To the Editor:
The translocation t(14;18)(q32;q21), characteristic for follicular lymphomas and a subset of diffuse large cell lymphomas, juxtaposes the bcl-2 proto-oncogene with the Ig heavy-chain (IgH) gene. This results in deregulation of the bcl-2 gene expression and elevation of BCL-2 protein, which protects the cells against induction of programmed cell death and, thus, confers a survival advantage leading to immortalization of t(14;18)-carrying cells. The acquisition of additional chromosomal aberrations is necessary for the malignant transformation and clonal progression of BCL-2 overexpressing lymphocytes. These secondary changes can be observed by karyotyping in more than 90% of lymphomas with t(14;18).1 2
By karyotyping a series of 201 lymphomas at the Department of Human Genetics in Kiel, we detected deletions in the long arm of chromosome 10 in 6 of 57 (11%) B-cell lymphomas with a t(14;18), but in only 1 of 144 (1%) lymphomas without a t(14;18). Thus, loss in 10q seems to be a characteristic secondary abnormality for t(14;18)-positive lymphomas (P < .001). The frequency of del(10q) in t(14;18)-positive lymphomas observed in our series are in line with other cytogenetic studies reporting abnormalities of the long arm of chromosome 10 in 7 of 66 (11%)2 and 9 of 75 (12%) follicular lymphomas.3
The common region of cytogenetic loss in our series encompassed the chromosome band 10q24, suggesting the existence of a tumor suppressor gene involved in the pathogenesis of t(14;18)-positive lymphomas in this region. So far, two tumor suppressor genes have been identified within this region, namely PTEN/MMAC1 in 10q23.34-6 and MXI1 in 10q24-25.7 To determine whether these two genes might be targets of the 10q-deletion in lymphomas, we performed interphase fluorescence in situ hybridization (FISH) with CEPH-YACs 746H8 and 790H10 containing the PTEN/MMAC14 and MXI1 genes, respectively (Whitehead Institute,http://www-genome.wi.mit.edu/cgi-bin/contig/yac_info). Indirect FISH was performed as described recently8 on fixed cells from cytogenetic suspension from a series of primary follicular and diffuse large cell lymphomas and the cell line Karpas 422.9 In addition to a karyotypically detectable deletion in 10q, all lymphomas except one with a variant translocation t(2;18) contained a t(14;18). The false-positive rates for YACs 746H8 and 790H10 determined in extensive control studies were 0.125% (SD 0.25%) and 0.25% (SD 0.5%), respectively. In analogy to previous studies,8 the cut-off level for the diagnosis of a deletion in interphase cells was therefore set at 5%, which is above the frequency of false-positive cells in controls plus 3 SD. Relying on this threshold, a deletion of the PTEN/MMAC1 containing YAC 746H8 was detected in the cell line Karpas 422 as well as in the primary lymphomas from all 14 patients for which material suitable for our assay was available (Fig1A). The mean percentage of cells displaying a signal constellation indicative for loss of the PTEN/MMAC1 gene was 47% (range, 12.5% to 90%). A codeletion of the MXI1 containing YAC 790H10 was only detected in two primary lymphomas. Thus, the common region of molecular cytogenetic loss comprises a region proximal to the MXI1 gene, but encompassing the PTEN/MMAC1 locus.
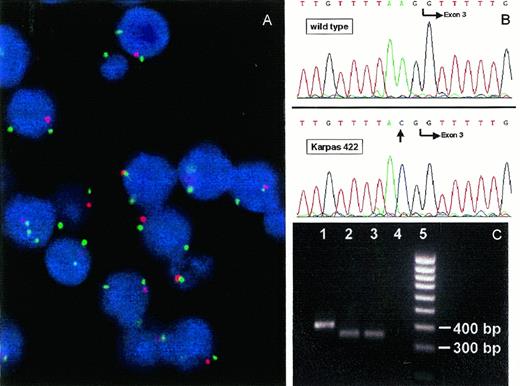
(A) FISH with YAC 746H8 (red fluorescence) containing the PTEN/MMAC1 gene and YAC 790H10 (green fluorescence) containing the MXI1 gene on the cell line Karpas 422. The interphase nuclei display two green signals indicating the two copies of the MXI1 gene but only one red signal indicating loss of one PTEN/MMAC1 allele. Some signals are out of focus. (B) Genomic sequencing exon 3 of PTEN/MMAC1 in a control (top) and Karpas 422 (bottom) revealing a splice acceptor site mutation (A → C) at position −2. (C) RT-PCR analysis of a control (lane 1) and Karpas 422 (lanes 2 and 3) with primers surrounding exon 311 showing a lack of the expected germline 397-bp fragment but a 45-bp shorter PCR product caused by skipping of exon 3. Lane 4, H2O-control. Lane 5, 100-bp ladder.
(A) FISH with YAC 746H8 (red fluorescence) containing the PTEN/MMAC1 gene and YAC 790H10 (green fluorescence) containing the MXI1 gene on the cell line Karpas 422. The interphase nuclei display two green signals indicating the two copies of the MXI1 gene but only one red signal indicating loss of one PTEN/MMAC1 allele. Some signals are out of focus. (B) Genomic sequencing exon 3 of PTEN/MMAC1 in a control (top) and Karpas 422 (bottom) revealing a splice acceptor site mutation (A → C) at position −2. (C) RT-PCR analysis of a control (lane 1) and Karpas 422 (lanes 2 and 3) with primers surrounding exon 311 showing a lack of the expected germline 397-bp fragment but a 45-bp shorter PCR product caused by skipping of exon 3. Lane 4, H2O-control. Lane 5, 100-bp ladder.
As to the consistent loss of one allele of the PTEN/MMAC1 gene in the lymphomas with 10q-deletions according to our FISH study we further investigated the role of this gene by complete sequencing of both strands of all nine exons as described by Steck et al4 with slight modifications. For these analyses, genomic DNA from primary lymphoma tissue was available for six patients shown to contain loss of one PTEN/MMAC1 allele by FISH. Additionally, a further t(14;18)-positive primary lymphoma not evaluable by FISH but containing a karyotypically detectable deletion in 10q and the cell line Karpas 422 were included. Except for well-known polymorphisms, no alterations of the PTEN/MMAC1 gene were detected in any of the primary lymphomas. Nevertheless, the cell line Karpas 422 was found to contain a splice acceptor site mutation (A → C) at position −2 of exon 3 of the PTEN/MMAC1 gene (Fig 1B), which was confirmed by sequencing a second sublineage of this cell line. Remarkably, the same mutation has been recently described in a primary glioblastoma.10 To determine the pathogenetic relevance of this alteration, reverse transcriptase-polymerase chain reaction (RT-PCR) analysis was performed on Karpas 422 with primers surrounding exon 3.11 This failed to amplify the expected germline 397-bp fragment but showed a shorter PCR product, which, by sequencing, was shown to be a splice variant caused by skipping of the 45-bp sized exon 3 (Fig 1C). Thus, the intact PTEN/MMAC1 transcript is completely absent in Karpas 422 due to deletion of the first and mutation of the second allele. The truncated transcript lacks part of conserved protein domain with high homology to tensin and auxillin and thus can be assumed to lack activity.12
In summary, our data provide evidence that deletions in the long arm of chromosome 10 are recurrent secondary changes in t(14;18) lymphomas. As to our FISH analyses, the distal border of the interstitial deletions in most cases is proximal to the MXI1-gene, rendering this gene as well as the candidate gene DMBT1, which is localized more distally in 10q25.3-26.1,13 highly unlikely as tumor suppressors in germinal center lymphomas. We detected consistent loss of the PTEN/MMAC1 gene by FISH in the cases investigated. Nevertheless, a mutation leading to inactivation of the second allele has been detected only in the cell line Karpas 422 but not in primary lymphomas. Inactivating mutations have also been described in a series of other cell lines derived from hematological neoplasms.12,14,15 In accordance with our results, alterations of the PTEN/MMAC1 have been reported by two groups to occur only in a small minority of unselected primary lymphoid malignancies. Grønbæk et al14 reported mutations of the PTEN/MMAC1 in 2 of 170 and Sakai et al15in 1 of 42 primary lymphoid malignancies. Considering the scarcity of PTEN/MMAC1 mutations in primary lymphoid malignancies, the existence of a thus far unknown tumor suppressor gene in 10q involved in the pathogenesis of at least germinal center lymphomas might be assumed.
ACKNOWLEDGMENT
The authors are grateful to Dr A. Fosså (Oslo, Norway) and the DSMZ (Braunschweig, Germany) for providing sublineages of Karpas 422, to the German Ressource Center (Berlin, Germany) for providing YACs, and to R. Zühlke-Jenisch, C. Becher, and D. Schuster for their excellent technical assistance. This work was supported by the Deutsche Krebshilfe, the Wilhelm-Sander-Stiftung, and the IZKF Kiel.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal