Abstract
Human K562 erythroleukemia cells were transfected with human band 3 (anion exchanger 1 [AE1]) cDNA, using the pBabe retroviral vector. Stable K562 clones expressing band 3 were isolated by flow cytometry, and surface expression was quantified by immunoblotting. The function of band 3 expressed at the cell surface was demonstrated in chloride transport assays. K562 cells expressing band 3 also displayed high levels of the Wrb blood group antigen, confirming the role of band 3 in Wrb expression, and an increase in the low levels of endogenous Rh antigen activity. We also performed coexpression experiments with K562 clones that had previously been transduced with cDNAs encoding RhD or RhcE polypeptides. The transfection and expression of band 3 in these clones substantially increased the levels of RhD and cE antigen activity expressed on the cells and also increased the reactivity of the cells with antibody to the endogenous Rh glycoprotein (RhGP, Rh50). The increased reactivity of Rh antigens may result from cell surface or intracellular interactions of band 3 with the protein complex which contains the Rh polypeptides and RhGP, or from indirect effects of band 3 on the membrane environment. This work establishes a system for cell surface expression of band 3 in a mammalian cell line, which will enable further studies of the protein and its interactions with other membrane components.
BAND 3, THE HUMAN RED blood cell (RBC) anion exchange protein (AE1), is the most abundant integral membrane protein found in erythrocytes and a well-characterized transporter.1-6 The expression of band 3 in heterologous systems7 is of interest for several reasons. First, the ability to express band 3 mutants is necessary for the development of improved structural and functional models for the protein. Second, it is important to understand how the biosynthesis of transport-active band 3 is regulated, allowing the maturing RBC to overcome any potential problems associated with the transient presence of a pH-modifying transporter in intracellular membranes. Third, the study of the interaction of band 3 with other proteins present in the erythrocyte plasma membrane and in the skeleton is essential for our understanding of erythrocyte maturation. Functional band 3 has been expressed to the cell surface in Xenopus oocytes injected with mouse band 3 poly-A+mRNA,8 mouse band 3 cRNA,9 and human band 3 cRNA.10Transport-active human band 3 membrane domain has been expressed to the cell surface in Saccharomyces cerevisiae,11 whereas the full-length protein expressed in yeast is retained in internal membranes.12 Functional cell surface expression of band 3 in mammalian cell lines has not been reported to date. Transfection of HEK 293 cells by Ruetz et al13 resulted in the synthesis of significant levels of functional band 3, but the protein was retained in intracellular membranes. Transfected cells showed abnormal morphology, possibly as a result of intracellular band 3 anion transport activity. The work of Gomez and Morgans14 using HEK 293 cells transfected with band 3 cDNA suggested that ankyrin binds to band 3 in the endoplasmic reticulum of these cells and may facilitate the exit of band 3 from this compartment, but not its movement to the cell surface.
An important aspect of band 3 expression is its interaction with glycophorin A (GPA). Evidence for this comes from the study of two rare types of human RBCs; MkMk cells, which lack glycophorins A and B, and homozygous En(a−) cells, which lack only GPA.15,16 The band 3 found in these cells migrates more slowly on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) than normal band 3, due to increased N-glycosylation.17-21 The increased glycosylation is indicative of a slower movement of the protein through the Golgi complex.22 Bruce et al21 also studied the anion transport activity of band 3 in MkMk RBCs and showed that the sulphate transport activity of the protein in these cells is reduced to about 60% of that of normal RBC band 3. In MkMk cells, the Vmax for sulphate influx remained unaltered, but the apparent KM was substantially higher than in normal cells. Furthermore, Groves and Tanner23,24 showed that GPA increases band 3-mediated chloride transport in the Xenopus oocyte system by increasing the rate at which band 3 accumulates in the oocyte plasma membrane. For these reasons, it has been suggested that GPA acts like a chaperone for band 3 by forming an intracellular complex with the newly synthesized protein. The formation of this complex is thought to promote structural and conformational changes to band 3, making it fully transport active, and increasing its rate of translocation to the cell surface.7
Here we report the functional expression of human band 3 at the cell surface in the human K562 erythroleukemia cell line. K562 cells25 do not express endogenous band 3, lack major erythroid surface antigens such as ABH and Wrb,26,27 and either do not express or express only low levels of the Rh polypeptides28,29 and the LW30 and Fy31 blood group active proteins. However they do express GPA,32 RhGP,28CD47,29 and, on induction,33 some other erythroid proteins, including hemoglobin. The K562 cell line is therefore potentially suitable as an expression system for band 3. We transfected K562 cells with band 3 cDNA, using the pBabe retroviral vectors,34 which have recently been used to express the RhD and G or the c and E antigens in K562 clones.35 Our work resulted in the isolation of stable K562 clones expressing high levels of functional band 3 on the cell surface and thus presents a novel expression system for band 3. We examined the effects of band 3 on the expression of other antigens in K562 cells and showed that band 3 surface expression coincides with the appearance of high levels of the Wrb antigen. We also detected an increase in the low level of endogenous Rh antigen activity. To further investigate the effect of band 3 on the expression of Rh polypeptides, we studied K562 clones sequentially transfected with cDNAs encoding RhD or cE and then band 3 cDNA and found that the expression of band 3 substantially increases the reactivity of the K562/Rh clones with antibodies to the RhD, c, and E antigens, as well as antibody to the RhGP glycoprotein. This may arise from indirect effects of band 3 on the membrane environment. However, it also suggests the possibility that band 3 may interact with the complex containing the Rh polypeptides and RhGP either at the cell surface or during the folding and translocation of newly synthesized Rh components to the cell surface.
MATERIALS AND METHODS
Monoclonal antibodies and RBCs.
Murine antibodies to band 3 (BRIC 6,36 IgG3; BRIC 155,37 IgG2b; BRIC 169,36 IgG1, and BRIC 170, IgG1), GPA (R10, IgG1 and R18,38 IgG2b), Wrb (BRICs 14,39 IgG2a and 201,40IgG1), Rh polypeptides (BRIC 69,41 IgG1), CD47 (BRIC 126,41 IgG2b) and glycophorin C (GPC; BRIC 4,42IgG1) were as described. Purified human anti-D, Brad 543(IgG1), was used at 50 μg/ml−1, diluted in phosphate-buffered saline (PBS)/1% bovine serum albumin/0.1% NaN3 (PBS-A). Human anti-c, MS37 (IgG3), was provided by Dr K. Thompson (IGRL, Oslo, Norway). Murine anti-Fy3, CBC-512 (IgG) was provided by Dr M. Uchikawa (Japanese Red Cross, Shibuya-ku, Japan). Murine anti-RhGP, LA18.18 (IgG1), was provided by Professor A.E.G. Kr. von dem Borne (C.L.B., Amsterdam, The Netherlands). Murine anti-LW, BS46 (IgG1), was provided by Dr H.H. Sonneborn (Biotest AG, Dreieich, Germany). RBCs of known phenotype were available in house.
Cloning of band 3 cDNA into the pBabe puro and pBabe neo retroviral vectors.
Polymerase chain reaction (PCR) was used to amplify band 3 cDNA from the BSXG1.B3 construct previously described.23 This band 3 cDNA encodes a common, asymptomatic band 3 polymorphism, Memphis I (Lys56 → Glu44,45). PCR reactions were performed in a DNA thermal cycler (Perkin Elmer, Norwalk, CT) usingPwo polymerase (Boehringer Mannheim, Lewes, UK). The primers were based on the band 3 cDNA 5′ and 3′ coding nucleotides and incorporated Mfe I and Sal I restriction sites, respectively. PCR products were cloned into the pBabe puro retroviral vector (pBp), kindly provided by Dr H. Land (ICRF, London, UK), using the compatible EcoRI and Sal I restriction sites, to yield the construct pBpB3. The band 3 cDNA was then subcloned into the pBabe neo vector (pBn), also provided by Dr H. Land, using theBstEII and Sal I restriction sites, thus generating the construct pBnB3. The band 3 coding region was confirmed in all constructs by DNA sequencing, using T7 DNA Sequenase 2.0 (Amersham, Little Chalfont, UK) and a 377 Applied Biosystems automated DNA sequencer (Warrington, UK), according to manufacturer’s instructions. Electroporation competent Escherichia coli XL-1 Blue cells (Stratagene, La Jolla, CA) were prepared using a method similar to that described by Hanahan,46 transformed with pBabe puro or neo constructs, and used for subsequent DNA purification (Qiagen, Dorking, UK). All constructs were linearized using the Sca I site in pBabe before transfection.
Transfection of K562 cells.
Human erythroleukemic K562 cells were obtained from the European Collection of Animal Cell Cultures (Porton Down, Salisbury, Wiltshire, UK). Cells were cultured in Iscove’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (IMEM/FBS), at 37°C, in a 5%CO2 humidified incubator. Approximately 2 × 105 cells were transfected by electroporation with approximately 10 μg of vector DNA in a Biorad gene pulser (Hercules, CA) at 200 V/1,000 μF. Electroporated cells were cultured in 10 mL of IMEM/FBS for 2 days and then transferred to 96-well plates (0.2 mL per well), in medium supplemented with 3 μg/mL puromycin (Sigma, Poole, UK), or 3 μg/mL puromycin and 2 mg/mL Geneticin G418 (Calbiochem, Beeston, UK) if subject to sequential transfection as described below. After 2 to 3 weeks, antibiotic-resistant, stable transfected clones from wells containing only a single discrete colony were transferred to 24-well plates and expanded before flow cytometric analysis. The clones expressing the highest levels of band 3 were selected using the anti–band 3, BRIC 6. We also transfected K562 clones already expressing RhD or cE polypeptides (generated by retroviral transduction of K562 cells with pBabe puro/Rh constructs as previously described35) with the pBnB3 construct. These sequential transfections generated K562/RhD+B3 clones and K562/RhcE+B3 clones, expressing both band 3 and RhD or cE polypeptides, respectively. Untransfected K562 cells or K562 cells expressing Rh polypeptides were transfected with empty pBabe puro or neo vectors to generate control cell clones.
Flow cytometric analysis.
K562 clones transfected with band 3 cDNA were analyzed by flow cytometry (FACStar Plus, Becton Dickinson, Mountain View, CA). Mean fluorescence intensity (FL1) was used as a measure of antibody binding. The specificity of the antibodies was confirmed using RBCs of the appropriate phenotype (data not shown). A total of 2 × 105 K562 cells were washed once and suspended in 50 μL of PBS-A. Cells were incubated with an equal volume of antibody for 1 hour at room temperature, then washed and resuspended in PBS-A. The cells were then incubated with affinity-purified fluorescein isothiocyanate (FITC)-labeled (Fab′)2 fragments of rabbit antihuman IgG or rabbit antimouse IgG (50 μL, 1:20 dilution, DAKO, Glostrup, Denmark), for 1 hour at room temperature. Cells were then washed once in PBS-A, and the sample volume was adjusted to 300 μL before analysis. A K562 clone transfected with empty pBp (K562/pBp) was used as control for K562 clones transfected with band 3. K562/RhD or K562/RhcE clones were transfected with empty pBabe neo vector, and the resulting K562/RhD+pBn and K562/RhcE+pBn clones were used as controls for the analysis of K562/RhD and K562/RhcE clones transfected with band 3.
Cell growth rates.
Clones were seeded at 5 × 104 cells per mL in 10 mL of culture medium, and cell numbers were determined (using a hemocytometer) over a 6-day period by counting four samples per clone per time point. Specific growth rate constants μ = ln((Nt− N0)/t) and doubling times td = ln2/μ were calculated from plots of ln (mean cell count) versus time.
Immunoblotting.
K562 cells were prepared for immunoblotting by lysing whole cells in a detergent buffer, followed by SDS-PAGE. Cells were first washed in PBS pH 7.5 at 4°C, and centrifuged. For samples subjected to chymotrypsin digestion, the cells were incubated with Nα-p-tosyl-L-lysine chloromethyl ketone (TLCK)-treated chymotrypsin (Sigma), at 2 mg/mL in PBS pH 7.5, at 4°C for 6 hours. Examination of the time course of digestion showed that there was no increase in the degree of digestion after this time (data not shown). Chymotrypsin-treated cells were washed three times in PBS pH 7.5, containing the protease inhibitors phenylmethyl sulfonyl fluoride (PMSF) (2 mmol/L), 4-(2-aminoethyl)benzenesulphonyl fluoride (AEBSF) (1 mmol/L), leupeptin (140 mmol/L), antipain (80 mmol/L) pepstatin A (44 mmol/L), bestatin (40 mmol/L), E-64 (14 mmol/L), and aprotinin (0.8 mmol/L) (PBS-PI), supplemented with 5 mg/mL bovine serum albumin (BSA). Cells were then washed once in PBS-PI, without BSA. For detergent extraction, 50 μL cell pellets of chymotrypsin-treated cells or untreated cells were suspended in an equal volume of PBS-PI, 100 μL of PBS-PI containing 1% Triton X-100 were added, and the suspensions were incubated on wet ice for 20 minutes. Samples were centrifuged at 13,000g for 10 minutes at 4°C to pellet out nuclei and cell debris. Ghosts of chymotrypsin-treated and untreated RBCs (homozygous for the band 3 Memphis I mutation) were used as controls on immunoblots. Triton extracts of K562 cells and RBC ghosts were both solubilized in SDS-PAGE buffer, and some chymotrypsin-treated samples were treated with PNGase F (Oxford Glycosystems, Oxford, UK) in NEBuffers (New England Biolabs, Hitchin, UK), for 1 hour at 37°C, to remove N-glycan chains. Samples were separated on 8% acrylamide gels,47immunoblotted,37 and visualized using the ECL method (Amersham) and Biomax MR film (Kodak, Rochester, NY). For quantitative analysis, blots were scanned using a high resolution color scanner (Sharp, Chiba-City, Japan) and IMAGEMASTER scanning software (Pharmacia LKB, Uppsala, Sweden).
Chloride-36 efflux assays.
Our transport studies were based on the method described by Dissing et al.48 Approximately 5 × 108 K562 cells were washed once and resuspended in degassed transport buffer (140 mmol/L KCl, 1 mmol/L MgCl2, 5 mmol/L glucose, 5 mmol/L KH2PO4, pH7.2), and cytocrits were determined using microhematocrit tubes. For 36Cl loading, 200 μL of cells were incubated in transport buffer (2% cytocrit) containing a tracer quantity of 36Cl at 20°C for 15 minutes, with occasional swirling, and then centrifuged at 1,600g for 5 minutes at 4°C. Loaded cells were resuspended at 2% cytocrit in ice-cold chloride-free degassed wash buffer (140 mmol/L potassium gluconate, 1 mmol/L MgSO4, 5 mmol/L glucose, 5 mmol/L KH2PO4, pH 7.2), split into two and centrifuged to give two 100-μL cell pellets. One pellet was washed in ice-cold wash buffer containing 50 mmol/L 4,4’-dinitrostilbene-2,2′-disulphonate (DNDS, a band 3 inhibitor) and the other pellet in ice-cold wash-buffer alone. Cell pellets were rapidly resuspended in transport buffer at 2% cytocrit, in the presence or absence of 50 mmol/L DNDS, at 5°C or 20°C. At intervals, 500 μL aliquots of the constantly mixed cell suspension were transferred into 1.5 mL microcentrifuge tubes, containing 500 μL of ice-cold stop buffer48 (transport buffer supplemented with 50 mmol/L DNDS and 2 mmol/L furosemide). The quenched samples were immediately centrifuged at 6,000g for 30 seconds, and 500 μL of the resulting supernatants were added to 10 mL of Emulsifier-Safe (Packard Instrument B.V., Groningen, The Netherlands) for liquid scintillation counting. The values for infinite time were established by counting 0.25 mL of unquenched cell suspension, and background counts were determined by counting three samples of transport buffer. The Cl- efflux rate constant,okCl−, was calculated according tookCl− = ln[(cpmi − cpm0)/(cpmi − cpmt)] t−1, where cpm0, cpmt, and cpmi are counts per minute released into the medium surrounding the cells after time zero, time t, and infinite time, respectively. Transport data were obtained from plots of ln(1−(cpmt/cpmi)) versus time, in which cpmt and cpmi had been corrected for cpm0. On these plots,okCl− equals the gradient of linear regression lines. The relative chloride efflux,oMCl−, was calculated according to the equation oMCl− = (mcv/A) ×okCl− × [Cl−]i, where mcv is the mean cell volume (determined as below) and A the mean cell surface area (which was calculated assuming that K562 cells are spherical). The intracellular chloride concentration [Cl−]i is 84 ± 2 mmol/L for K562 cells incubated in the transport buffer we have used.48
Determination of mean cell volume.
Approximately 5 × 107 cells were washed once and resuspended in 0.5 mL of transport buffer. Cell suspensions were counted in triplicate, using a Haematology Analyser (Sysmex, Milton Keynes, UK). The cytocrit of the same suspensions was then immediately determined and the mean cell volume calculated, allowing for 2% trapped medium.48
RESULTS
Analysis of K562 clones transfected with band 3 cDNA by flow cytometry.
Several K562 clones transfected with the pBpB3 construct were generated and analyzed by flow cytometry, using a panel of monoclonal antibodies recognizing band 3, Wrb, GPA, GPC, Rh polypeptides, CD47, and the RhGP, LW, and Fy glycoproteins. A K562 clone transfected with empty pBp vector (K562/pBp) was used as control; flow cytometric analysis showed almost identical expression of cell surface antigens for this clone and untransfected K562 cells (data not shown). Table 1 shows a comparison of antibody-binding to one K562/band 3 clone (K562/B3 clone 1) and K562/pBp cells. The tabulated results are the absolute values obtained on the same day for antibody-binding to the two clones, and the average increase in antibody-binding to K562/B3 clone 1 relative to K562/pBp, measured daily over 3 consecutive days. The histograms obtained on day 1 are shown in Fig 1. Most significantly, an antibody to an extracellular domain of band 3 (BRIC 6) bound much more strongly to K562/B3 clone 1 than to K562/pBp cells, with a 10-fold increase in mean fluorescence intensity. In addition, K562/B3 clone 1 showed high levels of de novo expression of the Wrbantigen; this was observed for all of the analyzed K562 clones which expressed band 3 to the cell surface (data not shown). One anti-Wrb (BRIC 14) showed a 42-fold increase in binding to K562/B3 clone 1 cells over K562/pBp cells, while another antibody (BRIC 201) showed an eightfold increase. Antibodies to an intracellular epitope of band 3 (BRIC 169, used as negative control), to GPC (BRIC 4), to Fy3 (CBC-512), and to CD47 (BRIC 126) showed only minimal differences in binding between K562/B3 clone 1 and control cells; these differences are probably a reflection of a slightly higher affinity of K562/B3 clone 1 for the secondary antibody. Two antibodies to GPA (R10 and R18) bound strongly to both K562/B3 clone 1 and K562/pBp cells. Both of these antibodies showed a small (twofold) increase in binding to K562/B3 clone 1 relative to the control clone. However, this increase was not found in all K562/band 3 clones tested and may be due to the heterogeneity of K562 cells with regard to GPA expression.49 50 For K562/B3 clone 1, we also observed a threefold increase in the background expression of Rh polypeptides (BRIC 69), and a twofold increase in binding of antibodies to the RhGP and LW glycoproteins (LA18.18 and BS46, respectively). Similar observations were made for most of the K562/band 3 clones. To further investigate the effect of band 3 expression on the expression of Rh components, we used K562 clones sequentially transfected with cDNAs encoding RhcE or D and then band 3 cDNA; the results of these experiments are described later.
Flow Cytometric Analysis of K562/B3 Clone 1
| Antibody . | Directed Against . | Fluorescence Intensity for K562/pBp (day 1) . | Fluorescence Intensity for K562/B3 Clone 1 (day 1) . | Mean (±SD) Ratio of Fluorescence Intensity for K562/B3 Clone 1 over K562/pBp (measured daily over 3 consecutive days) . |
|---|---|---|---|---|
| BRIC 169 | B3 (intracellular) | 3.0 | 4.0 | 1.4 ± 0.1 |
| BRIC 6 | B3 (extracellular) | 3.9 | 40 | 9.9 ± 0.7 |
| BRIC 14 | Wrb | 4.3 | 206 | 42 ± 5.1 |
| BRIC 201 | Wrb | 2.8 | 15 | 8.1 ± 2.2 |
| R10 | Glycophorin A | 117 | 259 | 2.5 ± 0.3 |
| R18 | Glycophorin A | 123 | 266 | 2.7 ± 0.6 |
| BRIC 4 | Glycophorin C | 144 | 152 | 1.2 ± 0.1 |
| BRIC 69 | Rh polypeptides | 5.7 | 18 | 3.0 ± 0.2 |
| LA 18.18 | Rh glycoprotein | 38 | 61 | 2.2 ± 0.6 |
| BS46 | LW | 5.6 | 10 | 2.1 ± 0.2 |
| BRIC 126 | CD 47 | 34 | 41 | 1.5 ± 0.3 |
| CBC-512 | Fy3 | 2.8 | 3.7 | 1.4 ± 0.1 |
| Antibody . | Directed Against . | Fluorescence Intensity for K562/pBp (day 1) . | Fluorescence Intensity for K562/B3 Clone 1 (day 1) . | Mean (±SD) Ratio of Fluorescence Intensity for K562/B3 Clone 1 over K562/pBp (measured daily over 3 consecutive days) . |
|---|---|---|---|---|
| BRIC 169 | B3 (intracellular) | 3.0 | 4.0 | 1.4 ± 0.1 |
| BRIC 6 | B3 (extracellular) | 3.9 | 40 | 9.9 ± 0.7 |
| BRIC 14 | Wrb | 4.3 | 206 | 42 ± 5.1 |
| BRIC 201 | Wrb | 2.8 | 15 | 8.1 ± 2.2 |
| R10 | Glycophorin A | 117 | 259 | 2.5 ± 0.3 |
| R18 | Glycophorin A | 123 | 266 | 2.7 ± 0.6 |
| BRIC 4 | Glycophorin C | 144 | 152 | 1.2 ± 0.1 |
| BRIC 69 | Rh polypeptides | 5.7 | 18 | 3.0 ± 0.2 |
| LA 18.18 | Rh glycoprotein | 38 | 61 | 2.2 ± 0.6 |
| BS46 | LW | 5.6 | 10 | 2.1 ± 0.2 |
| BRIC 126 | CD 47 | 34 | 41 | 1.5 ± 0.3 |
| CBC-512 | Fy3 | 2.8 | 3.7 | 1.4 ± 0.1 |
Abbreviation: B3, band 3.
FACS analysis of K562/pBp and K562/B3 clone 1. All data are from Table 1, day 1. Antibodies used in each histogram were (1) BRIC 6, anti–band 3; (2) BRIC 14, anti-Wrb; (3) BRIC 201, anti-Wrb; (4) R10, anti-GPA; (5) R18, anti-GPA; (6) BRIC 4, anti-GPC; (7) BRIC 69, anti-Rh30; (8) LA18.18, anti-RhGP; (9) BS46, anti-LW; (10) BRIC 126, anti-CD47, and (11) CBC-512, anti-Fy3. GPA, glycophorin A; GPC, glycophorin C; Rh30, Rh polypeptides; RhGP, Rh glycoprotein.
FACS analysis of K562/pBp and K562/B3 clone 1. All data are from Table 1, day 1. Antibodies used in each histogram were (1) BRIC 6, anti–band 3; (2) BRIC 14, anti-Wrb; (3) BRIC 201, anti-Wrb; (4) R10, anti-GPA; (5) R18, anti-GPA; (6) BRIC 4, anti-GPC; (7) BRIC 69, anti-Rh30; (8) LA18.18, anti-RhGP; (9) BS46, anti-LW; (10) BRIC 126, anti-CD47, and (11) CBC-512, anti-Fy3. GPA, glycophorin A; GPC, glycophorin C; Rh30, Rh polypeptides; RhGP, Rh glycoprotein.
Morphology and cell growth rates of K562 clones.
Under a light microscope, K562 cells expressing band 3 appear to be morphologically very similar to both K562 cells and K562/pBp cells, which are themselves morphologically indistinguishable. However, the flow cytometric results showed that the K562/band 3 clones are slightly more heterogenous in cell size and granularity, as measured by forward and side scatter (data not shown). The mean cell volume of K562/pBp cells was found to be 2.61 × 103 μm3and that of K562/B3 clone 1 cells was 2.36 × 103 μm3. Assuming that K562 cells are spherical, the mean cell surface area is 1,967 μm2 for K562/pBp and 1,840 μm2 for K562/B3 clone 1. We also measured the growth rates of untransfected K562 cells, K562/pBp cells, and three K562 clones expressing band 3 at the cell surface (Table 2). The doubling time of band 3–expressing clones was found to be about twofold greater than that of K562 and K562/pBp cells.
Growth Rates of K562 Clones
| Clone . | Specific Growth Rate Constant μ (h−1) . | Doubling Time td (h) . |
|---|---|---|
| Untransfected K562 | 5.43 × 10−2 | 12.8 |
| K562/pBp | 4.97 × 10−2 | 14.0 |
| K562/B3 clone 1 | 2.63 × 10−2 | 26.3 |
| K562/B3 clone 2 | 2.73 × 10−2 | 25.4 |
| K562/B3 clone 3 | 2.87 × 10−2 | 24.2 |
| Clone . | Specific Growth Rate Constant μ (h−1) . | Doubling Time td (h) . |
|---|---|---|
| Untransfected K562 | 5.43 × 10−2 | 12.8 |
| K562/pBp | 4.97 × 10−2 | 14.0 |
| K562/B3 clone 1 | 2.63 × 10−2 | 26.3 |
| K562/B3 clone 2 | 2.73 × 10−2 | 25.4 |
| K562/B3 clone 3 | 2.87 × 10−2 | 24.2 |
Immunoblotting and quantification of expressed band 3.
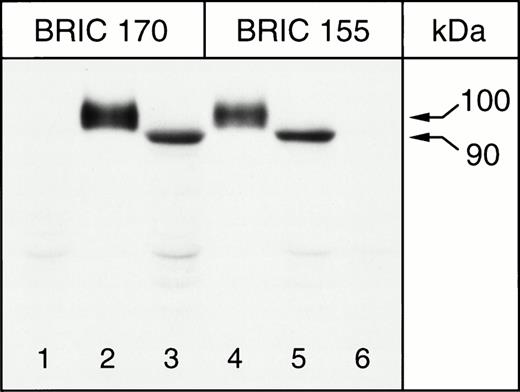
The band 3 protein expressed in K562 cells was characterized by SDS-PAGE and immunoblotting of K562/B3 clone 1 Triton X-100 extracts. Solubilized RBC ghosts and Triton extracts of K562/pBp cells were used as controls. Immunoblotting was performed with antibodies to band 3, BRIC 155 (recognizing an intracellular epitope near the C-terminus) and BRIC 170 (directed against an intracellular epitope towards the N-terminus), which have been shown to detect erythrocyte band 3.37 The results obtained for untreated K562/B3 clone 1 cells, untreated K562/pBp cells, and ghosts of untreated RBCs are shown in Fig 2. Both BRIC 155 and BRIC 170 detected band 3 polypeptide, with an apparent molecular weight of approximately 100 kD in erythrocyte membranes, and an apparent molecular weight of approximately 90 kD in K562/B3 clone 1 samples. No corresponding band was detected in K562/pBp samples. The same observations were made for all other analyzed K562/band 3 clones (data not shown). Quantitative scanning densitometry was used to calculate that the total amount of band 3 expressed in 55 μL of packed K562/B3 clone 1 cells is equivalent to that found in 1 μL of packed RBCs. To determine the surface fraction of band 3 expressed in K562 cells, we performed immunoblots of K562/B3 clone 1 cells which had been chymotrypsin-treated before Triton extraction; ghosts of chymotrypsin-treated erythrocytes were used as a control (see Fig 3). Chymotrypsin cleaves whole band 3 protein in the third extracellular loop at amino acid residues 553 and 558,51 giving a 60-kD N-terminal fragment and a 35-kD C-terminal fragment. In RBCs homozygous for the band 3 Memphis I mutation, present in K562/B3 clone 1 and in our RBC controls, the N-terminal chymotrypsin fragment has a reduced mobility on SDS-PAGE and runs at approximately 63 kD.52 The cell surface fraction of the band 3 expressed in K562/B3 clone 1 (determined by scanning densitometry of the bands shown in lane 2, Fig 3) was calculated to be 47% of the total expressed protein. The amount of cell-surface band 3 expressed in approximately 115 μL of packed K562/B3 clone 1 cells is therefore equal to that found in 1 μL of packed RBCs. Mature erythrocytes have a mean cell volume of 80 μm3 and a mean cell surface area of 142 μm2, while the mean cell volume for K562/B3 clone 1 is 2,360 μm3 and the mean cell surface area 1,840 μm2. Thus, the cell surface area per unit packed cell volume is 2.3 times larger in erythrocytes than in K562/B3 clone 1 cells. The density of cell surface band 3 molecules can therefore be estimated to be 50 times greater in RBCs than in K562/B3 clone 1.
Immunoblotting of K562 cells for band 3, 8% Laemmli gel. Lanes 1 and 6, K562/pBp; lanes 2 and 4, homozygous Memphis RBC ghosts; lanes 3 and 5, K562/B3 clone 1. BRIC 155 is a murine monoclonal antibody directed against an intracellular site at the C-terminus of band 3, while BRIC 170 binds to the N-terminal cytosolic domain of band 3.
Immunoblotting of K562 cells for band 3, 8% Laemmli gel. Lanes 1 and 6, K562/pBp; lanes 2 and 4, homozygous Memphis RBC ghosts; lanes 3 and 5, K562/B3 clone 1. BRIC 155 is a murine monoclonal antibody directed against an intracellular site at the C-terminus of band 3, while BRIC 170 binds to the N-terminal cytosolic domain of band 3.
Immunoblotting of chymotrypsin-treated K562 cells for band 3, 8% Laemmli gel. Lanes 1 and 4, ghosts of chymotrypsin-treated homozygous Memphis RBCs; lanes 2 and 3, chymotrypsin-treated K562/B3 clone 1 cells; lane 5, PNGase-treated ghosts of chymotrypsin-treated homozygous Memphis RBCs.
Immunoblotting of chymotrypsin-treated K562 cells for band 3, 8% Laemmli gel. Lanes 1 and 4, ghosts of chymotrypsin-treated homozygous Memphis RBCs; lanes 2 and 3, chymotrypsin-treated K562/B3 clone 1 cells; lane 5, PNGase-treated ghosts of chymotrypsin-treated homozygous Memphis RBCs.
Figure 3 also shows that both the C-terminal 35-kD chymotrypsin fragment (lane 3) and the N-terminal 63-kD chymotrypsin fragment (lane 2) of band 3 expressed in K562 cells migrated at a slightly lower apparent molecular weight than the corresponding band 3 fragments from RBCs. In addition, the N-terminal 63-kD fragment ran as a doublet rather than as a single band. The possible reasons for these observations are discussed later. However, it should be noted that the deglycosylated C-terminal chymotrypsin fragment of RBC band 3 (the PNGase-treated sample in lane 5) ran at a lower apparent molecular weight than the C-terminal fragment of band 3 from K562 cells. At least some of the bands seen in Fig 3, which do not correspond to whole band 3 or to the band 3 chymotrypsin fragments discussed above, probably represent proteolytic degradation products of band 3.
Chloride-36 transport in K562 clones.
DNDS-sensitive chloride efflux was measured for K562/B3 clone 1 at 20°C and 5°C, using K562/pBp as control. Figure 4 shows the chloride efflux at 20°C; the gradients of the linear regression lines are equivalent to efflux rate constants. In the absence of DNDS, efflux was much faster for K562/B3 clone 1 than for K562/pBp (14-fold). The increased efflux in K562/B3 clone 1 was inhibited by 50 mmol/L DNDS. A fraction of the slow efflux found in K562/pBp was also sensitive to the inhibitor; this efflux is mediated by transporters other than band 3.53 We therefore subtracted the efflux rate constant for K562/pBp without DNDS (okCl− = −0.158 × 10−2, r2 = .98) from the efflux rate constant for K562/B3 clone 1 without DNDS (okCl− = −2.224 × 10−2, r2 = .97) to calculate the efflux rate constant for band 3-mediated efflux in K562/B3 clone 1 (okCl− = −2.066 × 10−2). Because of the very fast efflux found for K562/B3 clone 1 at 20°C, we also measured transport at 5°C. At this temperature, efflux in the absence of DNDS was much slower than at 20°C (14 times) for both K562/B3 clone 1 (okCl− = −0.156 × 10−2, r2 = .99) and K562/pBp (okCl− = −0.012 × 10−2, r2 = .85). The band 3-mediated efflux in K562/B3 clone 1 was calculated as at 20°C (okCl− = −0.144 × 10−2). Table 3 compares the data for band 3-mediated chloride transport in K562/B3 clone 1 cells and RBCs at 5°C and 20°C; data for RBCs are according to Brahm.54 The relative chloride effluxoMCl− was calculated as described in Materials and Methods. For K562/B3 clone 1,oMCl− = 1.55 × 10−16 mmol Cl− per μm2 cell surface per second at 5°C, andoMCl− = 2.23 × 10−15 mmol Cl− per μm2 cell surface per second at 20°C. At both temperatures, the relative chloride flux is approximately 41 times greater in RBCs than in K562/B3 clone 1. This is consistent with our immunoblotting data, which suggest that the number of band 3 molecules per unit surface area is 50 times greater in RBCs than in K562/B3 clone 1 (see above).
Chloride efflux in K562 clones at 20°C. The gradients of linear regression lines are equivalent to efflux rate constants (okCl−). r2 values are K562/B3 clone 1 − DNDS, .97; K562/B3 clone 1 + DNDS, .96; K562/pBp − DNDS, .98; K562/pBp + DNDS, .88. The rate constant for band 3–mediated efflux in K562/B3 clone 1 was calculated as the difference between the rate constants for K562/B3 clone 1 − DNDS and K562/pBp − DNDS.
Chloride efflux in K562 clones at 20°C. The gradients of linear regression lines are equivalent to efflux rate constants (okCl−). r2 values are K562/B3 clone 1 − DNDS, .97; K562/B3 clone 1 + DNDS, .96; K562/pBp − DNDS, .98; K562/pBp + DNDS, .88. The rate constant for band 3–mediated efflux in K562/B3 clone 1 was calculated as the difference between the rate constants for K562/B3 clone 1 − DNDS and K562/pBp − DNDS.
Chloride Transport Data
| . | K562/B3 Clone 1 . | RBC . |
|---|---|---|
| Mean cell volume (μm3) | 2,360 | 78 |
| Mean cell surface area (μm2) | 1,840 | 142 |
| Intracellular Cl− concentration (mmol/L) | 84 | 115 |
| Rate constant for band 3-mediated chloride efflux °kCl− at 5°C (s−1) | −0.144 × 10−2 | −0.103 |
| Relative chloride flux °MCl− at 5°C (mmol · μm−2 · s−1 × 10−16) | 1.55 | 64.8 |
| Rate constant for band 3–mediated chloride efflux °kCl− at 20°C (s−1) | −2.066 × 10−2 | −1.430 |
| Relative chloride flux °MCl− at 20°C (mmol · μm−2 · s−1 × 10−16) | 22.3 | 901 |
| . | K562/B3 Clone 1 . | RBC . |
|---|---|---|
| Mean cell volume (μm3) | 2,360 | 78 |
| Mean cell surface area (μm2) | 1,840 | 142 |
| Intracellular Cl− concentration (mmol/L) | 84 | 115 |
| Rate constant for band 3-mediated chloride efflux °kCl− at 5°C (s−1) | −0.144 × 10−2 | −0.103 |
| Relative chloride flux °MCl− at 5°C (mmol · μm−2 · s−1 × 10−16) | 1.55 | 64.8 |
| Rate constant for band 3–mediated chloride efflux °kCl− at 20°C (s−1) | −2.066 × 10−2 | −1.430 |
| Relative chloride flux °MCl− at 20°C (mmol · μm−2 · s−1 × 10−16) | 22.3 | 901 |
Data for RBCs are according to Brahm.54 The extracellular chloride concentration is approximately 145 to 150 mmol/L for all efflux measurements. The rate constant for band 3–mediated efflux in K562/B3 clone 1 was calculated as the difference between the rate constant for K562/B3 clone 1 without DNDS and the rate constant for K562/pBp without DNDS.
Flow cytometric analysis of K562/RhD and K562/RhcE clones transfected with band 3 cDNA.
To examine the potential interactions of band 3 and Rh polypeptides, we investigated the effects of band 3 expression on K562 cells previously transfected with cDNAs encoding Rh polypeptides and already expressing substantial levels of the polypeptides on the cell surface. Two K562 clones transduced with RhD or RhcE cDNAs in pBp (as previously described35) were transfected with the pBnB3 construct, thus generating K562/RhD+B3 clones and K562/RhcE+B3 clones. As a control, we transfected the same two K562/Rh clones with empty pBn vector, yielding 12 K562/RhD+pBn and 11 K562/RhcE+pBn control clones. Antibodies to band 3, Rh polypeptides, RhGP and the RhD antigen were used to compare the expression of these antigens in the 12 K562/RhD+pBn clones with their expression in several K562/RhD+B3 clones. Care was taken to perform all measurements on the same day and under the same conditions. An analogous experiment was performed to compare antigen expression in the 11 K562/RhcE+pBn clones with that in several K562/RhcE+B3 clones, using anti-Rhc instead of anti-RhD. Flow cytometric results for the experiments are listed in Table 4, and typical histograms are shown in Fig 5. We found that cotransfection of a K562/RhD clone with empty pBn vector alone did not result in any noticable changes in the antigen activity of Rh polypeptides, Rh glycoprotein, and RhD antigen for any of the resulting K562/RhD+pBn clones. However, K562/RhD+B3 clone 1 (which binds 12-fold more anti–band 3 than an average K562/RhD+pBn clone) showed a substantial increase in binding of antibodies to the Rh proteins (×3.6 for Rh polypeptides, ×5.9 for RhGP, ×2 for RhD). Similar results were obtained for K562/RhcE+B3 clone 1 (which shows 11-fold greater binding of anti–band 3 than an average control clone); binding of antibodies to Rh polypeptides was raised 5.8-fold, antibodies to RhGP 8.8-fold, and antibodies to Rhc threefold. The other clones in which band 3 was coexpressed with Rh polypeptides also showed substantially enhanced binding of antibodies to the appropriate Rh polypeptides and RhGP. Band 3 expression did not have a similar effect on LW antigen activity in the cotransfected clones (data not shown).
Flow Cytometric Analysis of K562/RhD + B3 and K562/RhcE [6e0d] + B3 Clones
| Antibody . | Directed Against . | Mean Fluorescence Intensity (±SD) for 12 K562/RhD + pBn Clones . | Fluorescence Intensity of K562/RhD + B3 Clone 1 . | Fluorescence Intensity of K562/RhD + B3 Clone 2 . | Fluorescence Intensity of K562/RhD + B3 Clone 3 . |
|---|---|---|---|---|---|
| TCM (i) | 2.7 ± 0.2 | 3.2 | 3.7 | 2.8 | |
| BRIC 6 | Band 3 | 2.8 ± 0.2 | 33 | 17 | 12 |
| BRIC 69 | Rh30 | 19 ± 5.3 | 69 | 54 | 37 |
| LA18.18 | RhGP | 19 ± 3.6 | 113 | 122 | 88 |
| TCM (ii) | 2.8 ± 0.2 | 3.4 | 3.6 | 2.6 | |
| Brad 5 | RhD | 23 ± 6.6 | 45 | 36 | 32 |
| Mean Fluorescence Intensity (±SD) for 11 K562/RhcE + pBn Clones | Fluorescence Intensity of K562/RhcE + B3 Clone 1 | Fluorescence Intensity of K562/RhcE + B3 Clone 2 | Fluorescence Intensity of K562/RhcE + B3 Clone 3 | ||
| TCM (i) | 3.5 ± 0.6 | 3.4 | 3.4 | 3.6 | |
| BRIC 6 | Band 3 | 4.0 ± 0.7 | 42 | 14 | 13 |
| BRIC 69 | Rh30 | 24 ± 8.1 | 140 | 107 | 107 |
| LA18.18 | RhGP | 24 ± 7.4 | 211 | 208 | 212 |
| TCM (ii) | 3.6 ± 0.4 | 3.9 | 4.1 | 4.3 | |
| MS37 | Rhc | 10 ± 2.6 | 30 | 34 | 29 |
| Antibody . | Directed Against . | Mean Fluorescence Intensity (±SD) for 12 K562/RhD + pBn Clones . | Fluorescence Intensity of K562/RhD + B3 Clone 1 . | Fluorescence Intensity of K562/RhD + B3 Clone 2 . | Fluorescence Intensity of K562/RhD + B3 Clone 3 . |
|---|---|---|---|---|---|
| TCM (i) | 2.7 ± 0.2 | 3.2 | 3.7 | 2.8 | |
| BRIC 6 | Band 3 | 2.8 ± 0.2 | 33 | 17 | 12 |
| BRIC 69 | Rh30 | 19 ± 5.3 | 69 | 54 | 37 |
| LA18.18 | RhGP | 19 ± 3.6 | 113 | 122 | 88 |
| TCM (ii) | 2.8 ± 0.2 | 3.4 | 3.6 | 2.6 | |
| Brad 5 | RhD | 23 ± 6.6 | 45 | 36 | 32 |
| Mean Fluorescence Intensity (±SD) for 11 K562/RhcE + pBn Clones | Fluorescence Intensity of K562/RhcE + B3 Clone 1 | Fluorescence Intensity of K562/RhcE + B3 Clone 2 | Fluorescence Intensity of K562/RhcE + B3 Clone 3 | ||
| TCM (i) | 3.5 ± 0.6 | 3.4 | 3.4 | 3.6 | |
| BRIC 6 | Band 3 | 4.0 ± 0.7 | 42 | 14 | 13 |
| BRIC 69 | Rh30 | 24 ± 8.1 | 140 | 107 | 107 |
| LA18.18 | RhGP | 24 ± 7.4 | 211 | 208 | 212 |
| TCM (ii) | 3.6 ± 0.4 | 3.9 | 4.1 | 4.3 | |
| MS37 | Rhc | 10 ± 2.6 | 30 | 34 | 29 |
Abbreviations: TCM (i), negative control, incubating cells first in tissue culture medium, and then in rabbit anti-mouse/FITC; TCM (ii), negative control, incubating cells first in tissue culture medium, and then in rabbit anti-human/FITC; Rh30, Rh polypeptides; RhGP, Rh glycoprotein.
FACS analysis of K562 clones transfected with cDNAs corresponding to Rh and/or band 3. (1) to (3) are data for K562/pBp and K562/B3 clone 1, Table 1, day 1. (4) to (6) are data for a K562/RhD+pBn clone (with expression levels close to the calculated average) and K562/RhD+B3 clone 1; all data are from Table 4. (7) to (9) are data for a K562/RhcE+pBn clone (with expression levels close to the calculated average) and K562/RhcE+B3 clone 1; all data are from Table 4. Antibodies used were: BRIC 6 (anti–band 3) in histograms 1, 4, and 7; BRIC 69 (anti-Rh30) in histograms 2, 5, and 8; LA18.18 (anti-RhGP) in histograms 3, 6, and 9. Rh30, Rh polypeptides; RhGP, Rh glycoprotein.
FACS analysis of K562 clones transfected with cDNAs corresponding to Rh and/or band 3. (1) to (3) are data for K562/pBp and K562/B3 clone 1, Table 1, day 1. (4) to (6) are data for a K562/RhD+pBn clone (with expression levels close to the calculated average) and K562/RhD+B3 clone 1; all data are from Table 4. (7) to (9) are data for a K562/RhcE+pBn clone (with expression levels close to the calculated average) and K562/RhcE+B3 clone 1; all data are from Table 4. Antibodies used were: BRIC 6 (anti–band 3) in histograms 1, 4, and 7; BRIC 69 (anti-Rh30) in histograms 2, 5, and 8; LA18.18 (anti-RhGP) in histograms 3, 6, and 9. Rh30, Rh polypeptides; RhGP, Rh glycoprotein.
DISCUSSION
The purpose of this work was to develop a system for the heterologous expression of erythrocyte band 3 which would allow us to study (1) the biosynthesis of band 3, (2) its interactions with other erythrocyte proteins, and (3) its structure and function, by expressing band 3 mutants. Stable cell surface expression of functional band 3 in a cell line with protein expression similar to that found in maturing RBCs would clearly be useful for such investigations. However, previous cell surface expression of band 3 in mammalian cells has not been reported. In this study, we used the pBabe retroviral vectors to transfect K562 erythroleukemia cells with cDNA corresponding to the human AE1gene. Flow cytometric analysis was used to compare cell surface expression of band 3 in the resulting K562 clones with expression in control clones, transfected with empty pBabe. Monoclonal antibody BRIC 6 (which binds to an extracellular domain of band 3) detected high levels of band 3 expression in K562 cells transfected with band 3 cDNA, but no expression was detected in control cells. The expressed band 3 protein was also detected by immunoblotting, with monoclonal antibodies to intracellular epitopes. Our results suggest that approximately half of the total band 3 protein expressed in K562/B3 clone 1 is present at the cell surface. The amount of band 3 expressed per unit area of plasma membrane was estimated to be 50 times less in this clone than in RBCs. A simple chloride efflux assay was used to confirm the functionality of expressed band 3. The transport was found to be DNDS-sensitive, confirming it to be band 3–mediated. In K562/B3 clone 1, the band 3–mediated chloride flux was 41 times smaller than in RBCs, in agreement with our immunoblotting data. In conclusion, our results suggest that there are approximately 150 transport-active sites present per μm2 of K562/B3 clone 1 plasma membrane, assuming a density of 7,000 molecules of band 3 per μm2of erythrocyte membrane.9
Immunoblots showed that the band 3 expressed in K562 cells was not identical to the erythrocyte protein in that it migrated at an apparently lower molecular weight on SDS-PAGE (approximately 90 kD rather than 100 kD). Blotting for band 3, cleaved by chymotrypsin in the third extracellular loop, showed that the C-terminal 35-kD fragment of band 3 from K562 cells had an apparently lower molecular weight than the corresponding fragment found in RBC ghost samples. The presence of a smaller N-glycan chain may explain why the fragment from K562 cells ran faster than that from RBCs. PNGase treatment of the RBC sample showed that the deglycosylated C-terminal chymotrypsin fragment of RBC band 3 migrated faster than the K562 cell band 3 fragment. This suggests that the protein expressed in K562 cells is N-glycosylated to some extent. We were unable to successfully carry out PNGase-treatment of the band 3 expressed in K562 cells because of difficulties caused by endogenous proteolysis of the band 3 in the samples. The N-terminal 63-kD chymotrypsin fragment of band 3 from K562 cells also migrated at a slightly lower apparent molecular weight than the corresponding erythrocyte protein fragment. The reason for this observation may be that the band 3 expressed in K562 cells is not posttranslationally processed in the same way as RBC band 3, for example differently phosphorylated55 at Tyr8/Tyr21 or differently acetylated.56 In addition, the N-terminal K562 cell band 3 fragment migrated as a doublet rather than a single band. Although the examination of the time course of chymotrypsin-digestion showed that cleavage of the protein into two fragments was complete, it is possible that a proportion of the band 3 expressed in K562 cells undergoes only a single cleavage at residue 558, rather than cleavage at both residues 553 and 558. Incomplete digestion at residue 553 could be the result of different N-glycosylation of some of the protein. We also cannot eliminate the possibility that some of the band 3 expressed in K562 cells is proteolytically cleaved at the N-terminus.
The flow cytometric analysis of K562/band 3 clones demonstrated that all clones expressing band 3 also displayed high levels of Wrb antigen which is not expressed on untransfected K562 cells and K562/pBp cells. The Wrb antigen57 is a major blood group antigen which requires both GPA (residues 61-70) and band 3 (Glu658) for expression. The appearance of the Wrb antigen in K562/band 3 cells demonstrates that the expressed band 3 and the endogenous GPA present in these cells are able to interact as they do in RBCs. The two antibodies to Wrbwe used (BRICs 14 and 201) have previously been shown to recognize different epitopes within the antigen.40 The observation that both of these antibodies bind to K562/band 3 cells confirms that the band 3 and GPA interact normally in K562 cells. The large difference in reactivity of the two anti-Wrb antibodies with K562/B3 clone 1 may reflect differences in the accessibility of the two epitopes to the antibodies in the transfected K562 cells. The appearance of Wrb antigen in K562 cells transfected with band 3 cDNA also provides direct evidence that the band 3 polypeptide is required for formation of the Wrb epitopes.
Fluorescence-activated cell sorting (FACS) analysis with BRIC 69 (which binds to Rh polypeptides) demonstrated that K562/band 3 clones consistently showed an increase in the low level of endogenous Rh antigen reactivity. For K562/B3 clone 1, this increase was threefold relative to K562/pBp. The Rh polypeptides are the products of at least two highly homologous genes RHD andRHCE.58-60 The RHD gene gives rise to the D antigen, while the RHCE gene gives rise to the allelic antigens C/c and E/e, which are likely to be located on the same polypeptide chain.35 The antibody BRIC 69 recognizes both the RhD and RhcE polypeptides. The expression of Rh antigens on RBCs requires the presence/association of the Rh polypeptides with the RhGP glycoprotein, and there is evidence that the two groups of protein form a complex in the membrane.61,62 The Rh polypeptides are also thought to interact with other proteins, including CD47 and LW.58 K562 cells have substantial amounts of intrinsic RhGP expression, but only very low levels of Rh polypeptide expression. To investigate the effect of band 3 expression in K562 cells on expression of the Rh system antigens, we used two K562 clones previously transduced with pBpRhcE or pBpRhD constructs which express substantial amounts of RhD or cE polypeptides respectively.35 We cotransfected these K562/RhD and K562/RhcE clones with the pBnB3 construct. The resulting K562/RhD+B3 clones showed a substantial increase in binding of anti–band 3 in conjunction with marked increases in binding of antibodies to Rh polypeptides, RhGP, and RhD antigen. The binding of antibodies to the Rh components was increased in a similar manner in the K562/RhcE+B3 clones. It is unlikely that the increased antigen activity of Rh components in the K562/band 3 clones is a result of their reduced growth rates, as the increased reactivity was specifically observed for the Rh proteins and not for any of the endogenously expressed antigens GPC, CD47, or Fy3. The increased reactivity of the band 3–expressing clones with antibodies to Rh could originate in several ways. Band 3 may alter the membrane environment or interact with the proteins of the Rh complex in the plasma membrane, so that conformational or other rearrangements of the Rh protein complex result in increased reactivity with anti-Rh antibodies and BRIC 69. Alternatively, intracellular interactions of band 3 with the Rh proteins could increase the amount of Rh components present at the cell surface, by increasing the efficiency of folding of the Rh proteins, or by enhancing their translocation to the plasma membrane. All of these possibilities would be consistent with evidence that Rh antigen expression is reduced in RBCs which are heterozygous for the mutated band 3 present in Southeast Asian Ovalocytosis (deletion of residues 400-408).63 36 We have so far been unsuccessful in resolving the above possibilities by using immunoblotting to quantitatively detect the polypeptide chains of the Rh proteins in the transfected K562 cells with the antibody reagents available to us. More work will be necessary to establish the extent to which interactions occur between band 3 and the Rh protein complex, and the expression system presented here will be useful for these studies.
Under a light microscope, band 3-expressing K562 cells look morphologically similar to nontransfected K562 cells and K562 cells transfected with empty pBabe vectors, but FACS analysis demonstrated that the band 3-expressing clones are slightly more heterogenous in cell size and granularity than clones not expressing band 3. To further characterize the effects of band 3 expression on K562 cells, we measured growth rates of untransfected K562 cells, K562 cells transfected with empty pBabe vector, and three K562/band 3 clones. The doubling time of K562/band 3 clones was about twofold greater than that of nonexpressing clones. The increased demand on the protein synthetic apparatus of the cells may contribute to this effect, but it is likely that band 3 expression also has toxic effects on K562 cells. The transport activity of band 3 located transiently in the membranes of intracellular organelles could result in changes to the pH and ion contents of these compartments, adversely affecting their function. It is not clear whether the transport activity of internally localized, newly synthesized band 3 is regulated in K562 cells, but further investigations may yield information on this question.
ACKNOWLEDGMENT
We thank G.K. Jones for help with chloride transport assays, L.J. Bruce for ghosts of chymotrypsin-treated and untreated RBCs, H. Land for the pBabe puro and pBabe neo retroviral vectors, and K. Thompson and M. Uchikawa for monoclonal antibodies.
Supported in part by the Wellcome Trust. R.B. was the recipient of a University of Bristol Postgraduate Scholarship.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Michael J.A. Tanner, PhD, Department of Biochemistry, School of Medical Sciences, University of Bristol, Bristol BS8 1TD, UK; e-mail: M.Tanner@bris.ac.uk.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal