Abstract
Cytokines and growth factors induce activation of the family of signal transducers and activators of transcription (Stats) that directly activate gene expression. Recently, constitutively activated Stat1, Stat3, and Stat5 were identified in nuclear extracts of acute myeloid leukemia (AML) patients, suggesting involvement of constitutive Stat activity in the events of leukemogenesis. In the present study, blasts of nine AML cases were investigated for the constitutive binding activity of the recently identified transcription factor LIL-Stat (LPS- and IL-1-inducible Stat). Band-shift assays were performed using the LPS-and IL-1-responsive element (LILRE) oligonucleotide, a gamma interferon activation site-like site that is present in the human IL-1β promoter. Constitutive LIL-Stat binding activity was observed in three leukemic cell lines and in seven out of nine AML cases. Transient transfection studies with a reporter plasmid containing three sequential LIL-Stat binding sites showed distinct transcriptional activity of LIL-Stat only in those AML blasts that constitutively expressed LIL-Stat. In CD34+ cells LIL-Stat also constitutively bound to its consensus sequence. However, when these cells were cultured in the presence of macrophage-colony stimulating factor (M-CSF) and stem cell factor (SCF) for differentiation along the monocytic lineage, the LIL-Stat binding activity disappeared totally. In agreement with these findings neither mature monocytes nor granulocytes showed constitutive or inducible LIL-Stat binding activity. We conclude that the LIL-Stat transcription factor is constitutively activated in undifferentiated and leukemic hematopoietic cells, but not in mature cells. This may suggest a role for this transcription factor in the process of differentiation.
© 1998 by The American Society of Hematology.
THE STAT PROTEINS (signal transducers and activators of transcription) are a family of transcription factors involved in cytokine, and growth factor signaling.1,2Cytokine-receptor interaction, followed by oligomerization of the receptor chains, activates the Janus Kinases (JAK), which in turn phosphorylate specific tyrosine residues within the intracellular part of the cytokine receptor.3,4 Subsequently, the STAT proteins are selectively recruited to various cytokine receptors by virtue of their peptide-binding specificities mediated by their SRC homology 2 (SH2) domains.5 Phosphorylation of receptor-associated STATs by JAKs results in dissociation of the STATs from the receptor, dimerization, and subsequent translocation to the nucleus. The DNA-binding target of STAT dimers is the consensus gamma interferon (IFN-γ) activation site (GAS), which was initially reported to bind the IFN-γ–induced Stat1.6 Until recently, six STAT factors had been identified, Stat1 through Stat6.7-11
Recent identifications of constitutive Stat activation in malignant cells are the first attempts to link the aberrations in the JAK/Stat pathway to the malignant phenotype. One such example is the infection of T cells with the human T-cell leukemia virus type I (HTLV-I) that results in the activation of Stat5.12 In anaplastic large T-cell lymphoma (ALCL) cell lines constitutive phosphorylation of JAK3, Stat5, and Stat3 were reported.13 Furthermore, an amino acid substitution in the Drosophilia homolog of the JAK kinase (DrosophiliahopTum-1) gene causes leukemia-like hematopoietic defects.14 Overactivity of the mutated protein was responsible for the malignant neoplasia. In the BA/F3 hematopoietic cell line constitutive phosphorylation of JAK2 mediated induction of bcl-2, which in turn resulted in delayed cell death.15 So far, two reports have described the constitutive activition of Stat factors in cells of patients suffering from acute myeloid leukemia (AML).16 17 Band-shift assays combined with specific antibody supershifts showed that spontaneously active Stat1, Stat3, and Stat5 were present in a considerable number of AML cases, with Stat3 expression being most pronounced.
Recently, Tsukada et al18 reported the existence of a putative novel Stat factor, termed LIL-Stat (Lipopolysaccharide (LPS)- and Interleukin-1 (IL-1)-inducible Stat) in a murine thymoma and two human monocytic cell lines. LIL-Stat binds to a GAS-like element as it is found in the human prointerleukin-1β gene, named LILRE, for LPS- and IL-1–responsive element.19 20 The GAS-like binding activity of LIL-Stat contrasts with those of the other Stats in the requirement for a G residue at position 8 (TTCCTGAGA). Furthermore, LIL-Stat was recognized by an antibody specific for the N terminus of Stat1, but not by those specific for the C terminus of Stat1 or any other GAS-binding Stats. LIL-Stat was rapidly induced after stimulation with either LPS, IL-1, or IL-6. These features, together with the requirement of tyrosine phosphorylation for DNA-binding, led to the aknowledgement of this novel Stat factor, LIL-Stat.
In the present study we investigated blasts of AML patients for the presence and functional activity of LIL-Stat. The potential relevance of spontaneous LIL-Stat expression in AML is underscored by the fact that LIL-Stat binds the IL-1β promoter. Constitutive expression of this transcription factor may thus be involved in the autocrine production of IL-1β, a feature observed in 30% to 40% of AML cases.21
We show that the novel transcription factor LIL-Stat binds spontaneously to its consensus sequence in seven of nine AML cases investigated. Transfection studies indicated that LIL-Stat is transcriptionally active in AML blasts that express activated LIL-Stat. CD34+ cells also show constitutive binding activity of LIL-Stat, but LIL-Stat binding disappears on differentiation along the myelomonocytic lineage. In fully differentiated monocytes and granulocytes, LIL-Stat is not expressed. Taken together, these data suggest that LIL-Stat may play a role in the differentiation process of hematopoietic cells. Furthermore, constitutive LIL-Stat activation may be involved in constitutive IL-1β production in AML blasts.
MATERIALS AND METHODS
Patient population and isolation of AML cells.
Peripheral blood cells or bone marrow cells from nine patients with AML were studied. The AML cases were defined according to the classification of the FAB committee as M1 to M6.22 AML blasts were isolated by density-gradient centrifugation as described.23 The cells were cryopreserved in aliquots of 20 to 30 × 106 cells in 10% dimethylsulfoxide (DMSO; Sigma, St Louis, MO) and 10% fetal bovine serum (FBS; Hyclone, Logan, UT), employing a method of controlled freezing and storage in liquid nitrogen. After thawing, T lymphocytes were depleted by 2-aminoethylisothioronium bromide (AET)-treated sheep red blood cell (SRBC) rosetting. The cell population consisted of more than 98% AML blasts as determined by May-Grünwald-Giemsa staining. Fluorescence-activated cell sorting (FACS) analysis showed <1% CD3 (Becton Dickinson, Sunnyvale, CA) -positive cells. AML blasts were cultured at 37°C at a density of 1 × 106/mL in RPMI 1640 media (Flow, Rockville, MD) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 6 ng/mL of colistine, and 0.1% FBS for nuclear extract preparation or 10% FBS for transfection experiments.
Preparation of CD34+ cells, monocytes, granulocytes, and cell culture.
CD34+ cells were isolated over a column containing magnetically labeled anti-CD34+ antibodies (Isolex 300 system; Baxter, Deerfield, IL). For maturation experiments, isolated CD34+ cells were grown in RPMI 1640 medium supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 6 ng/mL colistine, macrophage-colony stimulating factor (M-CSF)(10 ng/mL)(Genetics Institute, Cambridge, MA), and stem cell factor (SCF)(10 ng/mL)(Amgen Inc, Thousand Oaks, CA). After 5 days of culture cells were analyzed for CD34+ and CD14+ expression by FACS analysis (Becton Dickinson). Morphology of the cell population was assessed by May-Grünwald-Giemsa staining.
Peripheral blood cells were obtained from healthy blood donors, and granulocytes and mononuclear cell suspensions were prepared by Ficoll-Hypaque density-gradient centrifugation. Granulocytes were enriched by lysis of the red blood cells with 155 mmol/L NH4CL, 10 mmol/L KHCO3, 0.1 mmol/L EDTA and after washing the granulocytes, cells were plated at a density of 1 × 106 cells/mL in RPMI 1640 supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 6 ng/mL colistine, and 10% FBS. Granulocytes were cultured for 2 hours before stimulation. Mononuclear cells were isolated from the interphase and washed twice in phosphate-buffered saline (PBS). T lymphocytes were depleted by AET-treated sheep red blood cell (SRBC) rosetting. Monocytes were further enriched by plastic adherence (1h, 37°C). Monocytes were cultured overnight at 37°C at a density of 1 × 106 cells/mL RPMI 1640 supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 6 ng/mL colistine, and 0.1% FBS, before stimulation.
The cell lines employed in this study included B1 and U937, human monocytic cell lines, and TF-1, a human erythroleukemic cell line.24-26 Both B1 and U937 were cultured in RPMI 1640 supplemented with 10% FBS, and TF-1 cell line was cultured in RPMI 1640 supplemented with 5% FBS and 10 ng/mL IL-3.
After overnight culture in 0.1% FBS, cells were stimulated for the appropriate time courses with IL-1β (100 U/mL)(Boehringer Mannheim, Mannheim, Germany), IL-6 (10 ng/mL)(Sandoz, Basel, Switzerland), IFN-γ (20 U/mL)(Alpha Therapeutic Corp, Los Angeles, CA), and LPS (1 μg/mL)(Sigma).
Electrophoretic mobility shift assay (EMSA).
After an overnight culture period (for cell lines, monocytes, and AML cells) or after a 2-hour culture period (granulocytes), cells were stimulated with IL-1β (100 U/mL), IL-6 (10 ng/mL), IFN-γ (200 U/mL), M-CSF (10 ng/mL), and SCF (10 ng/mL) for various periods of time. Nuclear extracts were prepared according to the miniscale procedure as described, with the addition of protease inhibitor cocktail (Complete, Boehringer Mannheim).27 Nuclear extracts were divided into small aliquots and stored at −80°C. Double-stranded synthetic oligonucleotide probes were used in the gel retardation assay. Oligonucleotide specific for LIL-Stat is derived from the human IL-1β promoter, LIL-RE19: 5′-AGCTTATAAG-AGGTTTCACTTCCTGAGAGTCGA-3′; Stat1 oligonucleotide is the GAS-site from the FcgammaRI gene: 5′-GTATTCCCAGAAAAGGAAC-3′; hSIE (serum-inducible element)-site from the c-fos promoter is specific for Stat3: 5′-GAGCAGTTCCCGTCAATCCCTcccg-3′; Stat5 oligonucleotide is derived from the bovine β-caseı̈ne gene promoter: 5′-AGATTTCTAGGAATTCAAATCCccct-3′. Fifty ng of high-performance liquid chromatography (HPLC) -purified single-stranded oligonucleotide were labeled with T4-polynucleotide kinase and [γ32P] adenosine triphosphate (ATP) (3000 Ci/mmol; Amersham, Buckinghamshire, UK), separated from nonincorporated radiolabel by sephadex G50 chromatography, ethanol precipitated, dried, and dissolved in 20 μL of 10 mmol/L Tris-HCl (pH 7.5), 50 mmol/L NaCl, 10 mmol/L MgCl2, 1 mmol/L EDTA, and 1 mmol/L dithiothreitol (DTT), containing a fourfold excess of the opposite strand. Annealing of the two strands was performed by heating the mixture for 2 minutes at 90°C and slow cooling to room temperature. Five μg nuclear extract and 0.1 ng double-stranded labeled oligonucleotide were incubated in 20 mmol/L HEPES (pH 7.9), 60 mmol/L KCl, 0.06 mmol/L EDTA, 0.6 mmol/L DTT, 2 mmol/L spermidine, 10% glycerol, supplemented with 2 μg poly(dI-dC). The binding reaction was performed at 26°C for 25 minutes. Competition experiments were performed by adding a 100-fold molar excess of unlabeled self or nonself double-stranded oligonucleotides.
Supershift experiments were performed by incubating the nuclear extracts (1 hr, 4°C) with polyclonal antibodies (1μg) against the C terminus of Stat1 (Stat1 p84/p91 antibody; Santa Cruz Biotechnology, Santa Cruz, CA), against the N Terminus of Stat1 (anti-ISGF3; Transduction Laboratories, Lexington, KY), against Stat3 (Stat3(C20)-antibody; Santa Cruz Biotechnology), or against phospho-tyrosine residue (PY20 antibody; Santa Cruz Biotechnology), before the addition of the labeled oligonucleotide. Nonspecific supershift experiments were performed with a polyclonal antibody against the p65 subunit of NF-κB (Santa Cruz Biotechnology).
The samples were loaded on prerun (30 minutes, 100 V) 4% (30:1) polyacrylamide gels and run for 1 hour at 150 V in 0.5 × Tris/borate/EDTA electrophoresis buffer (TBE) at room temperature. Gels were dried and exposed to Kodak XAR films (Sigma, St Louis, MO) at −80°C with an intensifying screen. Quantification of protein binding was performed by densitometry using a Phospho Imager (Molecular Dynamics, Sunnyvale, CA).
Transfection procedure.
The pGL3ti luciferase reporter plasmid was obtained by cloning the Adeno Major Late TATAA-box and the murine Terminal deoxynucleotidyl Transferase into the BglII and HindIII sites of the pGL3basic vector (Promega Corporation, Madison, WI). Subsequently, 3 consecutive LILRE sequences (5′-CGGGGTACCAGGTTTCACTTCCTGAGAGTCGAAGGTTTCACTTCCTGAGAGTCGAAGGTTTCACTTCCTGAGAGTCGAG-CTAGCTAGGCC-3′) were cloned into Kpn1 and Nhe1 sites of the pGL3ti plasmid, which was thus termed pGL3ti-LIL3. AML blasts were cultured in RPMI 1640 medium supplemented with 10% FBS for 16 hours and subsequently transfected with 25 μg of pGL3ti, or with 25 μg of pGL3ti-LIL3, containing 3 LIL-Stat binding sequences. In addition, the blasts were cotransfected with 2.5 μg of a CMV-CAT plasmid, to normalize for transfection efficiency. Electroporation (10 × 106 cells/200 μL RPMI) was performed at 240 V and 960 μF, and transfected cells were replated in 10 mL RPMI 1640 with 10% FBS. Twenty-four hours after electroporation, cells were harvested and assayed for both luciferase and CAT activity. Luciferase activity was measured by preparation of cell lysates using a commercially available luciferase assay kit (Promega) and quantification on the Anthos LUCY1 luminometer (Anthos Labtec Instruments Ges.m.b.H, Salzburg, Austria). CAT activity was determined with the commercially available CAT Elisa kit (Boehringer Mannheim).
Statistical analysis.
The student t-test for paired samples was used to determine significance of the transfection data.
RESULTS
Binding activity of LIL-Stat is induced in leukemic cell lines BI, U937 and TF-1.
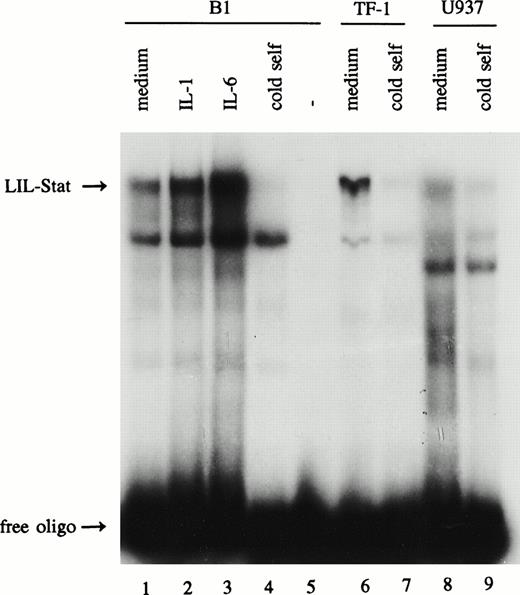
The leukemic cell lines B1, TF-1, and U937 were studied for the expression of the novel Stat factor LIL-Stat. After overnight incubation with 0.1% FBS, nuclear extracts were prepared from unstimulated cells, and from cells that were treated with IL-1 (100 U/mL) or IL-6 (10 ng/mL) for 10 minutes. Nuclear extracts were subsequently incubated with the LIL-RE oligonucelotide, the GAS-like sequence as it is found in the human IL-1β promoter for electrophoretic mobility shift assays (EMSAs).19 20Figure 1 shows that the putative Stat factor LIL-Stat is spontaneously expressed in all three leukemic cell lines (lanes 1, 6, and 8). Both IL-1 and IL-6 treatment resulted in an enhanced DNA-binding activity of the LIL-Stat factor in all three cell lines as determined by phosphoimager analysis. For the B1 cell line this is represented in Fig 1 (lanes 2 and 3).
LIL-Stat is constitutively activated in three leukemic cell lines. Five micrograms of nuclear protein derived from control, IL-1β(10’)-, and IL-6(10’)-treated B1 cells (lanes 1-3) and from control TF-1(lane 6) and U937 (lane 8) cells were analyzed for LIL-Stat binding activity by EMSA. Nuclear extracts were incubated with the32P-labeled oligonucleotide LILRE, the LIL-Stat consensus sequence from the IL-1β promoter. For each cell line competition experiments were performed with a 100-fold molar excess of unlabeled LILRE oligonucleotide (lane 4, 7, and 9, respectively). Labeled LILRE oligonucleotide alone is represented in lane 5.
LIL-Stat is constitutively activated in three leukemic cell lines. Five micrograms of nuclear protein derived from control, IL-1β(10’)-, and IL-6(10’)-treated B1 cells (lanes 1-3) and from control TF-1(lane 6) and U937 (lane 8) cells were analyzed for LIL-Stat binding activity by EMSA. Nuclear extracts were incubated with the32P-labeled oligonucleotide LILRE, the LIL-Stat consensus sequence from the IL-1β promoter. For each cell line competition experiments were performed with a 100-fold molar excess of unlabeled LILRE oligonucleotide (lane 4, 7, and 9, respectively). Labeled LILRE oligonucleotide alone is represented in lane 5.
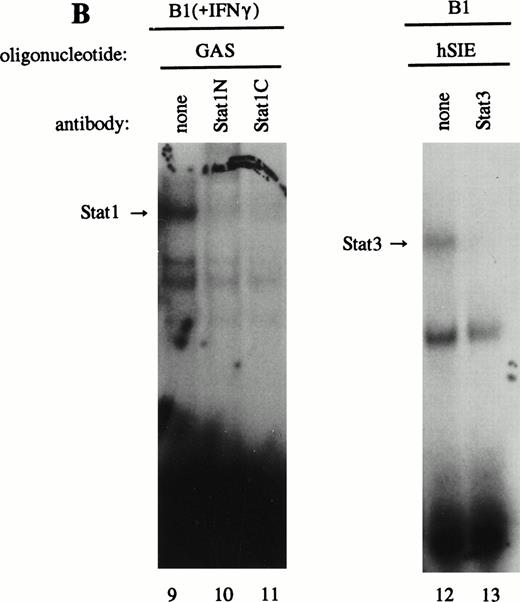
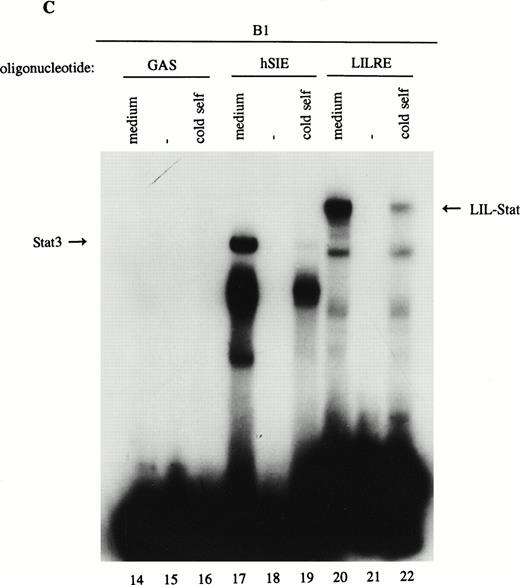
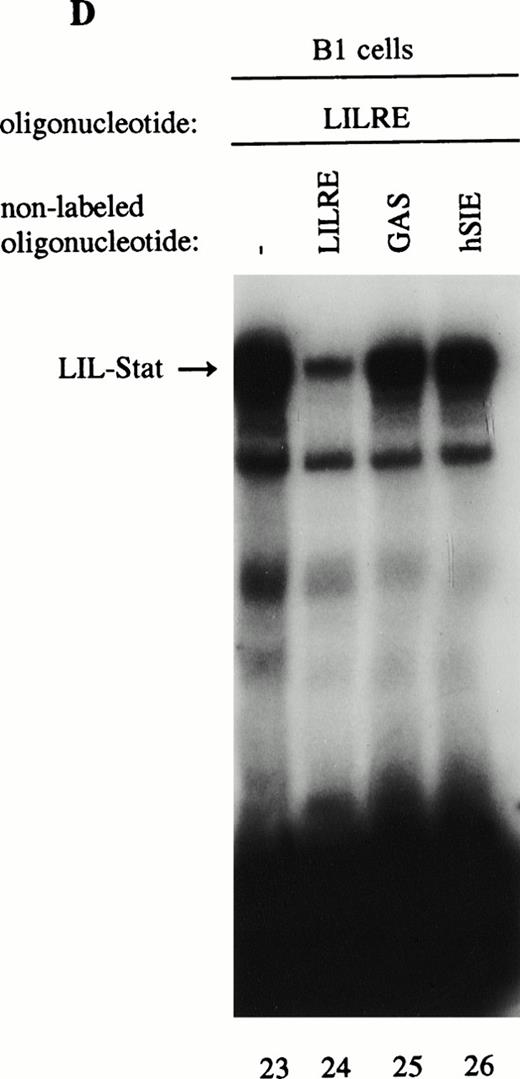
As previously described by Tsukada et al18 and further confirmed by our experiments, several features of this transcription factor indicated that this indeed most likely concerns the novel Stat transcription factor. Firstly, as is depicted in Fig 2A (lanes 1 and 2), nuclear extracts incubated with an antiphosphotyrosine antibody showed drastically reduced binding activity of the transcription factor. This would indicate that tyrosine phosphorylation of this factor plays a role in either dimerization and/or DNA-binding, a common feature attributed to the known Stat factors.6 8 Secondly, the binding activity of this putative LIL-Stat factor is inhibited when the nuclear extract is incubated with an antibody specific for the N-terminal region of the Stat1 transcription factor, suggesting binding of this antibody to the LIL-Stat transcription factor (Fig 2A, lanes 3 and 4). Supershift experiments with antibodies against the C terminus of Stat1 or against Stat3 did not affect the binding activity of LIL-Stat as quantified by phosphoimaging, indicating that this protein/DNA complex does not consist of either Stat1 or Stat3 (Fig 2A, lanes 5-7). Functionality of the antibodies against Stat1 and Stat3 was shown in Fig 2B, in which IFN-γ–treated (lanes 9-11) and control (lanes 12 and 13) nuclear extracts of the B1 cell line were incubated with the oligonucleotides for Stat1 (GAS-site from FcγRI gene) and Stat3 (SIE-site from c-fos promoter), respectively, and supershifted with the appropriate antibodies. Incubation of nuclear extracts from untreated B1 cells with the oligonucleotides specific for LIL-Stat, Stat1, and Stat3 showed differential constitutive binding activity of the transcription factors LIL-Stat, Stat1, and Stat3 in these cells (Fig 2C, lanes 14-22), with high constitutive activity of LIL-Stat and Stat3 and no spontaneous binding activity of Stat1. Moreover, Stat3 showed an electrophoretic mobility different from LIL-Stat, supporting the observation that LIL-Stat is a transcription factor different from Stat1 and Stat3. Finally, 100-fold molar excess of unlabeled oligonucleotides specific for Stat1 and Stat3, respectively, did not affect binding of the LIL-Stat protein to its consensus sequence LILRE, whereas unlabeled LILRE served as a trueful cold competition (Fig 2D, lanes 23-26).
LIL-Stat transcription factor is tyrosine phosphorylated and recognized by anti-Stat1N antibody. (A) Five micrograms of nuclear extract derived from control B1 cells was supershifted with antibodies against phospho-tyrosine residue (lane 2), Stat1 N-terminal region (lane 4), Stat1 C-terminal region (lane 6), and against Stat3 (lane 7). Quantification of LIL-Stat binding activity was performed by phosphoimager analysis. (B) Functionality of Stat1 and Stat3 antibodies was shown as follows: IFN-γ–treated B1 nuclear extracts were band-shifted with 32P-labeled Stat1 oligonucleotide from the FcγR1 promoter (lane 9) and supershifted with the antibodies against the N- (lane 10) and C- (lane 11) terminal Stat1 domains. Five micrograms of nuclear proteins derived from control B1 cells was band-shifted with the 32P-labeled Stat3 oligonucleotide from the c-fos promoter (lane 12) and supershifted with the Stat3 anitbody (lane 13). (C) Band-shift assays of control B1 nuclear extracts with the oligonucleotides for Stat1 (GAS), Stat3 (hSIE), and LIL-Stat (LILRE) showed differential binding to the Stat1, Stat3, and LIL-Stat oligonucleotide (lanes 14, 17, and 20, respectively). Lanes 15, 18, and 21 represent the respective oligonucleotides alone, whereas cold competition is represented in lanes 16, 19, and 22. (D) Band-shift assay of control B1 nuclear extract with the oligonucleotide for LIL-Stat (lane 23). Cold competition is performed with 100-fold molar excess of unlabeled oligonucleotide LILRE (lane 24), GAS (lane 25), and hSIE (lane 26).
LIL-Stat transcription factor is tyrosine phosphorylated and recognized by anti-Stat1N antibody. (A) Five micrograms of nuclear extract derived from control B1 cells was supershifted with antibodies against phospho-tyrosine residue (lane 2), Stat1 N-terminal region (lane 4), Stat1 C-terminal region (lane 6), and against Stat3 (lane 7). Quantification of LIL-Stat binding activity was performed by phosphoimager analysis. (B) Functionality of Stat1 and Stat3 antibodies was shown as follows: IFN-γ–treated B1 nuclear extracts were band-shifted with 32P-labeled Stat1 oligonucleotide from the FcγR1 promoter (lane 9) and supershifted with the antibodies against the N- (lane 10) and C- (lane 11) terminal Stat1 domains. Five micrograms of nuclear proteins derived from control B1 cells was band-shifted with the 32P-labeled Stat3 oligonucleotide from the c-fos promoter (lane 12) and supershifted with the Stat3 anitbody (lane 13). (C) Band-shift assays of control B1 nuclear extracts with the oligonucleotides for Stat1 (GAS), Stat3 (hSIE), and LIL-Stat (LILRE) showed differential binding to the Stat1, Stat3, and LIL-Stat oligonucleotide (lanes 14, 17, and 20, respectively). Lanes 15, 18, and 21 represent the respective oligonucleotides alone, whereas cold competition is represented in lanes 16, 19, and 22. (D) Band-shift assay of control B1 nuclear extract with the oligonucleotide for LIL-Stat (lane 23). Cold competition is performed with 100-fold molar excess of unlabeled oligonucleotide LILRE (lane 24), GAS (lane 25), and hSIE (lane 26).
In summary, binding of the factor to a GAS-like sequence, rapid induction by IL-1 and IL-6, tyrosine phosphorylation of the transcription factor, and binding of the antibody specific for the N-terminal part of Stat1, confirmed that this transcription factor possesses the characteristics of a Stat factor. In further description of experiments this transcription factor will thus be termed LIL-Stat.
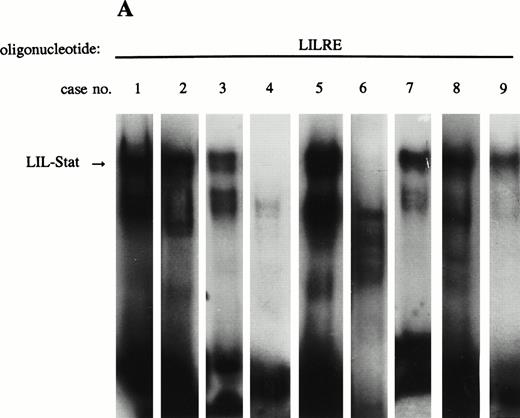
Constitutive binding activity of LIL-Stat is observed in 7 out of 9 AML cases. Comparison with constitutive activation of Stat1, Stat3, and Stat5.
Previous studies have identified the constitutive binding activity of Stat1, Stat3, and Stat5 in blasts of AML cases, although the frequency of occurrence is variable.16 17 Because LIL-Stat appeared to be abundantly expressed in the three leukemic cell lines we studied, we prepared nuclear extracts from blasts of nine AML cases to study the constitutive binding activity of LIL-Stat. FAB classification of the cases are shown in Table 1. Seven out of nine cases showed constitutive LIL-Stat binding to its consensus sequence (Fig 3A). Two cases (no. 4 and 6), FAB M4 and M5A, did not show binding activity of LIL-Stat, neither constitutive nor inducible.
Binding Activity of LIL-Stat, Stat1, Stat3, and Stat5 in 9 AML cases
| AML Case No. . | FAB Classification . | LIL-Stat . | Stat1 . | Stat3 . | Stat5 . |
|---|---|---|---|---|---|
| 1 | M2 | + | − | + | − |
| 2 | M5A | + | − | + | − |
| 3 | M2 | + | − | + | − |
| 4 | M4 | − | − | − | − |
| 5 | M4 | + | + | + | − |
| 6 | M5A | − | − | − | − |
| 7 | M4 | + | − | + | − |
| 8 | M2 | + | − | + | − |
| 9 | M2 | + | − | + | − |
| AML Case No. . | FAB Classification . | LIL-Stat . | Stat1 . | Stat3 . | Stat5 . |
|---|---|---|---|---|---|
| 1 | M2 | + | − | + | − |
| 2 | M5A | + | − | + | − |
| 3 | M2 | + | − | + | − |
| 4 | M4 | − | − | − | − |
| 5 | M4 | + | + | + | − |
| 6 | M5A | − | − | − | − |
| 7 | M4 | + | − | + | − |
| 8 | M2 | + | − | + | − |
| 9 | M2 | + | − | + | − |
Schematic representation of constitutive (+) or absence of constitutive (−) binding activity of LIL-Stat, Stat1, Stat3 and Stat5 in the nuclear extracts of 9 AML patients, as determined by EMSA.
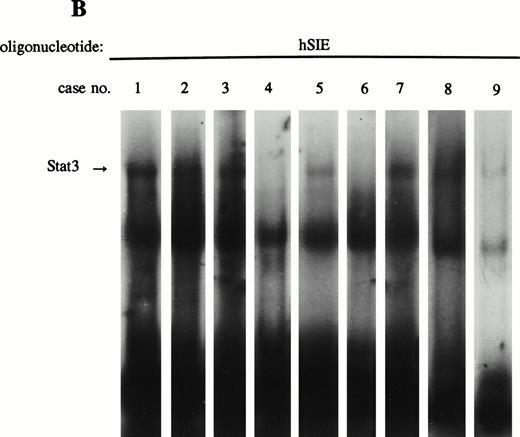
LIL-Stat is constitutively activated in nuclear extracts of seven out of nine AML cases. (A) Five micrograms of nuclear proteins from nine AML cases were incubated with 32P-labeled LILRE oligonucleotide for determination of constitutive LIL-Stat binding. (B) Similar to the above-mentioned experiment, nuclear extracts of nine AML cases were band-shifted with the Stat3-specific oligonucleotide.
LIL-Stat is constitutively activated in nuclear extracts of seven out of nine AML cases. (A) Five micrograms of nuclear proteins from nine AML cases were incubated with 32P-labeled LILRE oligonucleotide for determination of constitutive LIL-Stat binding. (B) Similar to the above-mentioned experiment, nuclear extracts of nine AML cases were band-shifted with the Stat3-specific oligonucleotide.
To compare the spontaneous binding activity of LIL-Stat with the constitutive binding of Stat1, Stat3, or Stat5, the nuclear extracts derived from the nine AML cases were also incubated with oligonucleotides specific for Stat1, Stat3, and Stat5. Supershift experiments with antibodies against Stat1, Stat3, and Stat5 confirmed that the binding to the respective oligonucleotides was specific (data not shown). Table 1 summarizes the constitutive DNA-binding activity of the different Stat factors in all 9 AML cases. All AML cases that showed spontaneous LIL-Stat binding activity also showed constitutively active Stat3 (Fig 3B). Stat1 only spontaneously bound to its consensus sequence in one (case 5, data not shown) out of nine cases, whereas Stat5 binding could not be detected in any of the cases.
Transactivational capacity of LIL-Stat only in AML cases which express LIL-Stat.
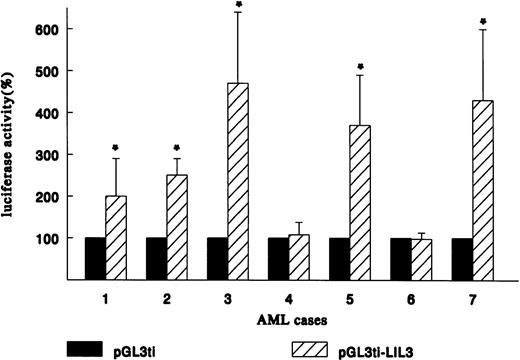
Because the LIL-Stat binding sequence (LILRE) is found in the human IL-1β promoter,19 we set out to study the possible transactivational capactity of the LIL-Stat/DNA complex in the AML cases. Three sequential LIL-Stat binding sites were cloned into a luciferase reporter construct containing the ti-minimal promoter (pGL3ti-LIL3), as described in Materials and Methods. Transfections were performed with 25 μg of the reporter construct pGL3ti-LIL3 or the empty vector pGL3ti and 2.5 μg of pCMV-CAT. We were able to succesfully transfect seven out of nine AML cases (cases 1 through 7). In Fig 4 the relative luciferase activity of each AML case after transfection with the empty vector pGL3ti was set at 100%. The results of at least three independent transfection experiments (mean ± SD) with pGL3ti-LIL3 are represented in the graph. In five out of seven cases a sizable induction in luciferase activity was observed after introduction of pGL3ti-LIL3 in the blasts. Case 1 showed a 2.0-fold ± 0.9 (P < .05) induction in luciferase activity with pGL3ti-LIL3 compared with pGL3ti. For cases 2, 3, 5, and 7 the upregulation of luciferase activities were 2.5-fold ± 0.4, 4.7-fold ± 1.7, 3.7-fold ±1.2, and 4.3-fold ± 1.7 (P < .05), respectively. For cases 4 and 6 no induction of transactivational activity could be observed after transfection with the reporter containing three LIL-Stat binding sites. The two cases that lacked enhancement of transcriptional activity of the LIL-Stat construct were identical to the two cases lacking constitutive DNA-binding activity of LIL-Stat.
LIL-Stat is transcriptionally active in 5 out of 7 AML cases. Blasts of AML cases 1 through 7 were transfected with the reporter constructs pGL3ti or pGL3ti-3LILRE together with pCMVCAT. CAT activity was used to normalize for transfection efficiency within 1 AML case. Luciferase activity obtained after transfection with pGL3ti control vector was set at 100%. Error bars represent standard deviation of at least three identical experiments (* P < .05).
LIL-Stat is transcriptionally active in 5 out of 7 AML cases. Blasts of AML cases 1 through 7 were transfected with the reporter constructs pGL3ti or pGL3ti-3LILRE together with pCMVCAT. CAT activity was used to normalize for transfection efficiency within 1 AML case. Luciferase activity obtained after transfection with pGL3ti control vector was set at 100%. Error bars represent standard deviation of at least three identical experiments (* P < .05).
LIL-Stat is not activated in fully differentiated monocytes or granulocytes.
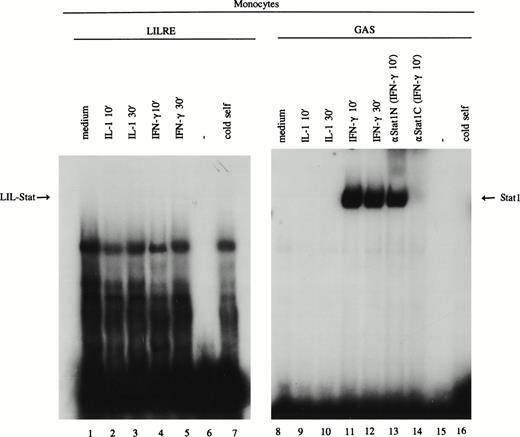
Because spontaneously activated LIL-Stat was found in the majority of the AML cases, we set out to determine the expression of LIL-Stat in freshly isolated monocytes and granulocytes. Monocytes were incubated at low-level serum (0.1% FBS) overnight before stimulation with medium, IL-1 (100 U/mL) or IFN-γ (20 U/mL for 10 minutes and 30 minutes. After preparation of nuclear extracts, EMSAs were performed using oligonucleotides specific for LIL-Stat and Stat1. As depicted in Fig 5 (lanes 1-7), neither untreated nor IL-1β- or IFN-γ–stimulated monocytes showed LIL-Stat binding activity. Treatment of monocytes with IL-6 or LPS, or stimulation with the abovementioned cytokines for longer periods of time could not elicit the DNA-binding of LIL-Stat (data not shown). The stimulation of the monocytes was, however, appropriate, because IFN-γ–stimulated monocytes showed a clear induction of Stat1 binding (Fig 5, lanes 11 and 12).
LIL-Stat binding activity is not observed in monocytes. Nuclear extracts derived from control, IL-1β–, and IFN-γ–treated monocytes were incubated with the 32P-labeled oligonucleotides specific for LIL-Stat (lanes 1-7) and Stat1 (lanes 8-16). Lanes 6 and 15 represent the respective labeled oligonucleotides alone. Competition was performed by addition of a 100-fold molar excess of unlabeled LIL-Stat oligonucleotide (lane 7) or unlabeled Stat1 oligonucleotide (lane 16). Specificity of Stat1 oligonucleotide was confirmed by supershift experiments with the anitbodies against Stat1N and Stat1C (lanes 13, and 14, respectively).
LIL-Stat binding activity is not observed in monocytes. Nuclear extracts derived from control, IL-1β–, and IFN-γ–treated monocytes were incubated with the 32P-labeled oligonucleotides specific for LIL-Stat (lanes 1-7) and Stat1 (lanes 8-16). Lanes 6 and 15 represent the respective labeled oligonucleotides alone. Competition was performed by addition of a 100-fold molar excess of unlabeled LIL-Stat oligonucleotide (lane 7) or unlabeled Stat1 oligonucleotide (lane 16). Specificity of Stat1 oligonucleotide was confirmed by supershift experiments with the anitbodies against Stat1N and Stat1C (lanes 13, and 14, respectively).
Similar to monocytes, LIL-Stat binding activity could not be observed in freshly isolated granulocytes in the presence of the abovementioned activators (data not shown).
LIL-Stat binding activity is observed in CD34+cells, but dissappears on differentiation.
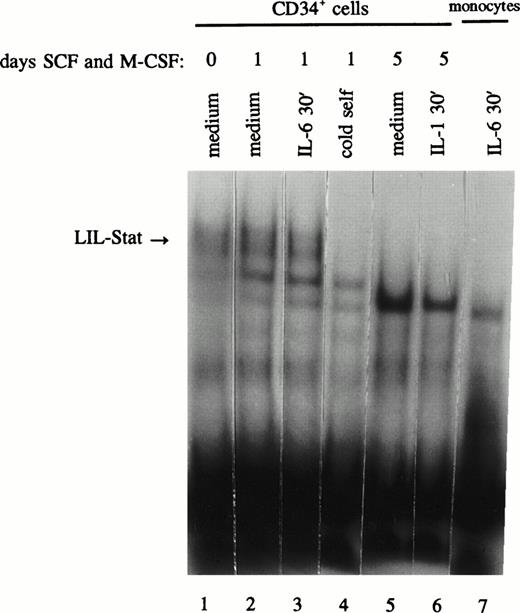
The observation of distinct LIL-Stat binding activity in AML blasts and leukemic cell lines and the lack of LIL-Stat expression in mature monocytes and granulocytes may suggest a possible role for LIL-Stat in the differentiation process. For this purpose we studied CD34+ cells for the presence of LIL-Stat binding. In addition, the CD34+ cells were allowed to differentiate along the monocytic lineage by the addition of both SCF (10 ng/mL) and M-CSF (10 ng/mL) to the medium. At day 0 of culture the percentage CD34+ cells was >98%, the percentage of CD14+ cells 0%. After 5 days of culture with SCF and M-CSF, 34% of the cells had turned CD14+, whereas merely 12% of the cells still expressed CD34, showing that differentiation along the monocytic lineage had occured. These data were confirmed by light microscopic, morphological observations of monocytic differentiation. At day 0, 1, and 5 of culture, nuclear extracts were prepared and studied for the presence of LIL-Stat. In Fig 6 (lanes 1-4) it is shown that CD34+ cells indeed express active LIL-Stat, both at day 0 and day 1 of culture. Treatment with IL-6, however, could not enhance the constitutive DNA-binding. At day 5 of culture, no LIL-Stat was detected anymore, neither constitutive, nor inducible. This may indicate a role for LIL-Stat in the differentiation of monocytic cells.
LIL-Stat is constitutively activated in nuclear extracts of CD34+ cells and disappears upon differentiation. CD34+ cells were incubated with SCF and M-CSF for 0 (lane 1), 1 (lanes 2-4), and 5 days (lanes 5 and 6) and treated with IL-6 (lane 3) or IL-1β (lane 6). Five micrograms of nuclear extracts were used in band-shift assays with the 32P-labeled LILRE oligonucleotide. Competition was performed using a 100-fold molar excess of unlabeled LILRE (lane 4). Absence of LIL-Stat binding in IL-6–treated monocytes is depicted in lane 7.
LIL-Stat is constitutively activated in nuclear extracts of CD34+ cells and disappears upon differentiation. CD34+ cells were incubated with SCF and M-CSF for 0 (lane 1), 1 (lanes 2-4), and 5 days (lanes 5 and 6) and treated with IL-6 (lane 3) or IL-1β (lane 6). Five micrograms of nuclear extracts were used in band-shift assays with the 32P-labeled LILRE oligonucleotide. Competition was performed using a 100-fold molar excess of unlabeled LILRE (lane 4). Absence of LIL-Stat binding in IL-6–treated monocytes is depicted in lane 7.
DISCUSSION
The JAK/Stat pathway is a unique pathway with respect to the rapid induction of gene transcription. After phosphorylation of Stat proteins and dimerization, the Stat dimers immediately migrate to the nucleus where they bind to the so-called GAS-sites [TT(C/A)CNNNAA].1-6 Many cytokines have been described to induce the activity of Stat proteins, of which six have been cloned.7-11 Tsukada et al18 were the first to describe a potential novel Stat factor that could be induced by IL-1, IL-6, and LPS, termed LIL-Stat (for LPS- and IL-1–inducible Stat). It was shown that this transcription factor showed several features common to all the known Stat factors. First, induction of LIL-Stat binding activity occured within minutes after stimulation with IL-1, LPS, or IL-6, and, second, it required tyrosine phosphorylation for DNA-binding. Third, the protein bound to a GAS-like site that, for LIL-Stat, is located in the human IL-1β gene enhancer region at position −2863 to −2841.19,20 LILRE, the LIL-Stat binding sequence, differs from the consensus GAS-site for the requirement of a G residue at position 8 (TTCCTGAGA). GAS-sequence variations have been reported to be essential for Stat selectivity.28 Stat3 for example, efficiently binds the SIE-site in the c-fos promoter.29 Although the sequence of SIE (TTCCCGTCA) is similar to the consensus GAS element, Stat3 cannot efficiently recognize FcγRI GAS (TTCCCAGAA). Stat1, on the other hand, binds FcγRI with high avidity.7 A G residue at position 7 has been reported to be essential for Stat5 binding, which preferentially binds to the GAS-site within the β-casein promoter.30 As control experiments we showed that Stat1, Stat3, and Stat5 did not bind the LILRE, nor did LIL-Stat bind to any of the other GAS-like oligonucleotides. Finally, supershift experiments showed that LIL-Stat was recognized by an antibody specific for the N-terminus of Stat1, but not for any other Stat-related antibody. Taken together, these findings showed that this transcription factor is the LIL-Stat factor.
It was suggested that LIL-Stat may be the product of a novel gene possessing homology with the Stat1 amino terminus, as is the case for Stat5a and Stat5b.10,31,32 A second possibility is that LIL-Stat is transcribed from the Stat1 gene followed by alternative splicing. Splice variants within the Stat family of proteins have been described for Stat1α and Stat1β.7 Recently, a truncated form of Stat3 was identified in a yeast two-hybrid screen of a mammalian cDNA library.33 The Stat3β interacted with the N-terminal segment of c-jun and is devoid of the 55 C-terminal amino acid residues. Nevertheless, additional studies are required for the full characterization of the LIL-Stat transcription factor.
In the present study we showed that LIL-Stat is constitutively active in seven out of nine AML cases, in three human leukemic cell lines, and in CD34+ cells. The presence of LIL-Stat binding activity in CD34+ cells disappeared in time when the cells differentiated along the monocytic lineage. After 5 days of incubation with SCF and M-CSF, the DNA-binding activity of LIL-Stat was totally absent. In agreement with these observations, LIL-Stat binding activity could not be detected in mature monocytes and granulocytes, independent on the stimuli applied. This coincides with the observation that the two AML cases lacking LIL-Stat expression belong to the FAB classification M4 and M5a, thus progressed in the differentiation cascade.22 Taken together, these data may suggest that LIL-Stat plays a role in the differentiation process of hematopoietic cells. However, further studies are necessary to elucidate this potential role.
Constitutive DNA-binding activity, represented by band-shift assays, does not necessarily imply a role of the respective transcription factor in the induction of gene transcription. For both Stat1 and Stat3 it was recently shown that the DNA-binding of these Stats was induced after tyrosine phosphorylation, but was not demonstrably affected by the presence or absence of serine phosphorylation.34 The serine phosphorylation, however, which occurs on a single residue (ser 727 for Stat1) only after binding to the consensus DNA sequence, greatly enhanced the transactivational capacity of these transcription factors.35 From our studies it is clear that LIL-Stat could potently drive transcription from the pGL3ti-LIL3 promoter construct containing 3 sequential LIL-Stat binding sites. Strong transcriptional activity of LIL-Stat was observed in the leukemic cell lines (unpublished data), as well as in the AML cases that spontaneously expressed LIL-Stat, suggesting a functional role of LIL-Stat in the control of gene expression. A LIL-Stat consensus sequence is present in the promoter of human IL-1β gene, a gene that is constitutively active in a sizable amount of AML cases.21,36-38 It is therefore tempting to speculate about the role of constitutive LIL-Stat binding in the spontaneous expression of IL-1β. Moreover, spontaneous IL-1β production by AML cells has been shown to be correlated with autocrine proliferation and poor clinical prognosis.36 37Thus, apart from a possible role in the differentiation of early hematopoietic cells, constitutive LIL-Stat binding activity may also support autocrine or paracrine proliferation of AML blasts through activation of the IL-1β promoter.
Several reports have previously described constitutive activation of the JAK/Stat pathway in malignant cells and cell lines, which coincided with increased proliferation of cells. For instance, inhibition of constitutive JAK2 phosphorylation in primary pre-B leukemia cells could inhibit cell proliferation, indicating a link between cell proliferation and JAK2 phosphorylation in acute leukemias.39 In lymphoblastoid cells constitutively activated Stat1 and Stat3 coincided with the presence of the Epstein-Barr virus (EBV) and with cellular IL-10 expression.17 In v-abl oncogene-transformed murine pre-B lymphocytes Stat5 and Stat6 were tyrosine phosphorylated in the absence of cytokine stimulation40, whereas in v-src-transformed 32Dcl3 myeloblastic cells Stat1, Stat3, and Stat5 exist in a constitutively phosphorylated state.41 Constitutive Stat activition in AML samples has been described previously on two occasions.16 17 In a considerable number of AML cases spontaneous binding activity of Stat1, Stat3, and Stat5 was observed. However, constitutive Stat activity in these studies could not be linked to increased proliferation of the AML blasts, nor to FAB classification.
It is uncertain whether LIL-Stat is activated as a result of JAK phosphorylation, because neither IL-1, nor LPS have been shown to use the JAK family of protein kinases for their signal transduction.42 However, constitutive Stat activation independent from JAK activation was shown previously in P210 BCR/ABL-transformed hematopoietic cells.43 Albeit the coexistence of constitutively activated JAK1, JAK2, and JAK3 with the spontaneous activity of Stat5, dominant-negative JAK mutant proteins could not inhibit the Stat5 binding activity. This may indicate that other upstream signaling molecules, other then the JAKs, may induce Stat binding activity, which could also be the case for LIL-Stat. Summarized, this study shows the spontaneous activation and transactivational capacity of a novel Stat factor in the majority of AML cases and immature hemopoietic cells. Further studies should show the possible role of LIL-Stat in the differentiation of hematopoietic cells and whether ongoing constitutive expression of this factor may play a role in the onset or process of leukemogenesis.
ACKNOWLEDGMENT
We are grateful to Dr Luigi Jonker (Division of Developmental Genetics, University of Groningen, The Netherlands) for supplying us with the pGL3ti plasmid.
Supported by grant no. RUG 94-788 from the Dutch Cancer Foundation.
Address correspondence to Edo Vellenga, MD, Division of Hematology, Department of Medicine, University Hospital Groningen, Hanzeplein 1, 9713 GZ Groningen, The Netherlands; e-mail: e.vellenga@int.azg.nl.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal