introducing the Kozak consensus leader sequence for maximum translational efficiency32 and the flanking restriction enzyme sites Kpn I and Not I for subsequent cloning into pCEP4 or pcDNA3 plasmids. The PCR primers in the 2 amplifications were upstream 5′ GA CTT CAG GGT ACC GCC GCC ACC ATG CAG CCA CTC CTG CTT CTG CTG G 3′ (no. 1), plus downstream 5′ CCG GCC TCC GAT GAT Δ TGC CTC AGC CCC AGT GG 3′ (no. 2), and upstream 5′ CC ACT GGG GCT GAG GCA Δ ATC ATC GGA GGC CGG GAG AGC 3′ (no. 3), plus downstream 5′ G ACT TCA GGC GGC CGCTCA CAG GGG GGT CTC CAT CTG ATC CAG C 3′ (no. 4), respectively (start and stop codons in bold, restriction enzyme sites underlined, deleted Gly19 and Glu20 indicated with triangle). The PCR products were isolated on agarose gel, mixed, and subjected to a 20-cycle splicing PCR amplification with primers no. 1 and 4, thus creating full-length preprocathepsin G cDNA with deleted Gly19 and Glu20(ΔGly19Glu20). The PCR product was digested by Kpn I and Not I, followed by isolation on agarose gel and cloning into plasmid. Individual clones were isolated and sequenced to verify the mutation and the integrity of the reading frame. cDNA with correct nucleotide sequence was cloned to create an expression vector for mutated preprocathepsin G. All PCR reactions were performed in a Perkin Elmer 480 Thermal Cycler usingPfu-polymerase (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. Analogously, a cDNA coding for a mutant form of preprocathepsin G lacking enzymatic activity (CatG/Gly201) was made by substituting Ser201 (numeration from the initial ATG translational initiation site) with Gly, using the mutant-primers 5′ GGG GCC TCC GCC ATC CCC CTT 3′ (no. 2) and 5′ AAG GGG GATGGC GGA GGC CCC 3′ (no. 3) (Gly201underlined) with wild-type preprocathepsin G as template. Identical mutagenesis, but with preprocathepsin G/ΔGly19Glu20 as template, resulted in a double mutant (CatG/Gly201/ΔGly19Glu20).
Cell culture.
The rat basophilic/mast cell line RBL-1 and murine myeloblast-like 32D cl3 (clone 3) cells were grown as described.28 Cell cultures were kept in 5% CO2 at 37°C in a fully humidified atmosphere. Exponentially growing cells were used in all experiments. COS-7, an African green monkey kidney cell line, was maintained in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO Ltd, Paisley, UK) supplemented with 10% heat-inactivated fetal calf serum.
Transfection procedure.
RBL-1 and 32D cl3 cells were transfected using the BioRad Gene Pulser (Bio Rad, Hercules, CA) with electrical settings 960 μF and 300 V, as previously described.19,28 After electroporation, geneticin or hygromycin B was added to select for recombinant clones expressing the geneticin-resistance gene of pcDNA3 or the hygromycin B-resistance gene of pCEP4. Individual clones growing in the presence of antibiotic were isolated, expanded into mass cultures, and screened for expression of cathepsin G by biosynthetic labeling. Clones with the most pronounced expression were chosen for further experiments. COS-7 cells were transiently transfected in 10-cm plastic dishes with 4 μg of plasmid DNA per 106 cells, using the diethyl aminoethyl (DEAE)-dextran method.33 Twenty-four to 48 hours after transfection, the expression of cathepsin G was detected by biosynthetic labeling of cells. In experiments with cotransfection of pCMVβ, COS-7 cells were transiently cotransfected in 2.3-cm wells with 0.2 to 0.3 μg of pCMVβ and equal or double amounts of plasmid DNA, encoding wild-type or mutated forms of preprocathepsin G in the pcDNA3 vector, per 50 × 103cells.
Biosynthetic labeling.
Biosynthetic radiolabeling of newly synthesized proteins was performed essentially as described elsewhere.19 Unless otherwise indicated, cells were starved for 30 minutes in methionine/cysteine-free medium followed by pulse-labeling with35S-methionine/35S-cysteine for 30 minutes. In chase experiments after pulse-labeling, cells were resuspended in complete medium. At timed intervals, cells were withdrawn and subjected to extraction of whole cells or homogenization and subsequent subcellular fractionation.
Radio sequence analysis.
For determination of the amino-terminal processing of procathepsin G, biosynthetic labeling with 3H-isoleucine, followed by amino acid sequencing, was performed essentially as described.20Briefly, cells were incubated in medium supplemented with3H-isoleucine (200 μCi/mL) to achieve metabolic labeling of synthesized proteins. After pulse-labeling for 30 minutes, cell lysates were prepared and immunoprecipitation was performed. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the proteins were transferred to a PVDF membrane, which was subjected to autoradiography. Radioactive bands were excised and subjected to amino acid degradation according to Edman (performed by the Biomedical Service Unit, Lund University, Lund, Sweden). The initial 9 to 10 degradation cycles were assayed for radioactivity by use of a scintillation counter.
Subcellular fractionation.
Subcellular fractionation was performed essentially as previously described.19 Briefly, the cell homogenate was fractionated in a Percoll density gradient after which 9 fractions were collected with all cytosol in fraction no. 9. The distribution of lysosomes and Golgi elements in the density gradient was determined by assaying β-hexosaminidase and galactosyl transferase. Peak activities of β-hexosaminidase and galactosyl transferase in subcellular fractions from RBL wild-type cells were localized in fractions no. 1 and 2 and no. 5 through 8, respectively, and from 32D wild-type cells in fractions no. 2 and 6.19 28
Immunoprecipitation.
For immunoprecipitation, whole cells or Percoll-containing subcellular fractions were solubilized essentially as previously described.19 Biosynthetically labeled cathepsin G was immunoprecipitated by addition of a polyclonal rabbit antiserum to cathepsin G and protein A-sepharose and was subjected to SDS-PAGE on a 5% to 20% (or a 10% to 20% precast Tris-Glycine gel; Novex, San Diego) gradient gel followed by fluorography essentially as described.19
Adsorption to aprotinin-agarose.
Adsorption to aprotinin-agarose was performed essentially as described by Salvesen and Enghild13 and modified.19Briefly, cells were lysed and the lysate was allowed to react with a suspension of aprotinin-agarose. The aprotinin with bound material was washed, followed by incubation with elution buffer to obtain release of bound material. Cathepsin G remaining in the lysate after adsorption to aprotinin-agarose (lacking affinity to aprotinin) or eluted (with affinity to aprotinin) was immunoprecipitated and subjected to SDS-PAGE and fluorography.
Digestion with endoglycosidase H and N-glycosidase F.
The susceptibility of procathepsin G to digestion with endoglycosidase H (Endo H) was determined essentially as described.23Digestion with N-glycosidase F was performed according to the manufacturer’s instructions. Briefly, immunoprecipitates, collected as described above, were dissolved in buffer and boiled for 5 minutes. After centrifugation, the supernatant was collected and Endo H or N-glycosidase F was added. Control incubations were treated identically, except that no enzyme was added. After incubation for 16 to 24 hours, the samples were subjected to SDS-PAGE and fluorography.
β-gal staining and statistical analysis.
The expression of β-gal was detected by β-gal staining performed with the β-gal staining kit, according to the manufacturer’s instructions. The numbers of positive cells in each well were counted in a light microscope. The statistical analysis was performed with a two-sided Wilcoxon signed rank test.
RESULTS
Establishment of stable cell clones expressing mutant forms of cathepsin G.
By use of site-directed mutagenesis as described in the Materials and Methods, a mutant form of preprocathepsin G lacking the amino-terminal propeptide (CatG/ΔGly19Glu20) was cloned into pCEP4 (pCEP4/CatG/ΔGly19Glu20) and into the pcDNA3 vector (pcDNA3/CatG/ΔGly19Glu20). However, despite 5 separate transfections of RBL-1 cells using these constructs, it was not possible to establish stable cell clones expressing detectable amounts of cathepsin G. Likewise, transfections of RBL cells using a muristerone-inducible expression system based on the expression plasmids pVgRXR and pInd (Invitrogen) did not result in any detectable synthesis when CatG/ΔGly19Glu20 was cloned into the pInd-vector (15 clones were screened), whereas 3 clones expressing cathepsin G were found when wild-type preprocathepsin G was cloned into vector (data not shown). Furthermore, it was not possible to detect any synthesis of cathepsin G in simian COS-7 cells transiently transfected with CatG/ΔGly19Glu20, although the protein was strongly expressed after transfection of wild-type preprocathepsin G to these cells (data not shown).
In contrast, transfection of mutant preprocathepsin G lacking functional catalytic site (CatG/Gly201) resulted in stable clones of both RBL and 32D cells expressing the protein. Because the deletion of the amino-terminal propeptide might result in a cotranslational activation of the protease, it was hypothesized that the difficulties in obtaining stable expression of CatG/ΔGly19Glu20 resulted from a premature emergence of catalytic activity. To test this hypothesis further, we transiently cotransfected wild-type preprocathepsin G or the 2 mutant forms of preprocathepsin G cloned into the pcDNA3 expression vector together with the expression vector pCMV β to COS-7 cells. The total numbers of β-gal–positive cells in each well were counted 1 to 5 days after transfection and mean values were calculated for each day. Three days after transfection, there was a significant difference (P < .05) between the number of cells expressing β-gal after cotransfection with CatG/ΔGly19Glu20compared with the cells cotransfected with wild-type or double-mutant preprocathepsin G (Fig 1). On day 5 after transfection, the number of β-gal–positive cells decreased in all wells, probably due to an unspecific decrease in the expression of pCMVβ (not shown in Fig 1). To circumvent the possible obstacle in the form of a premature catalytic activation, a double-mutant form of preprocathepsin G (CatG/Gly201/ΔGly19Glu20), lacking both Ser201, essential for catalytic activity, and the amino-terminal propeptide, was transfected to RBL or 32D cells. These transfections resulted in stable clones expressing the double-mutant form of procathepsin G.
β-gal positivity after cotransfection of pCMVβ and wild-type or mutated preprocathepsin G forms to COS-7 cells. COS-7 cells (50 × 103) were transiently cotransfected with 0.2 to 0.3 μg of pCMVβ and (•) pcDNA3/CatG, (▴) pcDNA3/CatG/Gly201/▵Gly19Glu20, or (▪) pcDNA3/CatG/▵Gly19Glu20 in equal or double amounts in six 12-well plates using the DEAE-dextrane method. Cells were stained using the β-gal kit and the absolute numbers of β-gal–positive cells were determined in a light microscope. The diagram shows the mean values of β-gal–positive cells 1 to 4 days after transfection. Values are from 4 separate experiments (with 8 wells counted on day 3).
β-gal positivity after cotransfection of pCMVβ and wild-type or mutated preprocathepsin G forms to COS-7 cells. COS-7 cells (50 × 103) were transiently cotransfected with 0.2 to 0.3 μg of pCMVβ and (•) pcDNA3/CatG, (▴) pcDNA3/CatG/Gly201/▵Gly19Glu20, or (▪) pcDNA3/CatG/▵Gly19Glu20 in equal or double amounts in six 12-well plates using the DEAE-dextrane method. Cells were stained using the β-gal kit and the absolute numbers of β-gal–positive cells were determined in a light microscope. The diagram shows the mean values of β-gal–positive cells 1 to 4 days after transfection. Values are from 4 separate experiments (with 8 wells counted on day 3).
The processing of the mutant forms of procathepsin G is similar to that of wild-type protein.
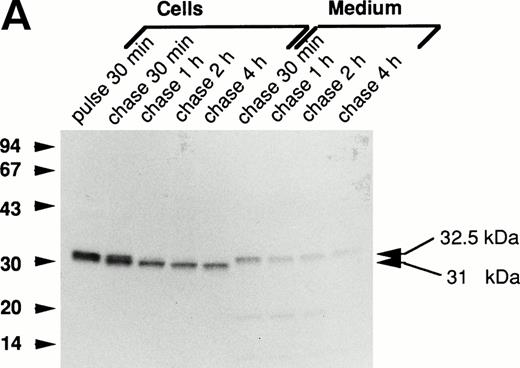
The processing of the mutant forms of procathepsin G was investigated by biosynthetic labeling and pulse-chase experiments as described in the Materials and Methods. Figure 2A demonstrates the biosynthesis of wild-type cathepsin G in RBL/CatG cells. A specifically immunoprecipitated protein with an apparent molecular mass of 32.5 kD, representing the proform of cathepsin G, was detected after 30 minutes of radio-labeling. During chase of the label, processing of the 32.5-kD proform to a 31-kD form was observed within 30 minutes, and after 2 hours of chase the conversion to the 31-kD form seemed complete. This reduction of molecular mass most probably represents the removal of the carboxyl-terminal peptide extension. In addition, a minor fraction was further processed to a 30-kD form (not indicated in Fig 2). Upon analysis of the incubation medium, 32.5-kD procathepsin G was observed. These results are consistent with those earlier published.13,19,20 28
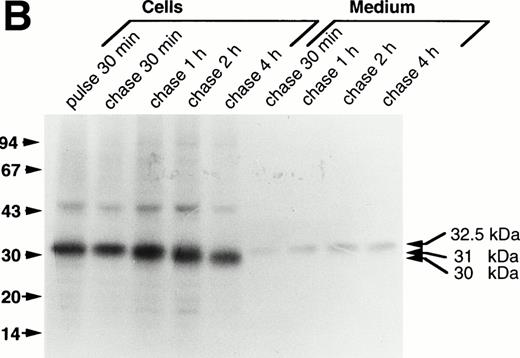
Processing of wild-type and double-mutant procathepsin G in RBL cells. (A) RBL/CatG and (B) RBL/CatG/Gly201/▵Gly19Glu20 cells were pulse-labeled with35S-methionine/35S-cysteine for 30 minutes followed by chase of the label for up to 4 hours. At indicated points, 20 × 106 cells were withdrawn and subjected to solubilization and immunoprecipitation with polyclonal anti-cathepsin G antiserum. In addition, cathepsin G was immunoprecipitated from the incubation medium after each period of chase. The immunoprecipitates were run in SDS-PAGE in a 10% to 20% and a 5% to 20% gradient gel, respectively, whereupon fluorography was performed. The fluorograms were exposed for 6 days and 2 weeks, respectively. The different processing forms of cathepsin G are indicated with arrows to the right. Numbers to the left in this and subsequent figures are the molecular weight values of molecular weight standards.
Processing of wild-type and double-mutant procathepsin G in RBL cells. (A) RBL/CatG and (B) RBL/CatG/Gly201/▵Gly19Glu20 cells were pulse-labeled with35S-methionine/35S-cysteine for 30 minutes followed by chase of the label for up to 4 hours. At indicated points, 20 × 106 cells were withdrawn and subjected to solubilization and immunoprecipitation with polyclonal anti-cathepsin G antiserum. In addition, cathepsin G was immunoprecipitated from the incubation medium after each period of chase. The immunoprecipitates were run in SDS-PAGE in a 10% to 20% and a 5% to 20% gradient gel, respectively, whereupon fluorography was performed. The fluorograms were exposed for 6 days and 2 weeks, respectively. The different processing forms of cathepsin G are indicated with arrows to the right. Numbers to the left in this and subsequent figures are the molecular weight values of molecular weight standards.
Upon analysis of the processing of mutant procathepsin G lacking functional catalytic site (CatG/Gly201) (data not shown) and of the double mutant (CatG/Gly201/ΔGly19Glu20) (Fig2B) transfected to RBL cells, similar results were obtained. Thus, both mutant forms of procathepsin G showed a processing pattern matching that of wild-type enzyme after transfection to RBL cells. Also, the amount of proform released into the medium, as compared with retained protein, was similar for the different forms of procathepsin G.
Results from transfection of 32D cells further substantiated that lack of both the amino-terminal propeptide and a functional catalytic site does not interfere with processing of the protein. Thus, both wild-type28 and the double-mutant form of procathepsin G was processed from a 32.5-kD proform into a 31-kD form and to a minor extent to a 30-kD form (data not shown). However, processing of both wild-type and mutated procathepsin G was not completed after 4 hours of chase, indicated by the presence of both the 32.5-kD proform and the 31-kD form, consistent with earlier results suggesting a retarded processing in 32D cells as compared with RBL cells.28 29Release of the proform into the medium was observed also in 32D cells.
The double-mutant procathepsin G acquires a mature amino-terminal cotranslationally.
By deletion of the two-residue amino-terminal propeptide, which normally is removed in pregranule or granule structures 60 to 90 minutes after translation,13,19 we assumed that the signal peptidase site34 would be preserved, leading to the cotranslational appearance of a mature amino-terminus lacking the propeptide. CatG/Gly201/ΔGly19Glu20 in RBL cells was subjected to radiosequencing after biosynthetic labeling with3H-isoleucine. Figure 3 shows that, after 30 minutes of radio-labeling of RBL/CatG/Gly201/ΔGly19Glu20cells, peak radioactivity appeared in the initial fractions of the amino acid-sequencing reaction, indicating the presence of a mature amino-terminus with 2 isoleucines. However, the expected amino-terminus starting with 2 isoleucines should result in the appearance of peak radioactivity only in the first 2 fractions. The fact that 3, and not only 2, of the initial fractions contained substantial amounts of radioactivity could indicate that more than one signal peptidase cleavage site is present, resulting in a mixture of newly synthesized mutant procathepsin G, with a portion of the protein having a remaining amino acid from the signal peptide before the 2 isoleucines of the normal, mature amino-terminus of cathepsin G. Alternatively, the appearance of radioactivity in fractions beyond the initial 2 fractions may be due to inefficient peptide degradation during the radiosequencing reaction. Similar results were obtained with 32D/CatG/Gly201/ΔGly19Glu20cells, demonstrating the presence of amino-terminal isoleucines after radio-labeling (data not shown), indicating the cotranslational appearance of a mature amino-terminal peptide sequence.
Amino-terminal radiosequencing of double-mutant procathepsin G in RBL cells. RBL/CatG/Gly201/▵Gly19Glu20 cells were pulse-labeled with 3H-isoleucine for 30 minutes as described in the Materials and Methods. After pulse-labeling, 100 × 106 cells were subjected to solubilization, immunoprecipitation, SDS-PAGE, and transfer to a PVDF membrane by Western blotting. Radioactive bands containing pulse-labeled 32.5-kD CatG/Gly201/▵Gly19Glu20 were excised and subjected to amino acid degradation. The amount of radioactivity in the initial 9 cycles of each sequence analysis is shown (dpm, disintegrations per minute).
Amino-terminal radiosequencing of double-mutant procathepsin G in RBL cells. RBL/CatG/Gly201/▵Gly19Glu20 cells were pulse-labeled with 3H-isoleucine for 30 minutes as described in the Materials and Methods. After pulse-labeling, 100 × 106 cells were subjected to solubilization, immunoprecipitation, SDS-PAGE, and transfer to a PVDF membrane by Western blotting. Radioactive bands containing pulse-labeled 32.5-kD CatG/Gly201/▵Gly19Glu20 were excised and subjected to amino acid degradation. The amount of radioactivity in the initial 9 cycles of each sequence analysis is shown (dpm, disintegrations per minute).
The double-mutant procathepsin G acquires a mature conformation cotranslationally.
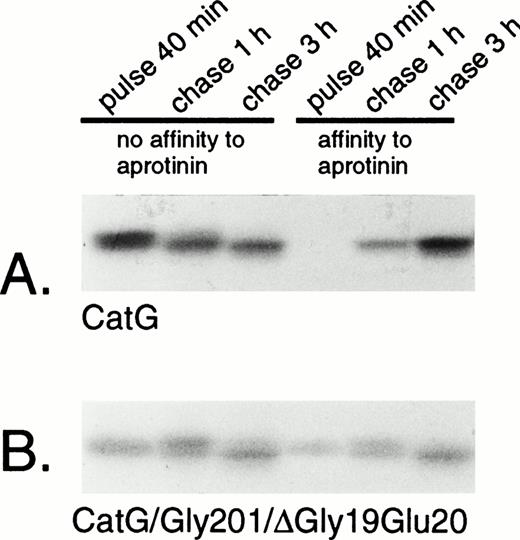
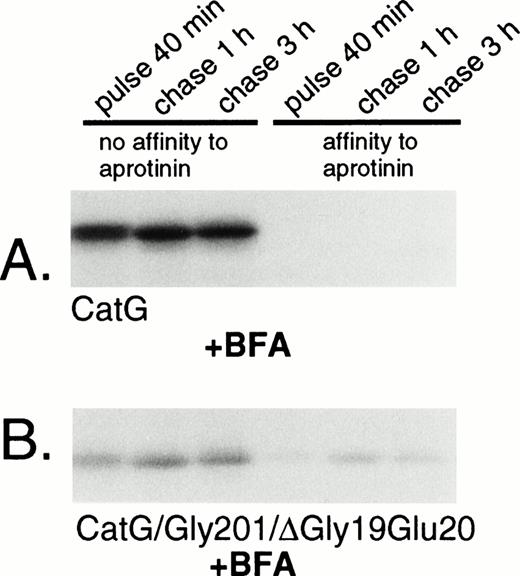
Removal of the amino-terminal propeptide of procathepsin G is a prerequisite for the conformational change leading to catalytic activation of the zymogen.13 The activation is reflected by the simultaneous acquisition of affinity to the serine protease inhibitor aprotinin.13,19 However, it should be noted that affinity for aprotinin is not necessarily equivalent to proteolytic capacity, as demonstrated by the strong affinity to aprotinin of azurocidin, a catalytically inactive serine protease homologue.35 Rather, the affinity to aprotinin parallels the conformational change due to removal of the propeptide,13 including formation of an internal salt bridge between the amino-terminal isoleucine and an aspartic acid close to the active catalytic site.36 To investigate the conformational status of the double-mutant procathepsin G, adsorption to aprotinin was performed. Wild-type cathepsin G, pulse-labeled for 40 minutes, did not show any affinity to aprotinin, indicating persistent proform-conformation (Fig 4A). However, after chase of the label for 1 hour, a substantial amount of the protein was bound to aprotinin, indicating conversion into mature conformation. The acquisition of affinity to aprotinin occurred concurrently with a reduction in the apparent molecular mass, representing the carboxyl-terminal processing. The expected cotranslational appearance of a mature conformation of CatG/Gly201/ΔGly19Glu20 was confirmed, as shown in Fig 4B. Already after 40 minutes of pulse-labeling, a considerable amount of the double-mutant procathepsin G was bound to aprotinin and the fraction of protein, with affinity for the protease inhibitor, increased further during chase of the label. This increase occurred concomitantly with the carboxyl-terminal processing. Normally, the removal of the amino-terminal dipeptide and the carboxyl-terminal peptide extension of procathepsin G occurs in post-Golgi structures, as demonstrated by the inhibition of processing by brefeldin A.18 To further substantiate that CatG/Gly201/ΔGly19Glu20 achieved a mature conformation without further proteolytic processing, we investigated the acquisition of affinity to aprotinin after biosynthetic radio-labeling in the presence of brefeldin A. Brefeldin A induces the disassembly of the Golgi complex, thus blocking the transport of proteins from the endoplasmic reticulum (ER) to the Golgi apparatus.37 Figure 5A demonstrates that brefeldin A completely abrogates the processing of wild-type procathepsin G, as judged by the lack of affinity to aprotinin and the absence of carboxyl-terminal processing, thus confirming previous data.18CatG/Gly201/ΔGly19Glu20, on the other hand, showed affinity to aprotinin despite the presence of brefeldin A with inhibition of proteolytic processing (Fig 5B). Thus, it can be concluded that CatG/Gly201/ΔGly19Glu20 obtains a mature conformation also in the absence of transfer to post-Golgi structures in which proteolytic processing normally occurs. Furthermore, removal of the carboxyl-terminal peptide extension is probably not necessary for enzymatic activation, because the double-mutant procathepsin G did bind to aprotinin in the absence of proteolytic processing.
Adsorption of wild-type and double-mutant cathepsin G to aprotinin-agarose. (A) RBL/CatG and (B) RBL/CatG/Gly201/▵Gly19Glu20 cells were labeled with 35S-methionine/35S-cysteine for 40 minutes and chased for 1 and 3 hours. At timed intervals, aliquots of labeled cells (20 × 106) were withdrawn for analyses. Cell lysis and adsorption to aprotinin-agarose were performed as described in the Materials and Methods. No affinity to aprotinin represents labeled protein that did not bind to aprotinin (non-active conformation), whereas material with affinity to aprotinin was eluted after binding (active conformation). After immunoprecipitation and SDS-PAGE, fluorography was performed. The fluorograms were exposed for 7 days.
Adsorption of wild-type and double-mutant cathepsin G to aprotinin-agarose. (A) RBL/CatG and (B) RBL/CatG/Gly201/▵Gly19Glu20 cells were labeled with 35S-methionine/35S-cysteine for 40 minutes and chased for 1 and 3 hours. At timed intervals, aliquots of labeled cells (20 × 106) were withdrawn for analyses. Cell lysis and adsorption to aprotinin-agarose were performed as described in the Materials and Methods. No affinity to aprotinin represents labeled protein that did not bind to aprotinin (non-active conformation), whereas material with affinity to aprotinin was eluted after binding (active conformation). After immunoprecipitation and SDS-PAGE, fluorography was performed. The fluorograms were exposed for 7 days.
Adsorption of wild-type and double-mutant cathepsin G to aprotinin-agarose in the presence of brefeldin A. (A) RBL/CatG and (B) RBL/CatG/Gly201/▵Gly19Glu20 cells were preincubated with brefeldin A (5 μg/mL) for 60 minutes, whereupon pulse-labeling and chase of the label were performed in the continued presence of brefeldin A. Lysis and adsorption to aprotinin was performed as described in the legend to Fig 4. The fluorograms were exposed for 7 days.
Adsorption of wild-type and double-mutant cathepsin G to aprotinin-agarose in the presence of brefeldin A. (A) RBL/CatG and (B) RBL/CatG/Gly201/▵Gly19Glu20 cells were preincubated with brefeldin A (5 μg/mL) for 60 minutes, whereupon pulse-labeling and chase of the label were performed in the continued presence of brefeldin A. Lysis and adsorption to aprotinin was performed as described in the legend to Fig 4. The fluorograms were exposed for 7 days.
The double-mutant procathepsin G acquires complex carbohydrates and is proteolytically processed.
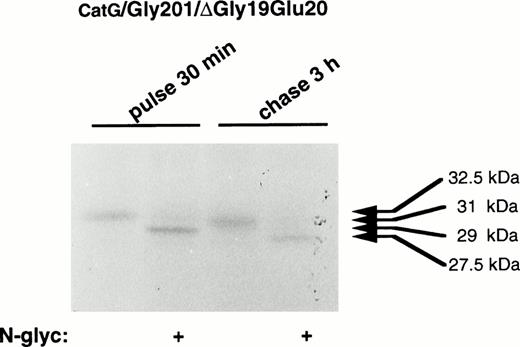
To characterize the processing of the present mutant forms of procathepsin G, digestion with N-glycosidase F was performed. As earlier demonstrated,19 pulse-labeled procathepsin G showed a molecular mass of 29 kD after digestion with N-glycosidase F, indicating the presence of asparagine-linked carbohydrates of approximately 3.5 kD (data not shown). After 3 hours of chase, the molecular mass of the processed protein was, upon digestion with N-glycosidase F, reduced from 31 to 27.5 kD, indicating that proteolytic processing of approximately 1.5 kD took place. When the double mutant was examined, similar results were found (Fig 6), demonstrating that CatG/Gly201/ΔGly19Glu20 is proteolytically processed to the same extent as the wild-type proenzyme. These results indicate that the proteolytic processing of procathepsin G is not dependent on autocatalysis, because both mutant forms, lacking enzymatic activity, were proteolytically processed similar to wild-type proenzyme.
Digestion of double-mutant cathepsin G with N-glycosidase F. RBL/CatG/Gly201/▵Gly19Glu20cells were pulse-labeled with35S-methionine/35S-cysteine for 30 minutes, whereupon the label was chased for 3 hours. At each time point, 50 × 106 cells were lysed and cathepsin G was immunoprecipitated. Half of the material was subjected to digestion with N-glycosidase F (indicated with “+”). Material without added N-glycosidase F, but otherwise treated identically, is shown as controls. The fluorogram was exposed for 5 days.
Digestion of double-mutant cathepsin G with N-glycosidase F. RBL/CatG/Gly201/▵Gly19Glu20cells were pulse-labeled with35S-methionine/35S-cysteine for 30 minutes, whereupon the label was chased for 3 hours. At each time point, 50 × 106 cells were lysed and cathepsin G was immunoprecipitated. Half of the material was subjected to digestion with N-glycosidase F (indicated with “+”). Material without added N-glycosidase F, but otherwise treated identically, is shown as controls. The fluorogram was exposed for 5 days.
Early after synthesis, procathepsin G acquires resistance to digestion with Endo H, demonstrating processing of its oligosaccharides into complex forms.19 23 Because this processing is known to take place in the Golgi, acquisition of resistance to Endo H demonstrates the translocation of the protein from the ER to the Golgi. When radio-labeled protein from RBL/CatG/Gly201/ΔGly19Glu20 cells was subjected to digestion with Endo H, it was demonstrated that almost all protein was Endo H-resistant after 15 minutes of chase (data not shown), thus indicating that the early processing of the double-mutant form was identical to that of wild-type procathepsin G.
The mutant forms of procathepsin G are translocated to granules in RBL and 32D cells.
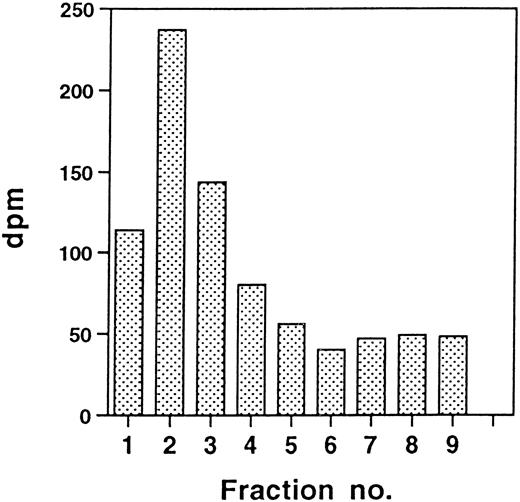
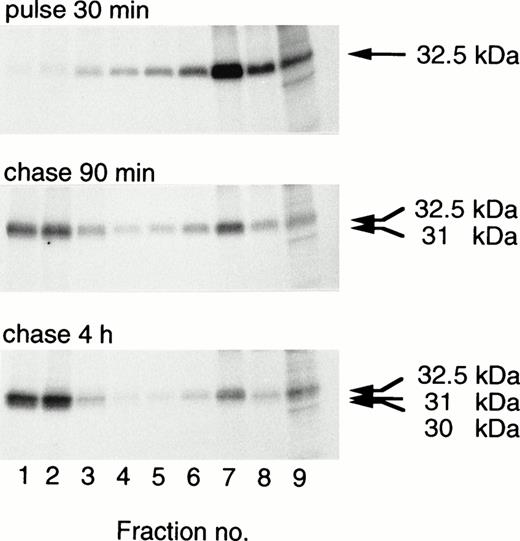
Transfected procathepsin G is translocated to granules, as indicated by the transfer with time to dense subcellular fractions containing granules.19,20 28 The intracellular distribution of CatG/Gly201/ΔGly19Glu20 in transfected RBL cells or 32D cells was investigated by use of subcellular fractionation. CatG/Gly201/ΔGly19Glu20 was translocated to dense fractions in RBL cells (Fig 7) and in 32D cells (data not shown). Similar results were obtained for CatG/Gly201 (data not shown). No obvious differences between the subcellular transfer of wild-type procathepsin G and that of the mutant forms of procathepsin G were evident. In contrast to what is seen in whole cell lysates (Fig2B), a minor amount of the 32.5-kD form is still visible after 4 hours of chase. The reason for this is not clear, but it could be the higher resolution in subcellular fractionation, compared with whole cells. The results demonstrate that procathepsin G lacking the amino-terminal dipeptide and thus cotranslationally adopting a mature conformation is efficiently sorted to granules.
Targeting of double-mutant procathepsin G to granules in RBL cells. RBL/CatG/Gly201/▵Gly19Glu20 cells were pulse-labeled for 30 minutes followed by chase for 90 minutes and 4 hours. At the times indicated, 100 × 106 cells were homogenized, after which subcellular fractionation was performed, with subsequent collection of eight 0.8 mL subcellular fractions with decreasing density and with fraction no. 9 containing all cytosol. Fractions were solubilized and subjected to immunoprecipitation with polyclonal anti-cathepsin G antiserum. Analyses of immunoprecipitates were as described in the legend to Fig 2. The different processing forms of cathepsin G are indicated with arrows to the right. The fluorograms were exposed for 3 weeks.
Targeting of double-mutant procathepsin G to granules in RBL cells. RBL/CatG/Gly201/▵Gly19Glu20 cells were pulse-labeled for 30 minutes followed by chase for 90 minutes and 4 hours. At the times indicated, 100 × 106 cells were homogenized, after which subcellular fractionation was performed, with subsequent collection of eight 0.8 mL subcellular fractions with decreasing density and with fraction no. 9 containing all cytosol. Fractions were solubilized and subjected to immunoprecipitation with polyclonal anti-cathepsin G antiserum. Analyses of immunoprecipitates were as described in the legend to Fig 2. The different processing forms of cathepsin G are indicated with arrows to the right. The fluorograms were exposed for 3 weeks.
DISCUSSION
In the present work, we have transfected mutant forms of human preprocathepsin G to 2 rodent hematopoietic cell lines (RBL-1 and 32D cl3) to investigate the importance of the proform-conformation of cathepsin G for targeting to granules. Our results suggest that premature absence of the amino-terminal propeptide impairs cell survival. This conclusion is based on the fact that we were not able to establish stable cell clones expressing a mutant form of procathepsin G lacking only the amino-terminal propeptide (CatG/ΔGly19Glu20). Moreover, transient transfection of this mutant to the simian kidney cell line COS-7 failed to result in detectable amounts of protein, in contrast to a comparable transfection of wild-type preprocathepsin G to COS-7 cells. Further, cotransfection of the mutant form of preprocathepsin G with a plasmid expressing β-gal resulted in a statistically significant lower number of β-gal–positive cells compared with controls 3 days after transfection. These results are partially in contrast to those from transient transfections of COS-7 cells with mutant granzyme B lacking the amino-terminal propeptide, showing synthesis of propeptide-deleted granzyme B.14,16 However, in these experiments, the levels of granzyme B were in fact lower in cells transfected with the propeptide-deleted form of this protease than in cells transfected with wild-type enzyme.16 It should be emphasized that our present results should be interpreted cautiously, because they only provide indirect lines of evidence for the adverse effect of prematurely activated procathepsin G. However, recent reports have shown that leukocyte elastase38 as well as cathepsin G39 can in fact induce apoptosis, in the latter case by activating pro-caspase-7.
In any case, our results led us to perform extended site-directed mutagenesis, also inactivating the functional catalytic site, and thereby stable clones expressing procathepsin G lacking the amino-terminal propeptide were easily obtained. This enabled us to investigate the role of the conformations of the protein for sorting to granules. Furthermore, the importance of the carboxyl-terminal processing for acquisition of a mature conformation could be studied.
Both azurophil granule and lysosomal proteins are often synthesized as larger precursors, which are subjected to late proteolytic processing in granule structures. Because the three-dimensional structures of the proforms probably differ from those of the mature proteins, recognition for sorting to granules could be limited to unprocessed forms that, upon arrival in granules, are converted to mature proteins. The proform-conformation would then be a prerequisite for sorting to granules. Some reports are compatible with this notion; in experiments with a mutant form of human prochymase lacking the amino-terminal dipeptide, it was suggested that the absence of the dipeptide rendered the nascent protein susceptible to intracellular proteolysis, possibly due to aberrant cellular trafficking, implying an important role of the dipeptide for subcellular sorting.15 Recent reports on neuroendocrine cells also indicate an important role of the proform-conformation for sorting; some neuroendocrine prohormones (eg, proinsulin and proenkephalin) bind specifically to a membrane-bound form of the processing enzyme carboxypeptidase E, which functions as a sorting receptor to regulated secretion.26 Also, recognition of the protease cleavage site in prorenin is sufficient to direct proteins to regulated secretion.27 Therefore, it could be that processing enzymes of neutrophil serine proteases, by binding to the proform of the enzyme, are important also for sorting of this family of proteins. However, the present data argue against this concept. It was shown that double-mutant procathepsin G, lacking the amino-terminal propeptide and functional catalytic site, was cotranslationally converted to a mature conformation, as judged by the mature amino-terminal sequence and early acquisition of affinity to the protease inhibitor aprotinin. Furthermore, this activation occurred also in the presence of brefeldin A, indicating that it took place in pre-Golgi or Golgi structures. Thus, the double-mutant procathepsin G did not adopt the proform-conformation, but was instead cotranslationally, or shortly after, adopting the mature conformation and should therefore not be a substrate for conformation-dependent processing enzymes. Nevertheless, the amount of protein released into the medium versus that transferred to granules was not increased, which let us conclude that sorting to granules is not restricted to the proform of cathepsin G. Neither was the amount of double-mutant procathepsin G retained intracellularly increased, arguing against the alternative hypothesis that conversion of proform into a mature conformation causes retention, by that constituting a mechanism for cellular retention and granule formation. Thus, the proform-conformation of procathepsin G seems not to have a role in sorting, but rather acts to keep the zymogen catalytically inactive during intracellular transfer of the protein.
Despite the cotranslational emergence of a mature amino-terminal sequence, the fraction of mutant protein with affinity to aprotinin, indicating conformational maturation, increased with time. This is unlikely to be due to further proteolytic processing, because this fraction also increased in the presence of brefeldin A, which inhibits transfer to compartments where proteolytic processing normally occurs. Processing of carbohydrates is also a far-fetched explanation, because the asparagine-linked oligosaccharides on cathepsin G are dispensable for conformational maturation.28 Furthermore, it was recently demonstrated by x-ray crystallography that the single carbohydrate chain on cathepsin G is distant from the active site and in fact points away from it. Possibly, the increased affinity to aprotinin could reflect the stabilization of the mature conformation by formation of internal salt bridges found in the mature protein.36
The crystal structure of cathepsin G confirms that mature cathepsin G is carboxyl-terminally truncated, ending with Ser or Phe, which could be a potential autocatalytic site.36 The carboxyl-terminal processing of cathepsin G might follow activation and result from intramolecular or intermolecular autocatalysis in analogy to the proteolytic processing of the lysosomal cysteine protease cathepsin B40,41 and mast cell tryptase,42 which involves the action of the hydrolase itself. However, the finding of similar processing of mutant procathepsin G deficient of a functional catalytic site and wild-type proenzyme in both RBL and 32D cells makes it unlikely that the carboxyl-terminal processing is mediated by cathepsin G itself. But, it cannot be entirely excluded that endogenous cathepsin G-like enzymes in RBL or 32D cells are responsible for an intermolecular carboxyl-terminal processing. A functional role for the carboxyl-terminal extension has not been proven. The presence of the carboxyl-terminal peptide extension of procathepsin G does not seem to be necessary for initial folding, activation, or sorting.20Because the double-mutant procathepsin G adopted a mature conformation (as judged by the affinity to aprotinin) in the absence of carboxyl-terminal processing, the present results demonstrate that enzymatic activation can occur without this processing.
ACKNOWLEDGMENT
The authors are most grateful for the expert technical assistance by Eva Nilsson and Karin Svensson.
Supported by the Swedish Medical Research Council (Project No. 11546), the Greta and Johan Kock Foundation, the Alfred Österlund Foundation, the Crafoord Foundation, the Anna-Greta Crafoord Foundation, Funds of Reumatikerförbundet, the John Persson Foundation, the Swedish Society for Medical Research, and Funds of Lunds sjukvårdsdistrikt.
Presented in part at the European Congress for Molecular Cell Biology (ECBO), Brighton, UK, March 22-25, 1997 (abstract no. H-5041) and at the European Molecular Biology Organization (EMBO)-Workshop “Protein sorting and processing in the secretory pathway,” Annaberg/St Martin, Austria, January 13-18, 1998.
Address reprint requests to Daniel Garwicz, MD, Research Dept. 2, E-blocket, University Hospital, SE-221 85 Lund, Sweden; e-mail: Daniel.Garwicz@hematologi.lu.se.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal