Abstract
In comparison with HLA-matched sibling bone marrow transplants, unrelated donor transplants are associated with increased graft-versus-host disease and graft failure. This is likely in part due to HLA incompatibilities not identified by current matching strategies. High resolution DNA-based typing methods for HLA class II loci have improved donor selection and treatment outcome in unrelated donor bone marrow transplantation. By using DNA-based typing methods for HLA-A and -B on a cohort of 100 potential bone marrow donor/patient pairs, we find that serological typing for HLA class I is limited in its ability to identify incompatibilities in unrelated pairs. Furthermore, the incompatibilities identified are associated with the presence at high frequency of alloreactive cytotoxic T-lymphocyte precursors. DNA typing also indicates that HLA-C mismatches are common in HLA-A and -B serologically matched pairs. Such mismatches appear to be significantly less immunogenic with respect to cytotoxic T-lymphocyte recognition, but are expected to influence natural killer cell activity. Thus, improved resolution of HLA class I shows many previously undisclosed mismatches that appear to be immunologically functional. Use of high resolution typing methods in routine matching is expected to improve unrelated donor selection and transplant outcome.
ALLOGENEIC BONE MARROW transplantation (BMT) involves a unique immunological assault on the recipient. HLA differences between individuals can lead to the expansion of high frequency alloreactive T cells, often leading to severe graft-versus-host disease (GVHD) in the allogeneic BMT setting.1 An HLA genotypically identical sibling is therefore the donor of choice. However, because such a donor is available to only about 30% of patients, alternative donors, such as partially HLA-matched related and phenotypically HLA matched unrelated donors, are increasingly used. Although single HLA differences may be tolerated in related donor transplants, especially in younger recipients,2 such donors are rare, and increased disparity is associated with greatly reduced transplant success.3Consequently, HLA-matched unrelated donor transplants are often used in the treatment of a range of hematological disorders.
HLA matching is complex involving at least three highly polymorphic loci, HLA-A, -B, and -DRB1, encoding more than 400 alleles.4 In siblings, this complexity is reduced, because matching simply involves distinguishing the inherited from noninherited haplotypes. However, unrelated donor/patient pairs must be matched for each antigen and/or allele and, due to the extensive polymorphism of the HLA system, the chance of unrelated individuals being HLA identical is remote.5 It is only with the establishment of large international registers of HLA-typed individuals (currently >4.6 million potential donors have been recruited worldwide) that identification of a suitable unrelated donor is practical. Compared with HLA genotypically identical sibling transplants, HLA phenotypically matched unrelated donor transplants are commonly associated with increased risk of GVHD posttransplant6 and failure to engraft. It is likely that the increase in such posttransplant complications is at least in part due to the presence of HLA alloantigens not discriminated in the current matching procedure.
HLA typing for DRB1 and DQB1 is now performed using DNA-based methods allowing the routine identification of most Caucasoid alleles. However, until recently, matching for HLA-A and -B has been dependent on serological techniques. Serological resolution was thought to be adequate, because, in contrast to the typing of HLA-DR and -DQ serology, serological typing for HLA-A and -B identifies a greater number of antigens and a high level of heterozygosity at each locus. Despite this, there are limitations in the use of HLA class I serological data for matching, as cellular,7biochemical,8 and sequencing strategies show.9Although the importance of such differences in BMT remains to be fully defined, case studies have implicated alloresponses due to a single amino acid difference in HLA-B44 subtypes in both graft rejection10 and GVHD.11
Although matching at HLA-A and -B loci is known to be important, the impact of HLA-C on transplantation is unclear. HLA-C is the third classical class I locus to be described, but, due to its low level of cell surface expression and reduced polymorphism, has been considered as less immunologically relevant.12 However, HLA-C–restricted virus-specific,13tumor-specific,14 and allospecific cytotoxic T lymphocytes (CTLs)15 are found, indicating that HLA-C has a role to play in a range of T-cell immune responses.
Interest in HLA-C and transplantation has been stimulated by recent exposition of its role in modulating natural killer (NK) cell activity. Interaction of self-MHC with receptors on a subset of NK cells is crucial for the protection from NK cell-mediated lysis.16In humans, this interaction has been mapped to a dimorphic epitope at residues 77 and 80 on the α1 domain of HLA-C, each epitope interacting with a distinct killer inhibitory receptor (KIR).17 Those cells expressing HLA-C molecules with a motif shared with HLA-Cw*0303 (Ser77/Asn 80) are protected from lysis by NK clones expressing KIR2DL2/3, whereas expression of a motif shared with HLA-Cw*0401 (Asn77/Lys80) inhibits lysis by NK clones expressing KIR2DL1.18 Alloreactive NK cells can be generated against stimulators expressing the opposite motif to that of the responder.19 The implications for BMT may be seen in mouse experiments in which NK cells of an irradiated F1 hybrid are able to reject parental bone marrow20 that lacks expression of self-MHC class I molecules.21 Recent reports suggest increased likelihood of graft failure in HLA-C–mismatched BMT,22 23 which may be analogous to the mouse hybrid resistance model.
The frequency of such serologically undetected mismatches and their impact on transplantation have yet to be fully assessed. In a previous report, we described that high CTL precursor (CTLp) frequencies were common in HLA-A, -B serologically and biochemically matched unrelated pairs, suggesting that undetected mismatches are common.24We have applied high resolution HLA class I typing techniques to study the true level of HLA compatibility in conventionally matched unrelated patient/donor pairs. Comparison of CTLp frequencies and DNA typing information may allow the assessment of the functional significance of such mismatches in allorecognition.
MATERIALS AND METHODS
Patients and donors.
Seventy-six patients and 100 potential unrelated bone marrow donors were included in this study. Donors were selected as the best available matches for patients on the basis of matching for HLA-A and -B by serology and DRB1 identity by DNA-based typing. All patient/potential donor pairs were also analyzed for alloreactive CTLp frequency in the graft-versus-host direction.
Initial HLA typing for donor selection.
HLA-A and -B typing was performed by the standard complement-dependent microcytotoxicity test, using a combination of local and commercial (Biotest, Germany) serological typing trays. HLA-DR and -DQ types were assigned initially using commercial serological typing trays (Biotest). DNA from 97 of 100 pairs was then typed by a combination of polymerase chain reaction–sequence-specific oligonucleotide probes (PCR-SSOP) and polymerase chain reaction–sequence-specific primers (PCR-SSP) for DRB1 and similarly 95 of 100 for DQB1, as previously described.25 Where fully HLA compatible unrelated donors were not found on international volunteer registers, donors with minor HLA mismatches were considered. Minor mismatches were defined as either an HLA-A or -B subtype of a serologically related group as defined by the World Health Organization (WHO) nomenclature committee or mismatched at the molecular level for serologically defined HLA-DR1-14 specificities. Three pairs were serologically mismatched for HLA-A and 9 pairs for HLA-B, and 14 pairs were originally mismatched at the molecular level for HLA-DRB1. In total, mismatches were detected between 23 pairs at HLA-A, -B, and/or -DRB1.
Limiting dilution analysis.
Limiting dilution analysis was performed in the graft-versus-host direction as described by Kaminski et al26 to determine the frequency of patient-specific donor CTL precursors. A high CTLp frequency was taken to be greater than 1:105 peripheral blood mononuclear cells (PBMCs), because this correlates with poor transplant outcome.27 Low CTLp frequency was taken as being between 1:105 and 5:106, with CTLp frequencies less than this being regarded as negative.
DNA typing for HLA-A, -B, and -C.
To identify the presence of serologically undetected incompatibilities, a combination of DNA-based typing methods were used. HLA-A and -B were typed by previously described PCR-SSOP methods28,29 to confirm serotypes. Locus-specific oligotyping enabled the subtyping of most common HLA-A and -B serotypes to varying levels of resolution. The resolution offered by these techniques was dependent on the combination of alleles present, because heterozygosity often hindered allele-specific assignment. B locus oligotyping resolution was improved by using 3′ primers D1 and D2 separately where appropriate to amplify and analyze each allele individually.29 In total, the B locus alleles were amplified separately in 30% of pairs.
Reference strand-mediated conformation analysis (RSCA) matching of patient/donor pairs.
Because PCR-SSOP typing resolution is limited both by the number of probes used and ambiguities caused by heterozygosity, a conformation-dependent technique has been used on selected pairs. RSCA has recently been described as a method of identifying HLA alleles on the basis of mobility of a heteroduplex formed between the sense strand of a reference allele and the antisense strand of the unknown allele.34
Briefly, HLA-A, -B, or -C amplification of a fragment containing exon 2, intron 2, and exon 3 was performed using locus-specific primers described previously.35 Reference strand amplification was performed from cell line DNA homozygous for the appropriate HLA allele using the same primers except with the sense primer labeled at the 5′ end with the Cy5 fluorochrome (Pharmacia Biotech, Uppsala, Sweden). Amplified reference and sample products were mixed at a 1:3 ratio and hybridized as previously described.34 Duplexes were separated by polyacrylamide gel electrophoresis (PAGE) using an ALFexpress automated sequencer (Pharmacia Biotech), and the mobility of the fluorescent duplex bands was analyzed using Fragment manager software (Pharmacia Biotech). By comparison with the mobilities of known heteroduplexes, it was possible to assign allelic specificities. Reference alleles used were as previously described,36 with the exception of HLA-B*1501 (amplified from DNA from IHW 9072 cell line) for confirmation of matching HLA-B62 seropositive donor/patient pairs.
RESULTS
Identification of serologically undetected HLA-A and -B incompatibilities.
To determine the level of HLA-A and -B undetected mismatching in serologically matched patient/donor pairs, samples were analyzed using higher resolution DNA-based methods. Patient/donor pairs were initially fully matched for HLA-A in 97% of cases (Fig 1). DNA typing of the patient/donor pairs confirmed the mismatches identified by serology. Two further patient/donor pairs were found to be incompatible using oligotyping for HLA-A, one indicating an HLA-A*02 subtype mismatch and the other an HLA-A*0301 versus -A*0302 mismatch (Table1). Specific subtyping confirmed the HLA-A*02 incompatibility as HLA-A*0201 versus -A*0205 and that 55 further HLA-A2 seropositive pairs were matched at the subtype level. That 91 of 92 HLA-A2 seropositive individuals were encoded by HLA-A*0201 reflects the dominance of this subtype in North European Caucasoids.37 RSCA matching indicated two further mismatches at A locus, an HLA-A*30 subtype mismatch and an HLA-A*03 heterozygote donor (A*0301, *03v) and HLA-A*0301 homozygous patient.
Level of matching achieved by conventional typing methods. Patients and potential donors were typed by serological methods for HLA-A and -B and DNA-based methods for HLA-DRB1 and -DQB1. Limiting dilution analysis was performed in the graft-versus-host direction to assess the frequency of host specific CTL precursors. Pairs with a high CTLp frequency (>1:105 PBMC) were judged to be mismatched at the cellular level.
Level of matching achieved by conventional typing methods. Patients and potential donors were typed by serological methods for HLA-A and -B and DNA-based methods for HLA-DRB1 and -DQB1. Limiting dilution analysis was performed in the graft-versus-host direction to assess the frequency of host specific CTL precursors. Pairs with a high CTLp frequency (>1:105 PBMC) were judged to be mismatched at the cellular level.
Frequency of Mismatching Is Serotype Dependent
| . | HLA Serotype . | ||||||
|---|---|---|---|---|---|---|---|
| A2*,† . | A3*,† . | A30† . | B35*,† . | B39† . | B44*,† . | B51† . | |
| Donor/patient pairs | 56 | 24 | 4 | 15 | 8 | 25 | 4 |
| Mismatches detected | 1 | 2 | 1 | 6 | 3 | 4 | 2 |
| Subtypes found | 2 | 3 | 3 | 4 | 2 | 2 | 3 |
| . | HLA Serotype . | ||||||
|---|---|---|---|---|---|---|---|
| A2*,† . | A3*,† . | A30† . | B35*,† . | B39† . | B44*,† . | B51† . | |
| Donor/patient pairs | 56 | 24 | 4 | 15 | 8 | 25 | 4 |
| Mismatches detected | 1 | 2 | 1 | 6 | 3 | 4 | 2 |
| Subtypes found | 2 | 3 | 3 | 4 | 2 | 2 | 3 |
Mismatch detected by PCR-SSOP.
Mismatch detected by RSCA.
Original serological testing showed that 91% of patient/donor pairs were matched for HLA-B. A greater level of mismatching was found for HLA-B using DNA-based typing techniques. Discrepancies may occur between serological techniques due to the cross-reactivity of alloantisera, limitations in the alloantisera used, or the lack of expression of an allele. HLA-B SSOP found 3 samples to be misassigned by serology. In one case, HLA-B58 was missed and in another the same antigen was misassigned HLA-B57. In both cases, HLA-B62 was the second serotype expressed and errors in serological typing were likely due to serological cross-reactivity for HLA-B15 and -B17 groups. In the third, an HLA-B38 was not originally identified by serology. However, retyping by serology confirmed the presence of HLA-B38 as being expressed and not a null allele.
By using group-specific oligotyping methods,30,31 we identified heterogeneity within the HLA-B35 and -B44 serotypes. Four HLA-B*35 subtypes were identified at varying frequencies within the 27 individuals tested (Table 1). Importantly, 40% of the HLA-B35 seropositive patient/donor pairs tested were mismatched at the subtype level. HLA-B*44 subtyping indicated two common subtypes, HLA-B*4402 and -B*4403, in a 3:2 ratio. Despite the presence of these two common subtypes, only 4 of 25 (16%) of HLA-B44 seropositive pairs were mismatched at the subtype level. This low percentage is likely due to the HLA-B*44 subtypes commonly segregating on different haplotypes38 and matching for other loci fortuitously results in frequent HLA-B*44 subtype compatibility.
Oligotyping was limited in its resolution by the number of probes used and the heterozygosity exhibited by most samples. Two PCR primer mixes were used for HLA-B SSOP, which enabled the separate typing of each allele in a proportion of allelic combinations. However, RSCA was useful in identifying polymorphisms not detected by oligotyping and also in confirming homogeneity in potentially heterogeneous serotypes. Three HLA-B*51 subtypes and two commonly occuring HLA-B*39 subtypes were identified by this technique (Table 1) as well as those HLA-B*35 and -B*44 subtypes identified by SSOP. In contrast, only one subtype was detected in 45 HLA-B7 and 40 HLA-B8 seropositive individuals. Furthermore, HLA-B62, a serotype shown to be encoded by many distinct alleles,4 was identified as B*1501 in all 16 cases tested.
After molecular typing methods were applied, mismatching for HLA-A and -B increased to 7% and 27% of pairs, respectively. In total, those matched for HLA-A and -B decreased from 89% as initially detected by serology to 70% of pairs (Fig 2). The higher level of HLA-B mismatching reflects the limitations in serological resolution at this locus.
HLA matching for bone marrow donor selection. DNA based typing (▪) showed an increased level of mismatching for HLA-A and -B over that defined by serology (░). Although no HLA-C serology was performed for original typing, DNA-based typing indicated a high level of incompatibility.
HLA matching for bone marrow donor selection. DNA based typing (▪) showed an increased level of mismatching for HLA-A and -B over that defined by serology (░). Although no HLA-C serology was performed for original typing, DNA-based typing indicated a high level of incompatibility.
Molecular HLA-C typing.
Unlike HLA-A and -B, no account was taken of HLA-C match status in the selection of donors for final stage testing. However, because HLA-B and HLA-C are in strong linkage disequilibrium, some level of matching was expected. To measure the level of matching at HLA-C and so determine its impact on the match status of patient donor pairs, PCR-SSP and PCR-SSOP typing methods were used. DNA-based typing identified HLA-C mismatching in 33 of 84 pairs tested (39%), a higher level than that seen for HLA-A or -B (Fig 2), with 8 pairs mismatched for both HLA-C alleles. Importantly, 24 of the 33 (73%) HLA-C mismatched pairs had further HLA-A and/or -B incompatibilities identified at the molecular level, indicating that HLA-C is a useful marker for other mismatches on the haplotype. The utility of HLA-C type as an indicator of HLA-B compatibility was allele dependent. HLA-B*4402 and B*4403 were predominantly associated with restricted HLA-C locus alleles, Cw*0501 and Cw*1601, respectively. In contrast, HLA-B*5101 was associated with a wide range of HLA-C alleles, as reported previously.15 39Thus, mismatching at HLA-C was not predictive of further mismatches for all haplotypes. Overall, 22 of 25 potential donor/patient pairs mismatched at HLA-B were also mismatched at HLA-C.
Mismatching for the HLA-C encoded motifs that interact with KIRs, thus influencing NK cell allorecognition, was next assessed. Each individual was classified as being positive for the Asn77 and Lys80 (group 1) and/or Ser77 and Asn80 (group 2) HLA-C molecules.18 We then calculated whether the HLA-C mismatched donor/patient pairs were also mismatched for this motif. Twenty-two of the 33 (67%) HLA-C mismatched pairs were also mismatched at the level of KIR binding motif. However, no mismatched pairs were homozygous for opposite KIR binding motifs, and so NK cell allorecognition would be expected to be unidirectional. Nine of the 22 mismatches may be expected to influence allorecognition in the graft-versus-host direction and 13 in the host-versus-graft direction.
Cellular recognition.
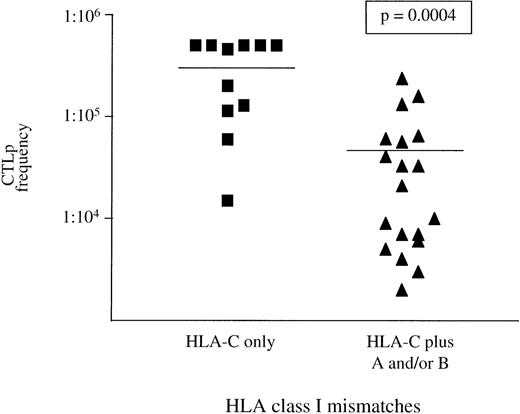
It has been reported that HLA-A and -B mismatches detected by serology and at the DNA level are recognized in vitro by high frequency CTL.24 40 In total, 86% of all donor/patient pairs mismatched for HLA-A or -B in the graft-versus-host direction fell into the high CTLp frequency group. Although the range of CTLp frequencies was broad, there was no evidence that mismatches detected by DNA typing were less immunogenic than those typed by serological methods, suggesting they may be as functionally relevant. In contrast to HLA-A and -B mismatches, 40% of HLA-C incompatible pairs fell into the low or negative CTLp frequency group. This suggests that HLA-C alloantigens may be less immunogenic than HLA-A or -B. To further investigate this, the donor/patient pairs were divided into those with only HLA-C mismatches and those with HLA-C plus other HLA class I incompatibilities. Mean CTLp frequencies differed significantly between both groups (P = .0004), with 82% of pairs with only HLA-C mismatches being in the low or negative CTLp frequency group (Fig 3). Only four donor/patient pairs were identified with HLA-A or -B mismatches in the graft-versus-host direction without HLA-C incompatibility. For these four pairs, a mean CTLp frequency of 1:30,000 was obtained (range, 1:23,000 to 1:36,000).
The effect of HLA-C mismatching on CTLp frequency. CTLp frequencies of 30 pairs with patient HLA-C locus incompatibilities are shown above. Mean CTLp frequencies differed significantly between those with only HLA-C mismatches (1:316,500) and those with further HLA class I mismatches (1:47,110). This indicates that HLA-C alloreactive T cells are found at a lower frequency than those at HLA-A and -B. The limit of sensitivity of this assay is 1 patient-specific donor CTLp/5 × 105 or higher. Mean CTLp frequencies differed significantly between groups according to the Mann-Whitney test.
The effect of HLA-C mismatching on CTLp frequency. CTLp frequencies of 30 pairs with patient HLA-C locus incompatibilities are shown above. Mean CTLp frequencies differed significantly between those with only HLA-C mismatches (1:316,500) and those with further HLA class I mismatches (1:47,110). This indicates that HLA-C alloreactive T cells are found at a lower frequency than those at HLA-A and -B. The limit of sensitivity of this assay is 1 patient-specific donor CTLp/5 × 105 or higher. Mean CTLp frequencies differed significantly between groups according to the Mann-Whitney test.
Previous studies have indicated undetected HLA class I mismatches to be responsible for high CTLp frequencies. Despite using DNA-based HLA-A, -B, and -C matching for serologically matched pairs, 24 of 47 pairs (51%) with a high CTLp frequency had no discernible mismatch at HLA class I (Fig 4). Furthermore, only 2 DRB1 and 4 DQB1 mismatches were detected in the 24 pairs in the high CTLp and no HLA class I mismatch group.
DNA-based typing shows many HLA class I mismatches in the high CTLp frequency group not identified using serological methods. (▪) Matched pairs; (░) those with detected HLA class I mismatches. Forty-seven donors had high (>1:105) patient-specific CTLp frequencies. Only 8 (17%) of these had HLA class I mismatches detected by serological typing, reflecting the low overall number of HLA class I serological mismatched pairs. The proportion of HLA class I mismatched pairs in the high CTLp frequency group increased to 23 (49%) after DNA-based typing, with 21 pairs mismatched at the HLA-B and/or -A locus. However, 24 pairs (51%) with high CTLp frequencies appear to be compatible for HLA class I.
DNA-based typing shows many HLA class I mismatches in the high CTLp frequency group not identified using serological methods. (▪) Matched pairs; (░) those with detected HLA class I mismatches. Forty-seven donors had high (>1:105) patient-specific CTLp frequencies. Only 8 (17%) of these had HLA class I mismatches detected by serological typing, reflecting the low overall number of HLA class I serological mismatched pairs. The proportion of HLA class I mismatched pairs in the high CTLp frequency group increased to 23 (49%) after DNA-based typing, with 21 pairs mismatched at the HLA-B and/or -A locus. However, 24 pairs (51%) with high CTLp frequencies appear to be compatible for HLA class I.
Overall level of matching.
Donors were selected on the basis of being closely matched for HLA-A and -B by serological methods and -DRB1 (and DQB1) by molecular methods. Using molecular typing methods for HLA-A and -B and including HLA-C typing data, the level of matching was seen to be greatly reduced. Of 89 pairs (89 donors for 76 patients) studied, 63% were originally regarded as fully matched. After DNA-based typing, this figure was reduced to 46%. However, single detected mismatches indicate mismatched haplotypes and increase the chances of there being incompatibilities at other loci. By using high resolution class I typing, we have found 43% of donor patient pairs with two or more mismatches and 17% with three or more (Fig5).
Multiple mismatches are shown by high resolution HLA class I typing. (▪) Original matching level; (░) the matching level obtained after DNA typing for HLA class I. After serological testing for HLA-A, -B and DNA typing for HLA-DRB1, -DQB1, 92% of pairs fell in the 0 or 1 mismatch group. This was reduced to 69% after DNA typing of HLA-A, -B, and -C. The increase in pairs with 3 or more mismatches increased from 2% to 16%, reflecting the association of HLA subtypes on different haplotypes.
Multiple mismatches are shown by high resolution HLA class I typing. (▪) Original matching level; (░) the matching level obtained after DNA typing for HLA class I. After serological testing for HLA-A, -B and DNA typing for HLA-DRB1, -DQB1, 92% of pairs fell in the 0 or 1 mismatch group. This was reduced to 69% after DNA typing of HLA-A, -B, and -C. The increase in pairs with 3 or more mismatches increased from 2% to 16%, reflecting the association of HLA subtypes on different haplotypes.
DISCUSSION
Unrelated donor BMT has only been made practical by the establishment of large volunteer registries and implementation of HLA typing techniques with sufficient resolution for matching. Although HLA-matched siblings can be identified by a combination of a mixed lymphocyte reaction (MLR) assay and serological typing, this combination of tests has not proven effective in predicting GVHD in the unrelated donor setting.41 DNA-based typing methods have therefore been developed to allow accurate matching of HLA class II loci and are now used routinely for selection of matched unrelated donors, with improved transplant results.42 In contrast, HLA-A and -B specificities have until recently only been defined using classical serological methods. We have implemented DNA typing methods for HLA-A, -B, and -C to study the deficiencies of current HLA class I typing in accurately matching unrelated pairs. It is only by identifying the precise level of matching in BMT that the importance of serologically undefined differences can be assessed.
By using DNA-based typing for HLA class I, we have been able to identify many more serologically undetected mismatches in HLA-B than HLA-A. It appears that most HLA-A serotypes in our population are encoded for by one dominant allele as indicated by HLA-A2 subtyping (Table 1). In contrast, several HLA-B serotypes were encoded by multiple alleles. Although the incompatibilities identified by DNA-based typing involved molecular differences of as little as one amino acid, these are likely to be in positions on the HLA molecule expected to interact with bound peptide and/or T-cell receptor.43,44 Such small differences have been shown to generate vigorous alloreactive CTL responses in the BMT setting leading to severe complications.10 11 To further emphasize the functional relevance of these mismatches, high CTLp frequencies were detected in 86% of HLA-A, -B mismatched pairs.
We have confirmed that HLA-C incompatibilities are frequent in serologically HLA-A, -B matched unrelated pairs.15,45However, due to the distribution of polymorphic residues within HLA-C antigens and their reduced cell surface expression, it has been suggested that HLA-C may not be as immunologically relevant as the other classical class I loci.46 It has previously been suggested that HLA-C incompatibilities do correlate with high CTLp frequency,47 although in that study HLA-A and -B typings had not been determined using high resolution methods. In contrast, we have demonstrated that, whereas HLA-A and -B mismatches correlate with a high CTLp frequency, this was not so for HLA-C mismatching. By using high resolution matching techniques, we show that, of all the C locus mismatched pairs with high CTLp frequency, 89% had other class I differences. Furthermore, 82% of those with no other HLA class I mismatch were found to have a low or negative CTLp frequency (Fig 4). This supports the view that HLA-C may be of less immunological relevance than HLA-A and -B with respect to CTL surveillance.
Whereas HLA-C incompatibilities may not be a major target for alloreactive CTLs, certain mismatches will result in differential expression of the motifs responsible for modulating alloreactive NK cell activity. Mismatching for the HLA-C encoded NK resistance motifs was seen in 67% of those mismatched at this locus and 26% of all pairs studied. Because HLA phenotype appears to play an important role in defining the KIR repertoire of an individual,19,48potentially alloreactive NK cells are likely to be present in a significant proportion of unrelated donor transplants and their possible impact should be considered. A phenomenon in the mouse known as hybrid resistance in which bone marrow from an MHC homozygous parent is rejected by its F1 hybrid offspring has been shown to be mediated by NK cells.20 It is now known that the lack of self-MHC molecules on murine donor bone marrow cells leaves them vulnerable to recipient NK-mediated lysis.21 An analogous situation has been demonstrated in vitro in the human, with HLA-C playing a key role in the protection from NK lysis.49
The detection of high CTLp frequency has been hypothesized to be a useful indicator of HLA class I incompatibilities not identified by serological methods.24,50,51 However, the range of CTLp frequencies for the same HLA mismatch may be large between unrelated individuals (Breur-Vriesendorp et al52 and unpublished data). Whether the absolute frequency or the qualitative properties of alloreactive CTLs calculated in vitro is important is not clear. Evidence suggests the CTLs involved in GVHD are not the same as those generated in vitro.51 Despite the use of high resolution HLA matching techniques, a significant number of pairs in the high CTLp frequency group (51%) had no detected mismatches in the GVHD direction. In such cases, full HLA-A, -B, and -C sequencing would be useful in confirming patient/donor HLA matching. A possible explanation for such CTL activity is that the peptide repertoire varies sufficiently between unrelated individuals to induce a strong alloreactive CTL response in vitro. In fact, serological reagents have been used to distinguish HLA-B antigens in the absence of allelic differences.53 54 CTLp frequency is often used as a criterion for selection of a suitable unrelated donor. It will therefore be important to assess the functional significance of high CTLp frequencies in the absence of HLA incompatibilities.
The term minor HLA mismatch is commonly used in reference to an HLA-A, -B mismatch of the same serological cross-reactive group or HLA class II allele encoding a product of the same serological group. Matching for HLA-B serological splits has been shown to be of little benefit in renal transplantation.55 However, evidence suggests that these incompatibilities are of major importance to the outcome of allogeneic BMT. Serologically undetected HLA class I mismatches are recognized efficiently by alloreactive CTL correlating with poor transplant outcome.27,40,56 57
By improving the resolution of HLA class I matching, the number of perfectly matched pairs has significantly decreased. Furthermore, many pairs were found to have multiple mismatches, which has been shown to increase the risk of posttransplant complications. Thus, the problem created by high resolution typing of HLA loci is a reduction in the number of matched donors that can be provided. However, this study has demonstrated that many of the HLA mismatches identified are on haplotypes frequent in the Caucasoid population. Therefore, the likelihood increases that several unrelated donors will be matched at the serological level at HLA class I. The development of techniques such as RSCA will allow the convenient, rapid, and accurate screening of a large number of such potential donors. By using such a strategy, HLA matching levels should improve. However, there will remain a significant number of patients without fully HLA matched donors and we need to assess carefully what level of HLA mismatching can be acceptable for a beneficial outcome to BMT. Retrospective analyses of unrelated donor transplants using high resolution typing have been performed by several groups in an attempt to identify those mismatches best tolerated,58 59 and indications that mismatched transplants can be successful, especially for younger patients, are encouraging. The hope is that such studies will allow the rational selection of the most appropriate mismatched bone marrow donor.
ACKNOWLEDGMENT
The authors thank I. Anthony Dodi and Steven G.E. Marsh for critical review of the manuscript and the staff of the Anthony Nolan Tissue-typing Laboratories for excellent technical assistance.
Supported by The Anthony Nolan Bone Marrow Trust. J.R.A. is a recipient of a fellowship from the Consejo Nacional de Ciencia y Tecnologia, Mexico and Overseas Research Students Awards (CVCP) U.K.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to J. Alejandro Madrigal, MD, PhD, The Anthony Nolan Research Institute, The Royal Free Hospital, Pond Street, London, NW3 2QG, UK; e-mail: madrigal@rfhsm.ac.uk.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal