Abstract
The locus control region (LCR) activates high-level human β-globin transgene expression. LCR cassettes composed of 5′HS2-4 linked to the 815 bp β-globin proximal promoter do not express fully. Here, we show that LCR (5′HS2-4) β-globin transgenes that also contain either 5′HS1 or the distal promoter fail to express fully in single- and low-copy transgenic mice. In contrast, full expression is obtained in the presence of both 5′HS1 and the distal promoter. Nine factor binding sites were identified in 5′HS1, using in vitro DNaseI footprint and gel retardation assays, and these include a strong Sp1/Sp3 site, four GATA-1 sites, and two sites that encompass an ACTAAC motif. LCR (5′HS1-4) β-globin transgene constructs with the distal promoter deleted or replaced by spacer DNA show that specific distal promoter sequences are required for full expression. An LCR (5′HS1-4) transgene construct with truncated downstream β-globin gene sequences indicates that 3′ sequences also play an important role. These results show that full expression of the β-globin gene directed by the LCR requires 5′HS1, the distal β-globin promoter, and 3′ sequences, and has implications for gene therapy construct design and models of LCR activation.
EXPRESSION OF THE HUMAN β-globin locus is regulated by an array of cis-acting DNA elements including: five DNaseI hypersensitive sites (HS) in the locus control region (LCR); five promoters that incorporate certain silencer elements for individual genes in the cluster; and at least three enhancers in the introns or 3′ of some of the genes.1 These elements have several functional roles. The first step in transcriptional activation is to open chromatin throughout the locus, which has been viewed as being accomplished solely by, and established at, the LCR.2 This chromatin opening event leaves the five genes of the cluster poised to express in a developmentally restricted pattern that is governed by the binding of specific factors to gene proximal promoter, silencer, and enhancer elements.1
The activity of each individual HS has been extensively studied. Transient transfection experiments show that only 5′HS2 has a strong classical enhancer activity.3 However, 5′HS2-4 all enhance transcription in stably transfected cells and direct copy number-dependent expression in multicopy β-globin transgenic mice.4-9 Only 5′HS3 can reproducibly activate single-copy transgene expression, suggesting that it contains a chromatin opening activity when linked to the β-globin gene.10,11 5′HS1 has no significant activity in transfection and transgenic assays,12-14 but appears to have some important undefined function because full expression in transgenic mice is only obtained in the presence of all four HS.11,12 5′HS5 has an insulator activity15 but is not required for full expression in transgenic mice. Therefore, it is clear that the HS have several different functions but interact in some way with each other to activate full expression.
The β-globin gene promoter and enhancers are also well characterized.16-19 The minimal β-globin promoter maps to a 103 bp fragment that is inducible by the LCR in stable transfection studies20; but in single-copy transgenic mice, the 815 bp promoter is more active than the 265 bp promoter commonly used in gene therapy vectors.21 LCR activation of γ-globin transgenes has also been shown to be dependent on the length of the γ-globin promoter,22 but similar experiments have not been performed on the β-globin promoter.
Two enhancers are localized in the second intron and 3′ of the β-globin gene.16,18,19 These enhancers have no role in LCR-mediated induction of the promoter in stable transfection studies14,20 and were therefore omitted from β-globin gene therapy vectors.23-25 However, deletion of the 3′ enhancer in yeast artificial chromosome (YAC) transgenic mice causes a reduction in β-globin gene expression indicating that the 3′ enhancer influences globin switching.26 In addition, the human LCR does not reproducibly activate γ-globin transgene promoters unless a fragment containing the 3′ γ-globin enhancer27 or the entire β-globin gene is included.28,29 Hence, more recently developed adeno-associated virus (AAV) vectors that transfer the A γ-globin gene include the 3′ enhancer.30
These results suggest that promoters and 3′ enhancers are involved in chromatin opening activity mediated by the human β-globin LCR. Similar conclusions were reported for chicken β-globin and lysozyme transgenes assayed in mice, which require the enhancer activities of their LCRs and linked promoters to open chromatin.31-33 Taken together, the above work gathered by many groups strongly suggests that HS elements of the LCR not only interact with each other, but also indicates that sequences 5′ to minimal globin promoters and in 3′ enhancers have important functions that cannot be supplied by the LCR alone at ectopic transgene sites.
We are particularly interested in defining the minimal combination of cis-acting regulatory elements that direct full and reproducible expression of the β-globin gene in single-copy transgenic mice.21 The purpose of the present study is to identify functional interactions between the LCR and β-globin gene regulatory elements and to create an optimal transgene construct for gene therapy of β-thalassemia or sickle cell anemia. Our findings show that reproducible full expression of transgenes regulated by the LCR requires the simultaneous presence of 5′HS1, the distal portion of the 1555 bp β-globin promoter, and a large fragment that includes the 3′ enhancer. These data have implications for the design of gene therapy constructs for treatment of β-thalassemia, and can be incorporated into models of LCR activation.
MATERIALS AND METHODS
Plasmid construction.
Transgene constructs were derived from the plasmids pGSE1359 and pBGT14. pGSE1359 contains a 6.5 kb LCR cassette and the 4.8 kbBglII-EcoRV β-globin gene fragment regulated by the 1555 bp promoter.34 pBGT14 contains a 3.0 kb LCR cassette and the 4.2 kb Hpa1-EcoRV β-globin gene fragment regulated by the 815 bp promoter.21 The 6.5 kb LCR cassette includes 5′HS1-4 as described previously.34 The 3.0 kb LCR contains the 1.15 kb Stu1-Spe1 fragment of 5′HS4, the 0.85 kb Sac1-PvuII fragment of 5′HS3, and the 0.95 kb Sma1-Stu1 fragment of 5′HS2.
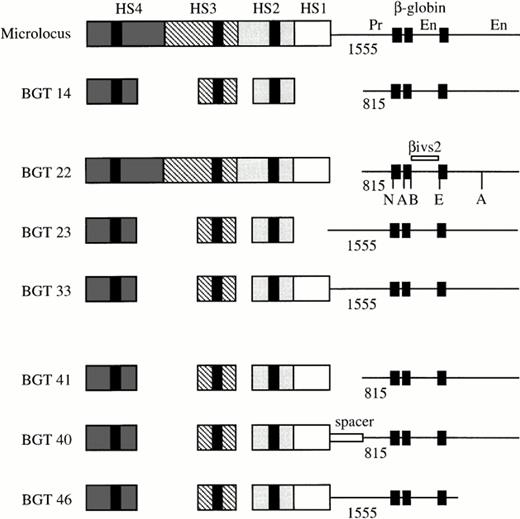
In short, BGT22 was constructed by linking the 6.5 kb LCR from GSE1359 to the 4.2 kb β-globin gene from BGT14, using the Sal1 linker present in both plasmids between the LCR and the promoter. BGT23 was made by linking the 3.0 kb LCR from BGT14 to the 4.8 kb β-globin gene from GSE1359, using the same Sal1 linker site. BGT33 was made by inserting a 2.6 kb Snab1 fragment from pGSE1359 (containing the 0.3 kb Snab1-BglII fragment of the 3′ part of 5′HS2, the 1.0 kb Sac1-HindIII fragment of 5′HS1, and the 1.3 kb BglII-Snab1 fragment of the promoter) into the Snab1 sites in 5′HS2 and the promoter of BGT23. This manipulation left a Sal1 site in the polylinker between the LCR and the promoter, and linked a 4.0 kb LCR (composed of 5′HS1-4) to the 4.8 kb β-globin gene fragment including a reconstructed 1555 bp promoter. BGT41 was cloned by deleting the 741 bpBglII-Hpa1 distal promoter fragment from BGT33 via digestion at the Sal1 site in the polylinker and theHpa1 site in the promoter and religation with a Sal1 linker. This resulted in the 4.0 kb LCR of BGT33 linked by aSal1 site to the 4.2 kb β-globin gene fragment. The spacer element of BGT40 was cloned by insertion of the 717 bpXho1-Hpa1 A γ-globin intron 2 fragment (provided by S.Philipsen) between the Sal1 polylinker site and theHpa1 site in the promoter of BGT33. This destroyed theSal1 site and recreated the Hpa1 site. Finally, the BGT46 construct bears a truncation of sequences downstream of theBspH1 site located 267 bp 3′ of the end of exon 3 in the β-globin gene. This was accomplished by cloning the 4.0 kb LCR from BGT41 linked via the Sal1 site to a 3.15 kb β-globin gene fragment from BGT33 in which the downstream BspH1 site was destroyed and replaced by an Nhe1 linker. Therefore, BGT46 is regulated by the 1555 bp promoter but lacks extensive 3′ sequences.
pGEM-HS1 was cloned by polymerase chain reaction (PCR) amplification of the β-globin 5′HS1 core using the 5′HS1 and 3′HS1 oligonucleotides as primers and the plasmid BGT22 as a template. Cycling conditions for PCR with Taq polymerase (Gibco BRL, Gaithersburg, MD) were: 94°C, 3 minutes, (1 cycle); 94°C, 1 minute, 58°C, 1 minute, 72°C, 2 minutes (30 cycles); 72°C, 5 minutes (1 cycle). The PCR product was gel purified and inserted into the plasmid pGEM-T (Promega, Madison, WI) using the 3′ thymidine overhangs. Sequence of the cloned insert was verified by dideoxy sequencing using Sequenase version 2 (Amersham, Oakville, Ontario, Canada)) and the primers 5′HS1, 3′HS1, and HS1FpGa.
Generation of transgenic mice.
Transgene DNA was prepared using Plasmid Maxi Kits (Qiagen, Santa Clara, CA). Transgene fragments were liberated from their plasmid backbones by digestion with EcoRV that cleaves in the polylinker 5′ of the LCR cassettes and at the 3′ end of the β-globin gene, with the exception of BGT46, which lacks the 3′ EcoRV site and was double digested with EcoRV and Nhe1. DNA fragments were recovered from 0.7% TAE agarose gel slices using GeneClean II or GeneClean Spin Column Kits (Bio101) and Elutip-d columns (Schleicher and Schuell, Mississauga, Ontario, Canada), and resuspended in injection buffer (10 mmol/L Tris-HCl, pH 7.5; 0.2 mmol/L EDTA). DNA concentration was determined by comparison with DNA standards run on agarose gels, and the injection fragment was diluted to 0.5-1 ng/μL in injection buffer. The diluted DNA was prespun for 20 minutes and aliquots removed for microinjection into fertilized FVB mouse eggs. Injected eggs were transferred into recipient CD1 female animals. Fetal mice were dissected and DNA extracted from head tissue, at 15.5 days postinjection, whereas the fetal livers were saved frozen in two halves for future analysis. Head DNA was extracted by Proteinase K digestion overnight, a single phenol/chloroform extraction and isopropanol precipitation. Transient transgenic fetuses were identified by slot-blot hybridization with the βivs2 probe using standard procedures.
DNA analysis.
Southern transfer and hybridization were by standard procedures. Copy-number determination was performed using a Molecular Dynamics PhosphorImager (Sunnyvale, CA). Single-copy animals showed a single random-sized end-fragment in BamH1 andEcoR1 digests hybridized with the βivs2 probe. With multicopy animals, the intensity of the end-fragment was defined as one transgene copy, and was used to calculate the copy number of the multicopy junction-fragment in the same lane. Mosaicism in the fetal liver of founder 15.5 day transgenic mice was calculated by quantifying the intensity of bands on a Molecular Dynamics PhosphorImager and using the following formula: (Tg Hβ / Tg mThy-1) / (B26 Hβ x Tg copy number / B26 mThy-1). Tg, transgenic; Hβ, human β-globin; mThy-1, mouse Thy-1; B26, 1 copy bred line B26.
RNA analysis.
Fetal liver (embryonic day 15.5) RNA was extracted using Trizol Reagent (Gibco BRL), 1 μg was hybridized to kinased double-stranded DNA probes, digested with 75 U S1 nuclease (Boehringer Mannheim, Laval, Quebec, Canada),), and run on a 6% sequencing gel as described.18 Probe excess was shown by including a sample containing 3 μg fetal liver RNA. Specific activities of human β-globin (Hβ) relative to the mouse βmajor (βmaj) probe were 2:1 unless otherwise noted. The protected 160 nt Hβ and 95 nt βmaj bands were quantified on a Molecular Dynamics PhosphorImager and the % expression levels calculated according to the formula (Hβ / 2βmaj) ×100 to account for the specific activity differences. Percent expression per copy was calculated as (2βmaj genes / number Hβ transgenes) × (% expression / % mosaicism) × 100.
Nuclear extract preparation.
Nuclear extracts were performed essentially as described.35In brief, 5 × 108 murine erythroleukemia (MEL) C88 (provided by L. Wall) or Jurkat cells (obtained from American Type Culture Collection [ATCC]) were grown in alpha MEM or RPMI 1640 media, respectively, supplemented with 10% fetal bovine solution, (FBS; Gibco BRL). MEL C88 cells were induced for 4 days with 2% dimethyl sulfoxide (DMSO; Sigma, St Louis, MO). Cells were washed in phosphate buffered saline (PBS), washed in cold Hypotonic Solution, and resuspended on ice in 2.5 mL cold Hypotonic solution for 10 minutes. Nuclei were released using 20 strokes of a cold Dounce Homogenizer, pelleted, and resuspended in a cold microfuge tube in 0.25 mL cold low-salt buffer; 0.75 mL cold high-salt buffer was added, chromatin was precipitated on ice for 30 minutes and pelleted in a refrigerated Sorval (Newtown, CT) RMC 14 microfuge at 14.5 Krpm for 30 minutes at 4°C. The supernatant was then dialysed for 45 minutes at 4°C in a Slide-a-Lyser cassette (Pierce, Rockford, IL) against dialysis buffer before centrifugation in the refrigerated microfuge for 20 minutes at 14.5 Krpm. Protein concentration was determined with the Bradford Assay Kit (Bio-Rad Laboratories, Mississauga, Ontario, Canada), and the nuclear extract frozen in small aliquots for later use.
In vitro DNaseI footprint assay.
In vitro footprint reactions were performed essentially as described.35 The pGEM-HS1 probes were prepared by digestion with either Nco I, which cuts in the 5′ polylinker, orSpe I which cuts the 3′ end of 5′HS1. The DNA ends were labelled with [α-32P] dCTP using Klenow enzyme (Boehringer Mannheim), and the labelled DNA cut with the second enzyme. The 556 bp Spe I-Nco I fragments labelled at either the 5′ or 3′ ends were isolated by excision of the band from a preparative gel and purified by centrifugation through siliconized glass wool to remove agarose and subsequent ethanol precipitation. As a marker lane, G-sequence ladders of the same probe DNA were prepared using the Maxam-Gilbert Sequencing kit (Sigma). Approximately 5000 cpm of the probe was treated with dimethyl sulphate followed by alkali hydrolysis with piperidine at 90°C.
Each 25 μL footprinting assay including extract contained approximately 5000 dpm of labeled probe DNA, 2 μg poly(dI):poly(dC) (Pharmacia, Uppsala, Sweden), and 3 to 18 μg protein extract in 20 mmol/L HEPES-KOH (pH 7.9), 8% glycerol (vol/vol), 100 mmol/L KCl, 0.25 mmol/L phenylmethylsulfonyl fluoride (PMSF), 1.25 mmol/L dithiothreitol (DTT), 0.1 mmol/L EDTA, and 2.0 mmol/L MgCl2. These reactions were incubated at room temperature for 30 minutes, followed by a 3-minute incubation on ice before addition of 0.18 μg of DNaseI (Sigma) on ice for 120 seconds. The assay was stopped with an equal amount of 1.2 mol/L NaCl containing 0.4% sodium dodecyl sulfate (SDS), 20 mmol/L EDTA, and 200 μg/ml tRNA. After phenol/chloroform extraction and ethanol precipitation, the samples were run in loading dye on 6% sequencing gels and exposed to XAR-5 film (Kodak, Rochester, NY) for 3 days.
Gel retardation assay.
Gel retardation assays were performed as described.16 Fifty ng of synthetic single-strand sense oligonucleotide was labeled with T4 polynucleotide kinase (Boehringer Mannheim) and [γ-32P] adenosine triphosphate (ATP) and annealed to 250 ng of antisense strand to a final double-strand concentration of 1 ng/μL. Competitor oligonucleotides were annealed in equal amounts of both strands to a total DNA concentration of 50 ng/μL. Each 10 μL reaction contained 1 ng of labeled double-stranded oligonucleotide probe, 2 ng of poly(dI):poly(dC) (Pharmacia), 3 to 6 μg of nuclear extract protein in 2 μL of buffer D, and 1 μL of Binding Buffer. In competition experiments, 50 ng of unlabeled double-stranded oligonucleotide (50 times molar excess) was added to the reaction mixture. For supershift assays, 1 μL of antibody was added. Antibodies (Santa Cruz, Santa Cruz, CA)) used for this purpose were: GATA-1 (N6) rat monoclonal IgG2a, Sp1 (1C6) mouse monoclonal IgG1, Sp3 (D20) rabbit polyclonal IgG. For supershift experiments, the reactions were incubated overnight at 4°C before addition of probe and a further 20-minute incubation, 1 μL loading dye was added and the reactions were run on 4% acrylamide gels for 4 hours at constant 13 mA in 1 × TBE at 4°C.
Oligonucleotide primers and probes.
Oligonucleotide Primers and Probes are as follows: 3′HS1, CTCAAGCCTCATTCAGACACTAG; 5′HS1, TTTCCTGGTATCCTAGGACCTGC; HS1FpAs, TCACGTTTTGATGATAATCACATATTTGTAAACACA; HS1FpAa, TGTGTTTACAAATATGTGATTATCATCAAAACGTGA; HS1FpCs, CATATTTATCGGGCATTTCTGAG; HS1FpCa, CTCAGAAATGCCCGATAAATATG; HS1FpEs, TAGCTAGGCCCCTCCCTCATCACAGCT; HS1FpEa, AGCTGTGATGAGGGAGGGGCCTAGCTA; HS1FpFs, CGAGCTCTTATCTATATCCACACA; HS1FpFa, TGTGTGGATATAGATAAGAGCTCG; HS1FpGs, GCCCAGCTATCACCATCCCAAGTC; HS1FpGa, GACTTGGGATGGTGATAGCTGGGC; GATA-Cruz-s, CACTTGATAACAGAAAGTGATAACTCT; GATA-Cruz-a, AGAGTTATCACTTTCTGTTATCAAGTG; Sp1-Cruz-s, ATTCGATCGGGGCGGGGCGAGC; Sp1-Cruz-a, GCTCGCCCCGCCCCGATCGAAT; 46int1, GCAAAGAATTCACCCCACCAG; and 46int2, ATGCACTGACCTCCCACATTC.
RESULTS
We previously established that the 6.5 kb microlocus LCR cassette composed of 5′HS1-4 linked to the 1555 bp β-globin promoter and gene sequences, including both enhancers expresses at 100% levels in single-copy transgenic mice.11 In addition, the BGT14 construct composed of a 3.0 kb LCR cassette containing 5′HS2-4 linked to the 815 bp promoter and both enhancers directs reproducible expression of 45% at single copy.21 Expression levels from BGT14 therefore are reduced by about twofold in comparison with the microlocus. To identify the minimal combination of cis-acting DNA elements capable of directing full expression from single- and low-copy transgenes, we created several transgene constructs (Fig 1). Initially, we wished to determine the relative importance of the distal promoter, 5′HS1, and auxiliary sequences in transgene expression. For this purpose, the BGT22 transgene combines the 6.5 kb LCR with the 815 bp promoter to test the importance of the distal promoter sequences. The BGT23 transgene links the 3.0 kb LCR to the 1555 bp promoter to examine the role of 5′HS1 and auxiliary sequences near 5′HS2-4, and the BGT33 transgene was designed to determine whether 5′HS1 and the distal promoter functionally interact.
Map of the human β-globin LCR transgene constructs designed to determine the importance of 5′HS1, distal β-globin promoter, and 3′ sequences in directing full transgene expression levels in single-copy transgenic mice. The activity of the microlocus and BGT14 constructs has been previously described,11 21and the specific sequences used in each construct are outlined in Materials and Methods. The black boxes in each HS correspond to the known core elements, and the black boxes in the β-globin gene correspond to the three exons. The length of each promoter (Pr) is given below the line, and the enhancers (En) located in the second intron and 3′ of the gene are indicated. A, Acc1; B,BamH1; E, EcoR1; N, Nco1.
Map of the human β-globin LCR transgene constructs designed to determine the importance of 5′HS1, distal β-globin promoter, and 3′ sequences in directing full transgene expression levels in single-copy transgenic mice. The activity of the microlocus and BGT14 constructs has been previously described,11 21and the specific sequences used in each construct are outlined in Materials and Methods. The black boxes in each HS correspond to the known core elements, and the black boxes in the β-globin gene correspond to the three exons. The length of each promoter (Pr) is given below the line, and the enhancers (En) located in the second intron and 3′ of the gene are indicated. A, Acc1; B,BamH1; E, EcoR1; N, Nco1.
Generation of transgenic mice.
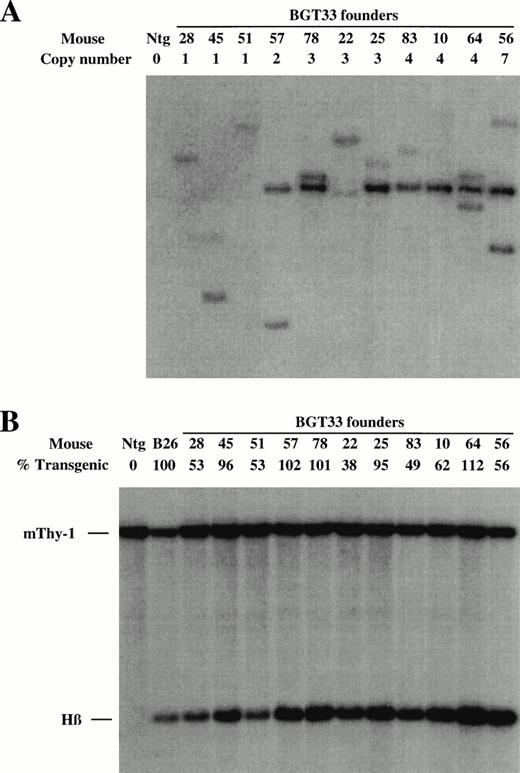
These DNA constructs were purified as linear fragments and microinjected into fertilized FVB mouse eggs to create transgenic mice. The fetuses derived from these eggs were dissected at embryonic day 15.5 and genomic DNA extracted from head tissue, whereas the fetal livers were frozen in two halves for future analyses. Positive transient transgenic founder (F0) animals were identified by slot-blot hybridization with the βivs2 probe, and transgene copy number subsequently deduced by genomic Southern blots after digestion with EcoR1 and BamH1, which we have previously shown can unambiguously identify junction fragments that define single-copy transgenic mice.21 A representative Southern analysis for the BGT33 construct is shown in Fig 2A. All founder animals were characterized to determine whether they harbored intact transgenes by Southern blot analyses with multiple diagnostic restriction enzymes and βivs2 or 5′HS3 probes (data not shown). Finally, the level of transgene mosaicism was determined by Southern blots of DNA derived from one half of the frozen fetal livers after digestion with Acc1, an enzyme which releases a 1.9 kb fragment detected by the βivs2 probe regardless of the integration site.21 A representative mosaic analysis for BGT33 is shown in Fig 2B. By comparison of the intensity of the transgene signal with that from the single-copy bred B26 line that by definition is 100% transgenic, it is possible to calculate the degree of transgenesis in the fetal liver for each founder animal. Nonintact and highly mosaic animals were excluded from this study. Identical screening procedures were performed on the BGT22 and BGT23 transgenic animals (data not shown).
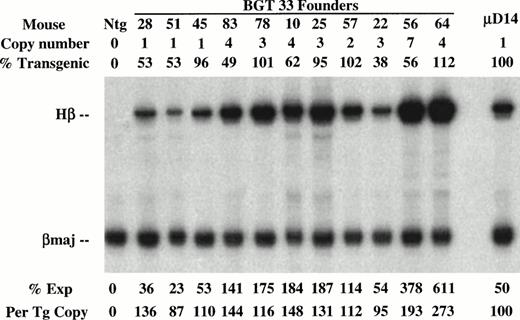
Determination of transgene copy number and mosaicism in embryonic day 15.5 F0 transgenic mice containing the BGT33 construct. (A) Copy numbers of BGT33 transgenic mice determined by Southern blot analysis of F0 fetal head DNA digested withBamH1 and hybridized with the βivs2 probe. (B) Transgene mosaicism levels determined by Southern blot analysis of Acc1 digested DNA extracted from BGT33 fetal livers and hybridized with the βivs2 (Hβ) and mouse Thy-1 (mThy-1) probes. The percentage of transgenic cells was calculated by comparison of the Hβ band intensity of each BGT33 transgenic mouse with the one copy-bred line (B26), taking into account copy number and the mThy-1 loading control (see Materials and Methods).
Determination of transgene copy number and mosaicism in embryonic day 15.5 F0 transgenic mice containing the BGT33 construct. (A) Copy numbers of BGT33 transgenic mice determined by Southern blot analysis of F0 fetal head DNA digested withBamH1 and hybridized with the βivs2 probe. (B) Transgene mosaicism levels determined by Southern blot analysis of Acc1 digested DNA extracted from BGT33 fetal livers and hybridized with the βivs2 (Hβ) and mouse Thy-1 (mThy-1) probes. The percentage of transgenic cells was calculated by comparison of the Hβ band intensity of each BGT33 transgenic mouse with the one copy-bred line (B26), taking into account copy number and the mThy-1 loading control (see Materials and Methods).
Requirement for 5′HS1 and distal promoter sequences.
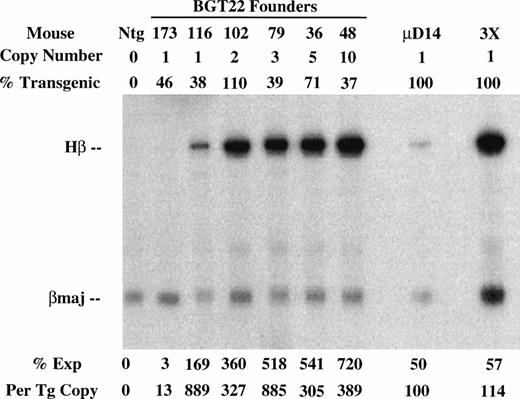
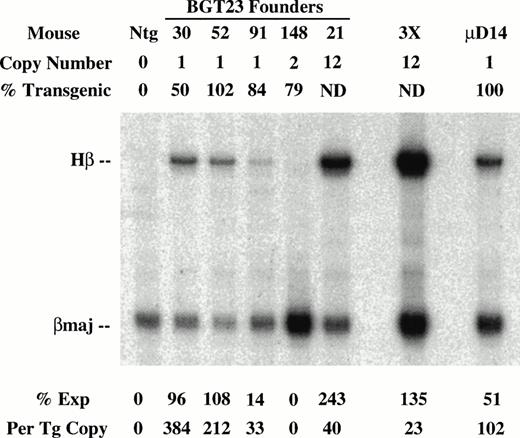
To determine the effect of these transgene constructs on expression levels, RNA was extracted from the other half of the frozen transgenic fetal livers for S1 nuclease protection assays using human β-globin and mouse βmajor probes (Fig 3). Expression from the BGT22 construct ranged from 13% to 889% in single-copy animals and remained variable even at low-copy numbers of 2 to 10 (Fig 3A). These data suggest that the distal promoter is important for obtaining reproducible full levels of expression directed by the LCR. Expression from the BGT23 construct ranged from 33% to 384% in single-copy animals, and a two-copy animal failed to express (Fig 3B). These data indicate that the distal promoter is not sufficient to maintain reproducible levels of expression, and that other sequences may also be required. By adding back both 5′HS1 and the distal promoter in the BGT33 construct, reproducible 87% to 136% expression levels were obtained from the single-copy transgenic animals, and the multicopy animals also express to the same magnitude (Fig 3C). Such consistent expression was only obtained in the construct that contains both 5′HS1 and the distal promoter. We therefore suggest that 5′HS1 has an important role in mediating the interaction between the LCR and the β-globin proximal promoter. The distal promoter sequences may specifically participate in this functional interaction with 5′HS1, or may act as a passive spacer element.
Expression of globin mRNA in transgenic mice containing 5′HS1 and/or β-globin distal promoter constructs. (A) S1 nuclease analysis of fetal liver RNA of 15.5 day F0BGT22 transgenic mice showing that the range of expression from single-copy transgenes is 13% to 889% per copy and indicating that the distal promoter is important for reproducible expression levels. (B) S1 nuclease analysis of fetal liver RNA of 15.5 day F0BGT23 transgenic mice showing that the range of expression from single-copy transgenes is 33% to 384% per copy and indicating that 5′HS1 is important for reproducible expression levels. (C) S1 nuclease analysis of fetal liver RNA of 15.5 day F0 BGT33 transgenic mice showing that the range of expression from single-copy transgenes is 87% to 136% per copy and showing that both 5′HS1 and the distal promoter are required to obtain reproducible full expression levels. Hβ, human β-globin protected probe fragment; βmaj, mouse β major protected probe fragment; Ntg, nontransgenic; μD, one copy μD14 microlocus line; 3X, probe excess control. {/ANNT;4224n;;left;71808n}C {/ANNT;4224n;;0n;0n}A {/ANNT;4224n;;84480n;0n}B
Expression of globin mRNA in transgenic mice containing 5′HS1 and/or β-globin distal promoter constructs. (A) S1 nuclease analysis of fetal liver RNA of 15.5 day F0BGT22 transgenic mice showing that the range of expression from single-copy transgenes is 13% to 889% per copy and indicating that the distal promoter is important for reproducible expression levels. (B) S1 nuclease analysis of fetal liver RNA of 15.5 day F0BGT23 transgenic mice showing that the range of expression from single-copy transgenes is 33% to 384% per copy and indicating that 5′HS1 is important for reproducible expression levels. (C) S1 nuclease analysis of fetal liver RNA of 15.5 day F0 BGT33 transgenic mice showing that the range of expression from single-copy transgenes is 87% to 136% per copy and showing that both 5′HS1 and the distal promoter are required to obtain reproducible full expression levels. Hβ, human β-globin protected probe fragment; βmaj, mouse β major protected probe fragment; Ntg, nontransgenic; μD, one copy μD14 microlocus line; 3X, probe excess control. {/ANNT;4224n;;left;71808n}C {/ANNT;4224n;;0n;0n}A {/ANNT;4224n;;84480n;0n}B
Molecular analysis of 5′HS1.
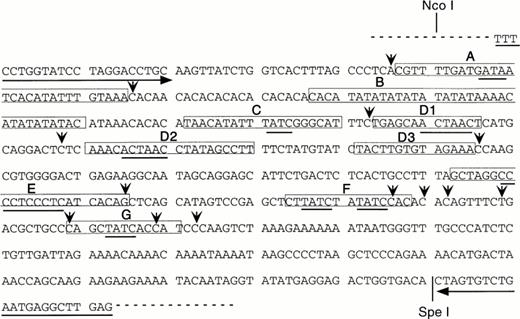
The role of 5′HS1 in activating expression from the β-globin proximal promoter is likely to be mediated by trans-acting factors. As the cis-acting binding sites in 5′HS1 have not been extensively characterized, we chose to identify them by employing in vitro DNaseI footprint analyses. The core fragment of 5′HS1 was cloned by PCR to facilitate these experiments. PCR primers were synthesized that flank the minimal HS site (Fig 4) and that incorporate all the phylogenetically conserved 5′HS1 sequences described in the globin server (http://globin.cse. psu.edu). The sequence of the resulting pGEM-HS1 plasmid was confirmed, although the expected (CA)12(TA)6 tract at the 5′ junction of footprint B was altered to (CA)10(TA)8 (Fig 4). The latter sequence was also present in the pBGT22 plasmid used as a template in the PCR reactions and is presumably a polymorphism (data not shown).
Human 5′HS1 core sequence and nine in vitro DNaseI footprints detected using MEL cell nuclear extracts. Footprints are boxed and labelled (A-C, D1-D3, and E-G). Consensus binding sites are underlined for Sp1 (footprint E), GATA-1 (footprints A, C, F, G), and the ACTAAC motif (footprints D1, D2). Hypersensitive bases are indicated by vertical arrows, horizontal arrows correspond to primers used for cloning 5′HS1 by PCR, dashed line represents the polylinker of the pGEM-T vector.
Human 5′HS1 core sequence and nine in vitro DNaseI footprints detected using MEL cell nuclear extracts. Footprints are boxed and labelled (A-C, D1-D3, and E-G). Consensus binding sites are underlined for Sp1 (footprint E), GATA-1 (footprints A, C, F, G), and the ACTAAC motif (footprints D1, D2). Hypersensitive bases are indicated by vertical arrows, horizontal arrows correspond to primers used for cloning 5′HS1 by PCR, dashed line represents the polylinker of the pGEM-T vector.
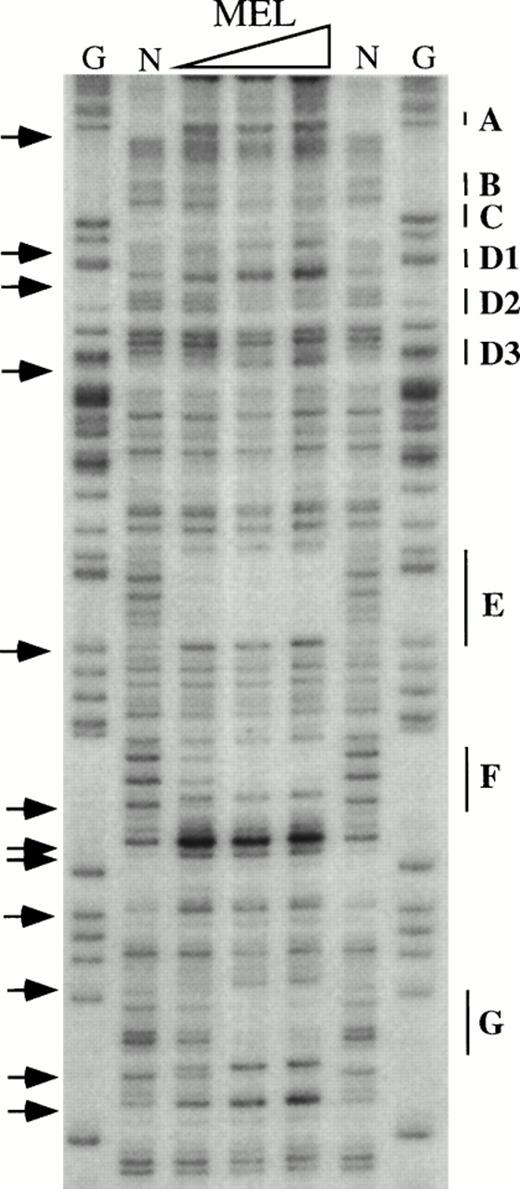
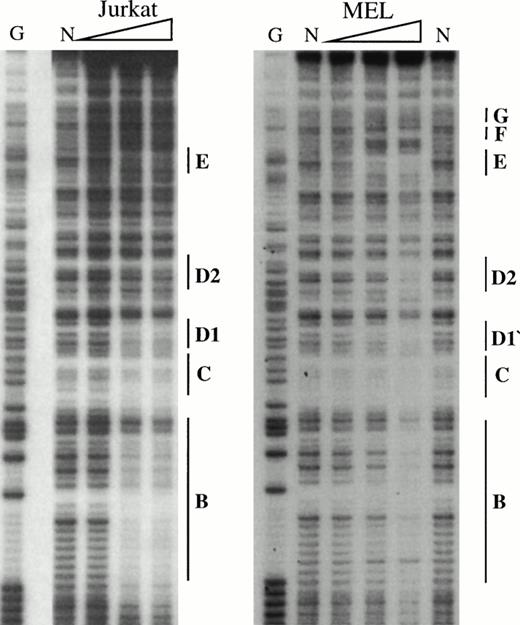
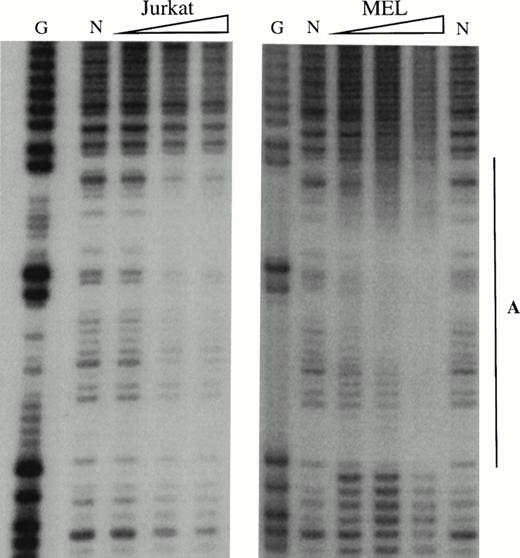
To obtain a probe for in vitro DNaseI footprint analyses, the 3′ end of the pGEM-HS1 sense strand was end-labelled at the Spe1 site, and the 3′ end of the antisense strand was end labelled at the Nco1 site. These two probe fragments were incubated with increasing amounts of induced MEL or Jurkat T cell nuclear extracts before DNaseI digestion, and run in parallel to lanes incubated without extracts (Fig 5). G ladders of the same probes serve as sequence markers. The DNase1 digestion ladders of the two probe fragments shown in Fig 5A-C are protected by MEL extracts at nine footprints labelled A-C, D1-D3, and E-G. Many of these footprints correspond to the DNA-binding sites of known trans-acting factors (Fig4). For example, the strongest footprint E (Fig 5A) contains a consensus site for Sp1 factor, and footprints A, C, F, and G appear to be good candidates for GATA-factor–binding sites (Fig 4). The weak footprints B and D1-D3 do not exhibit any characterized binding sites, but footprints D1 and D2 share a centrally located ACTAAC motif of unknown importance (Fig 4). In vitro DNaseI footprint analyses using nonerythroid Jurkat T-cell nuclear extracts bound to the antisense probe (Fig 5B-C), detect the footprints A to E but not F and G. These results support the conclusion that F and G are bound by an erythroid-specific factor, and that footprints A to E can be bound by factors that are ubiquitous or at least present in hematopoietic cells of nonerythroid origin.
In vitro DNaseI footprint analysis of the 5′HS1 core element. (A) Footprint of the 5′HS1 sense strand probe showing footprints A-C, D1-D3, and E-G. Hypersensitive bases indicated by arrows. (B-C). Footprint of 5′HS1 antisense strand probe protected by the indicated MEL or Jurkat cell nuclear extract. G, chemical sequencing G ladder of the same probe; N, no extract. {/ANNT;;4224n;36608n;0n}B {/ANNT;4224n;;103488n;0n}C {/ANNT;4224n;;0n;0n}A
In vitro DNaseI footprint analysis of the 5′HS1 core element. (A) Footprint of the 5′HS1 sense strand probe showing footprints A-C, D1-D3, and E-G. Hypersensitive bases indicated by arrows. (B-C). Footprint of 5′HS1 antisense strand probe protected by the indicated MEL or Jurkat cell nuclear extract. G, chemical sequencing G ladder of the same probe; N, no extract. {/ANNT;;4224n;36608n;0n}B {/ANNT;4224n;;103488n;0n}C {/ANNT;4224n;;0n;0n}A
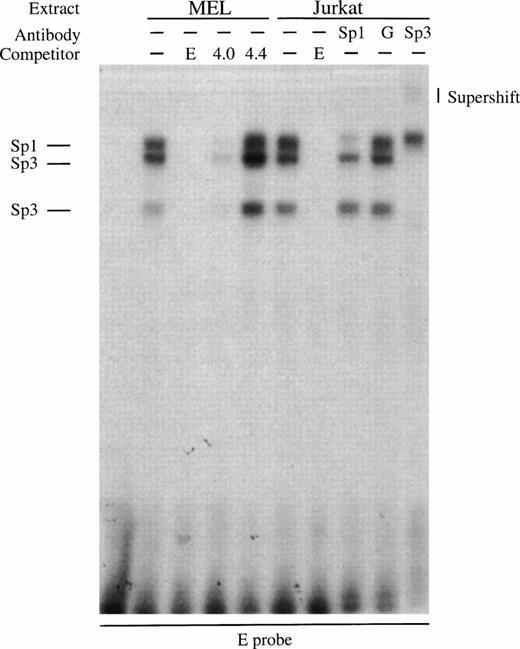
To better characterize the factors responsible for generating the footprints that do contain consensus sites, we performed gel retardation assays. An oligonucleotide probe for the strong footprint E is bound by factors in induced MEL extracts and these complexes are competed by an excess of wild-type Sp1 (4.0) but not mutant Sp1 (4.4) binding sites (Fig 6A). All the complexes are also present in Jurkat extracts, and the upper complex in the Jurkat extracts is blocked by Sp1 antibody but not by GATA-1 antibody (Fig 6A). The lower two complexes migrate at the approximate position of Sp3 and are supershifted by Sp3 antibody (Fig 6A). The erythroid-specific factor EKLF and the widely expressed factor BKLF can recognize some Sp1 consensus sites,36 but we have been unable to show EKLF binding to footprint E probes using our MEL extracts with EKLF or BKLF antibodies, or with GST-EKLF fusion proteins (data not shown). These data show that footprint E is bound by Sp1 and Sp3 factors.
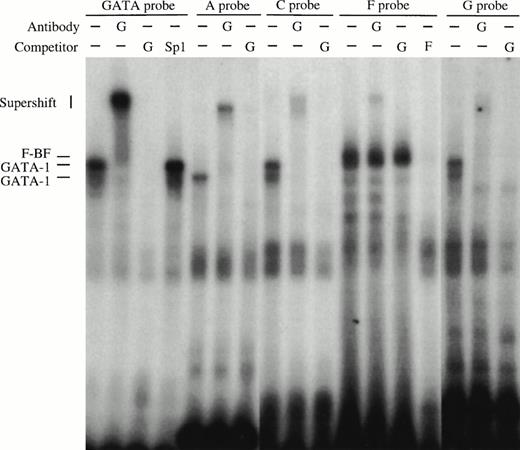
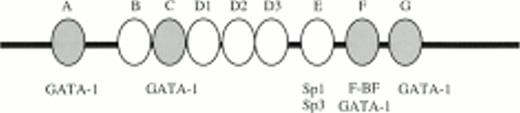
Gel retardation assays of the strong in vitro DNaseI footprinted regions in 5′HS1. (A) Gel retardation assay on Sp1/Sp3 factors bound to the E probe corresponding to the strongest footprint E. Competitors: E, self-competition with HS1FpE site; 4.0, wild-type Sp1 site from 5′HS2; 4.4, mutant Sp1 site from 5′HS2. Antibodies: G, Gata-1. B. Gel retardation assay on the GATA factors bound to the A, C, F, and G footprints. Competitors: Sp1, Sp1-Cruz consensus site; G, GATA-Cruz dimer GATA-1 site; F, self-competition with HS1FpF site. C. Summary of factors bound to each footprint as deduced by gel retardation assays and in vitro DNaseI footprinting. Filled circles, erythroid-specific factors; open circles, ubiquitous factors. {/ANNT;4224n;;0n;0n}A {/ANNT;4224n;;84480n;0n}B {/ANNT;4224n;;0n;103488n}C
Gel retardation assays of the strong in vitro DNaseI footprinted regions in 5′HS1. (A) Gel retardation assay on Sp1/Sp3 factors bound to the E probe corresponding to the strongest footprint E. Competitors: E, self-competition with HS1FpE site; 4.0, wild-type Sp1 site from 5′HS2; 4.4, mutant Sp1 site from 5′HS2. Antibodies: G, Gata-1. B. Gel retardation assay on the GATA factors bound to the A, C, F, and G footprints. Competitors: Sp1, Sp1-Cruz consensus site; G, GATA-Cruz dimer GATA-1 site; F, self-competition with HS1FpF site. C. Summary of factors bound to each footprint as deduced by gel retardation assays and in vitro DNaseI footprinting. Filled circles, erythroid-specific factors; open circles, ubiquitous factors. {/ANNT;4224n;;0n;0n}A {/ANNT;4224n;;84480n;0n}B {/ANNT;4224n;;0n;103488n}C
Oligonucleotide probes to footprints A, C, F, and G all bind complexes that migrate at the approximate size of GATA-1 in induced MEL extracts (Fig 6B). A consensus GATA-1 dimer site is bound by two complexes that can be supershifted with GATA-1 antibody. Complexes bound to each of the A, C, F, and G probes comigrate with either the upper or lower GATA-1 complex or with both, and these complexes supershift with GATA-1 antibody. In addition, footprint F contains a non–GATA-1 complex that is competed with F oligonucleotides but not by GATA-1 sites, and is not supershifted by GATA-1 antibodies. These data show that GATA-1 binds to footprints A, C, F, and G, but that an additional factor (F-BF) also binds to footprint F. The finding that Jurkat extracts protect sites A and C, as shown by the in vitro DNaseI footprints (Fig 5B), may be explained by complexes observed with Jurkat extracts in gel retardation assays (data not shown), perhaps due to binding by a GATA-related factor such as GATA-3 that is present in T cells. In summary, the footprint and gel retardation assays are consistent with the assignment of binding sites shown in Fig 6C, and these factors may participate in a functional interaction between 5′HS1 and the other HS of the LCR and/or the distal promoter.
Distal promoter is not a spacer element.
The distal promoter is required together with 5′HS1 in the BGT33 construct to direct full and reproducible transgene expression. This finding could be explained by either a requirement for specific sequences in the distal promoter to mediate this effect; or a passive spacer effect that merely distances the proximal promoter from the 5′HS1 element. To distinguish these possibilities we tested two new constructs in transient transgenic mice. The BGT41 construct contains a 5′HS1-4 cassette linked to the 815 bp β-globin promoter and serves as a baseline to assess the effect of distal promoter deletion within the context of the 4.0 kb LCR (Fig 1). The BGT40 construct essentially adds back neutral DNA into the BGT41 transgene to examine the role of spacing (Fig 1). If specific sequences within the distal promoter are required to obtain full expression levels and spacing has no effect, then expression levels of BGT41 and BGT40 should be roughly equivalent. In contrast, if spacing alone can account for the distal promoter effect then the BGT40 construct should express to reproducible and full levels equivalent to those previously determined for BGT33. As a neutral spacer for the BGT40 construct, we chose to employ the 717 bp Xba1-Hpa1 fragment containing the human A γ-globin intron 2 because: it does not contain any known regulatory elements unlike the β-globin intron 2; its endogenous location is between the LCR and the β-globin promoter, implying that these spacer sequences will not interfere with LCR activation; it is roughly the same size and has compatible ends for replacement of the distal β-globin promoter; and other mammalian or prokaroyotic DNA fragments may contain undescribed sequences that interfere with LCR function.
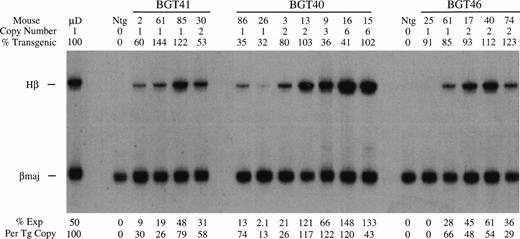
BGT40 and BGT41 transient transgenic mice were created and DNA analyses for copy number, intactness, and mosaicism were determined as before (data not shown). The RNA analysis shown in Fig 7 shows that expression from the BGT41 construct in which the distal promoter has been deleted is reduced to a range of 26% to 79% at single copy. Clearly, deletion of the distal promoter in the context of the 4.0 kb LCR reduces the average expression level. Likewise, the BGT40 construct that contains the spacer element instead of the distal promoter expresses to a similar level as BGT41, with a range of 13% to 74% in single-copy animals and levels approaching 100% in most of the higher-copy animals. The similarly reduced levels of expression in animals bearing these two constructs supports the conclusion that specific sequences are required in the distal promoter to direct 100% expression at single copy.
Expression of globin mRNA in transgenic mice containing the β-globin distal promoter deletion construct (BGT41), distal promoter spacer construct (BGT40), and the 3′ truncation construct (BGT46). (A) S1 nuclease analysis of fetal liver RNA of 15.5 day F0 BGT41 transgenic mice showing that the range of expression from single-copy transgenes is 26% to 79% per copy and indicating that the distal promoter is important for full expression levels within the context of the 4.0 kb LCR. (B) S1 nuclease analysis of fetal liver RNA of 15.5 day F0 BGT40 transgenic mice showing that the range of expression from single-copy transgenes is 13% to 74% per copy and indicating that a spacer element combined with the 815 bp promoter is not sufficient to reconstitute full expression levels equivalent to those directed by the 1555 bp promoter (BGT33). (C) S1 nuclease analysis of fetal liver RNA of 15.5 day F0 BGT46 transgenic mice showing that the range of expression from single-copy transgenes is 0% to 66% per copy and showing that 3′ sequences are required to obtain reproducible full expression levels from the 4.0 kb LCR combined with the 1555 bp promoter. Hβ, human β-globin protected probe fragment; βmaj, mouse β major protected probe fragment; Ntg, nontransgenic; μD, one copy μD14 microlocus line. {/ANNT;4224n;;35904n;0n}A {/ANNT;4224n;;76032n;0n}B {/ANNT;4224n;;133056n;0n}C
Expression of globin mRNA in transgenic mice containing the β-globin distal promoter deletion construct (BGT41), distal promoter spacer construct (BGT40), and the 3′ truncation construct (BGT46). (A) S1 nuclease analysis of fetal liver RNA of 15.5 day F0 BGT41 transgenic mice showing that the range of expression from single-copy transgenes is 26% to 79% per copy and indicating that the distal promoter is important for full expression levels within the context of the 4.0 kb LCR. (B) S1 nuclease analysis of fetal liver RNA of 15.5 day F0 BGT40 transgenic mice showing that the range of expression from single-copy transgenes is 13% to 74% per copy and indicating that a spacer element combined with the 815 bp promoter is not sufficient to reconstitute full expression levels equivalent to those directed by the 1555 bp promoter (BGT33). (C) S1 nuclease analysis of fetal liver RNA of 15.5 day F0 BGT46 transgenic mice showing that the range of expression from single-copy transgenes is 0% to 66% per copy and showing that 3′ sequences are required to obtain reproducible full expression levels from the 4.0 kb LCR combined with the 1555 bp promoter. Hβ, human β-globin protected probe fragment; βmaj, mouse β major protected probe fragment; Ntg, nontransgenic; μD, one copy μD14 microlocus line. {/ANNT;4224n;;35904n;0n}A {/ANNT;4224n;;76032n;0n}B {/ANNT;4224n;;133056n;0n}C
β-Globin 3′ sequences are crucial for LCR function.
As a final investigation to identify the minimal combination of regulatory elements required for full expression, we examined the role of sequences downstream of the β-globin gene. The BGT46 construct resembles the fully functional BGT33 construct except for a 1.65 kb truncation that includes the 3′ enhancer (Fig 1). Expression analysis of transient transgenic mice bearing the BGT46 transgene is shown in Fig 7. Transgene copy number, intactness, and mosaicism levels was determined as described earlier. Mosaicism was determined usingNco1-EcoR1 digested fetal liver DNA compared with the single-copy–bred line B26, and intactness examined with multiple diagnostic restriction sites in addition to PCR (using the 46int1 and 46int2 primers) for the 3′ transgene terminus (data not shown). Expression of most of the transgenes is in the 29% to 66% range, suggesting that 3′ sequences are required to obtain full levels of gene expression. More surprisingly, mouse 25 fails to express significant levels of β-globin mRNA, showing that at some single-copy integration sites 3′ sequences play an essential role in controlling expression of the β-globin promoter despite the presence of the LCR.
DISCUSSION
Definition of the minimal combination of regulatory elements capable of directing full expression of the human β-globin gene in transgenic mice serves the dual purpose of examining the functional and cooperative interactions of these elements; as well as creating a transgene cassette whose expression levels is well suited for gene therapy purposes. Our results show that such full expression is only obtained in the presence of 5′HS1, the distal promoter, and a 3′ fragment that includes a β-globin enhancer.
Requirement for 5′HS1 and distal promoter sequences.
We previously showed that truncation of the β-globin promoter from −815 bp to −265 bp compromised expression from single-copy transgenic mice.21 Here we refine those observations with regard to the distal promoter sequences located from −1555 bp to −815 bp and to its potential interactions with 5′HS1. The BGT22 transgenes show that the complete 6.5 kb microlocus LCR is not capable of reproducibly activating the 815 bp promoter, suggesting that the distal promoter plays an important role in LCR activation. However, the BGT23 transgenes show that in the absence of 5′HS1, the 1555 bp promoter is not sufficient to reproducibly activate full transgene expression directed by the 3.0 kb LCR. The range of expression for single copy BGT22 (13% to 889%) and BGT23 transgenes (33% to 384%) is surprising given that the highest level previously described for the microlocus construct (linked to the 1555 bp promoter) is 188% from a four-copy line.11 34 It is only the BGT33 transgene that is fully activated at all single-copy integration sites (87% to 136%) and that expresses to a range similar to that previously described for the microlocus construct (86% to 188%). BGT33 and the microlocus construct share the feature of containing both 5′HS1 and the distal promoter.
Further experiments were designed to determine whether the distal promoter was a spacer element required for DNA looping by the putative LCR holocomplex to the proximal promoter; or contained specific sequences that were required for full expression. Linkage of the 815 bp promoter to the 4.0 kb LCR in the BGT41 transgene led to a reduction in expression, supporting the earlier results obtained with the 815 bp promoter linked to the 3.0 kb LCR (BGT14). Finally, the BGT40 transgenes showed that neutral spacer DNA inserted upstream of the 815 bp promoter is not sufficient to reactivate full expression, and indicates that specific sequences present in the distal promoter are involved in LCR activation.
Taken together, these data are consistent with a cooperative effect between specific sequences in 5′HS1 and the distal promoter. The importance of 5′HS1 to LCR activity in vivo was also shown by using linked-cosmid transgenic mice to show that 5′HS1 deletion affects position independence directed by the entire human β-globin locus.37 The importance of the distal promoter in the context of the full LCR and the entire β-globin locus is not known, and requires additional mouse knockout or linked-cosmid/YAC transgenic mouse experiments.
Cis-acting sites in 5′HS1.
To complete the molecular characterization of the LCR HS cores, we sought to identify trans-acting factor binding sites in 5′HS1. In vitro DNaseI footprint analyses show that 5′HS1 contains nine binding sites, and gel retardation assays showed that they include a strong Sp1/Sp3 site and four GATA-1 sites. The B and D1 to D3 footprints bind factor(s) that are not erythroid-specific. These data generally agree with the findings of in vivo footprint analyses of 5′HS1 that detected protection of the Sp1 site at footprint E and GATA-1 sites at footprint F and G,38 as well as two weak AP-1 sites that might potentially bind the erythroid factor NF-E2. We have been unable to confirm in vitro footprints at the latter weak AP-1 sites, although NF-E2 and AP-1 are present in our MEL nuclear extracts (data not shown). These AP-1 sites are not phylogenetically conserved in 5′HS1, unlike the Sp1 and GATA-1 sites.39 We have also been unable to detect YY-1 factor binding to 5′HS1 (data not shown).
In summary, we have extended the in vivo analyses by identifying the specific factors that bind to sites in 5′HS1 using competition and antibody supershift protocols on gel retardation assays, and by identifying additional in vitro binding sites 5′ of the region that was footprinted in vivo. The cis-acting features of 5′HS1 differ from those found in 5′HS2-4 by the absence of NF-E2 and YY-1 sites and the presence of the ACTAAC motif in footprints D1 and D2, which is not found at any other location in the sequence of the human β-globin locus.
Cis-acting sites in the distal promoter.
The BGT40 spacer transgene construct indicates that specific sequences in the distal promoter are required for full LCR activity and suggest a functional interaction with 5′HS1. Footprint analyses have been performed on the minimal 265 bp human β-globin promoter,18 and binding sites for the negative factors BP-1 and BP-2 have been shown within the 600 bp promoter element.40 Our preliminary footprint experiments show that there are additional binding sites in the 1555 bp distal promoter (D.P., unpublished results) but these experiments have been hampered by extensive A:T tracts that are not well digested by DNaseI. It has not escaped our notice that the 1555 bp promoter colocalizes precisely with the minimal β-globin origin of replication that extends to exon 2.41 Because replication timing is linked to active expression and is dependent on the LCR,42 it is possible that 5′HS1 functionally interacts with replication factors present in the distal promoter.
Requirement for 3′ sequences.
The final BGT46 transgene construct bearing a 3′ truncation showed that the presence of the 4.0 kb LCR together with the 1555 bp β-globin promoter and intron 2 enhancer is not sufficient to obtain full expression at one or two copies. In fact, mouse 25 expressed to an insignificant amount indicating that this transgene was unable to open chromatin at its integration site, or lies in open chromatin but is unable to activate its promoter. The remaining animals expressed the transgene at a reduced level indicating that 3′ sequences play a role in transcriptional enhancement. This finding that 3′ β-globin sequences are required for LCR activity agrees well with reports that γ-globin transgenes are not highly activated by the LCR unless they contain downstream enhancers or a linked β-globin gene.26-29
Use in gene therapy vectors.
Our minimal combination of regulatory elements that direct full expression from all integration sites and transgene copy numbers includes 5′HS1-4 in a 4.0 kb LCR cassette in which the HS core elements are spaced apart by about 700 bp, and which is linked to a 4.8 kb β-globin gene composed of a 1555 bp promoter and both the intron 2 and 3′ enhancers. This BGT33 construct is 8.8 kb in length and therefore is too large for insertion into retrovirus or AAV vectors designed for gene therapy of β-thalassemia or sickle cell anemia. Moreover, the intron 2 element in this construct is deleterious to retrovirus replication.24,25 Nevertheless, the BGT33 construct has clear advantages for DNA-mediated gene transfer approaches that are not size limited. The size-limited viral approaches have tended to employ nanolocus LCR cassettes, the minimal 265 bp promoter element, and a β-globin gene that contains virtually no 3′ sequences.23-25 Contrary to this reductionist approach designed to optimize viral titers, better expression after viral gene transfer might require larger globin genes including an extended promoter and the 3′ enhancer, while avoiding the deleterious β-globin intron 2 sequences.30 Ideally, these globin transgenes should be regulated by individual HS or small LCR cassettes that function at single copy.
LCR-activation models.
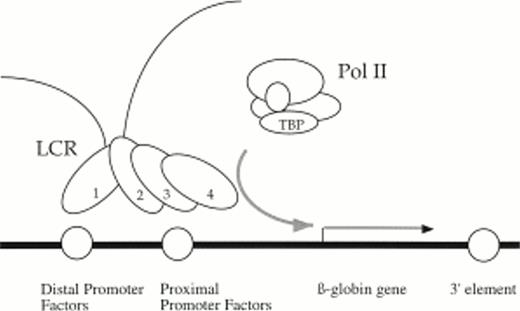
Existing models of LCR activity have no specific function assigned to 5′HS1, the β-globin distal promoter, and the β-globin enhancers. Our data is compatible with the Binary model that does not ascribe specific roles to the individual HS or enhancer elements.43 In addition, our findings can be incorporated into the holocomplex model of LCR activation.11 44 This model postulates that trans-acting factors bound to 5′HS1-4 interact with each other by protein:protein interactions to form a single LCR holocomplex that can then activate transcription from a nearby proximal promoter by DNA looping. We propose a modified version of the model in which 5′HS1 contacts the distal promoter and in the process tethers the enhancement activities of 5′HS2-4 to the proximal promoter element (Fig 8). This mechanism suggests that the role of 5′HS1 and the distal promoter is architectural and designed primarily for regulatory element alignment. A specific role for the 3′ sequences is more elusive. We infer that they either participate in chromatin opening and/or its maintenance, or they directly influence the activity of the proximal promoter.
Holocomplex model of LCR/promoter functional interactions illustrating a putative DNA looping event between 5′HS1 and the distal promoter element. In this model, 5′HS1 interacts with the distal promoter element and tethers the transcriptional enhancement activities of a 5′HS2-4 holocomplex to the proximal promoter element and recruits polymerase. The proposed architectural role of 5′HS1 and the distal promoter facilitates alignment of 5′HS2-4 with the proximal promoter. 3′ sequences are required either to establish and/or maintain open chromatin or directly influence promoter activity.
Holocomplex model of LCR/promoter functional interactions illustrating a putative DNA looping event between 5′HS1 and the distal promoter element. In this model, 5′HS1 interacts with the distal promoter element and tethers the transcriptional enhancement activities of a 5′HS2-4 holocomplex to the proximal promoter element and recruits polymerase. The proposed architectural role of 5′HS1 and the distal promoter facilitates alignment of 5′HS2-4 with the proximal promoter. 3′ sequences are required either to establish and/or maintain open chromatin or directly influence promoter activity.
ACKNOWLEDGMENT
We thank A. Huang for animal care, L. Posner for cloning the pGEM-HS1 plasmid and preliminary footprint experiments, and C. Osborne for critical reading of the manuscript. L.Wall provided MEL C88 cells, MEL cell nuclear extract used in preliminary experiments, and valuable advice on preparation of nuclear extracts and in vitro DNaseI footprinting. S. Philipsen provided the A γ-globin gene plasmid and advice on Sp1/Sp3 supershift experiments. M. Crossley also offered supershift advice and EKLF and BKLF antibodies, and J. Bieker made GST-EKLF fusion protein available.
Supported by a grant from the Medical Research Council (MRC) of Canada to J.E.
Address reprint requests to James Ellis, Developmental Biology Program, Hospital for Sick Children, 555 University Ave, Toronto, Ontario, Canada M5G 1X8; email: jellis@sickkids.on.ca.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.
© 1998 by the American Society of Hematology.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal