Abstract
The human myeloid-lymphoid leukemia gene, MLL (also calledALL-1, Htrx, or HRX ), maps to chromosomal band 11q23. MLL is involved in translocations that result in de novo acute lymphoblastic leukemia (ALL), acute myelogenous leukemia (AML), mixed lineage leukemia, and also in therapy AML (t-AML) and therapy ALL (t-ALL) resulting from treatment with DNA topoisomerase II (topo II) targeting drugs. MLL can recombine with more than 30 other chromosomal bands, of which 16 of the partner genes have been cloned. Breaks in MLL occur in an 8.3-kb breakpoint cluster region (BCR) encompassing exons 5 through 11. We recently demonstrated that 75% of de novo patient breakpoints in MLL mapped in the centromeric half of the BCR between two scaffold-associated regions (SAR), whereas 75% of the t-AML patient breakpoints mapped to the telomeric half of the BCR within a strong SAR. We have mapped additional structural elements in the BCR. An in vivo DNA topo II cleavage site (induced with several different drugs that target topo II) mapped near exon 9 in three leukemia cell lines. A strong DNase I hypersensitive site (HS) also mapped near exon 9 in four leukemia cell lines, including two in which MLL was rearranged [a t(6;11) and a t(9;11)], and in two lymphoblastoid cell lines with normalMLL. Two of the leukemia cell lines also showed an in vivo topo II cleavage site. Our results suggest that the chromatin structure of the MLL BCR may influence the location of DNA breaks in both de novo and therapy-related leukemias. We propose that topo II is enriched in the MLL telomeric SAR and that it cleaves the DNase I HS site after treatment with topo II inhibitors. These events may be involved in recombination associated with t-AML/t-ALL breakpoints mapping in the MLL SAR.
THE MYELOID-LYMPHOID leukemia (MLL)1 gene (also calledALL-1,2Htrx,3 andHRX4), which maps to human chromosome 11 band q23 (11q23), recombines with more than 30 different chromosomal partners (reviewed in Rowley5 and Bernard and Berger6),7-10 resulting in acute myelogenous leukemia (AML; usually monoblastic), acute lymphoblastic leukemia (ALL) and, more rarely, in lymphoma and myelodysplastic syndromes.11,12 The most common 11q23 translocations involving MLL are the t(9;11), t(6;11), and t(11;19)(19p13.1), usually resulting in AML de novo, and the t(4;11) and t(11;19)(19p13.3), usually resulting in ALL.5,6,11,12 Sixty to eighty percent of cases of acute leukemia in children less than 1 year of age show MLL rearrangements.13-16 Depending on the schedule and the total dosage, between 1% and 15% of cancer patients treated with chemotherapeutic drugs, particularly the epipodophyllotoxins that target topoisomerase II (topo II), develop therapy AML (t-AML) and, in rare cases, therapy ALL (t-ALL).17-21 The translocation t(9;11) is the most common of the 11q23 translocations in t-AML.11 22
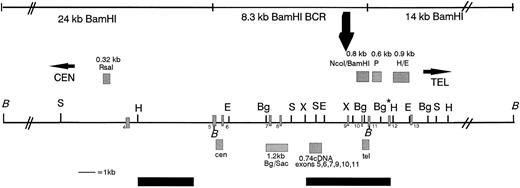
Although the MLL gene spans approximately 90 kb,23virtually all MLL breaks occur in an 8.3-kb BamHI fragment, the breakpoint cluster region (BCR).5,6 We and others have cloned and sequenced a total of 19 de novo6,22,24-28 and 3 t-AML21,29 patient breakpoint junctions. A variety of motifs, including topo II consensus cleavage sites, chi, and heptamer and nonamer consensus sequences have been noted at or near the breakpoint junctions. In addition, theMLL breakpoint region contains eight Alu repetitive elements, five of which occur in the first half of the BCR, the site of the majority of MLL breakpoints in de novo leukemias.24,30,31 However, breakpoints in only one translocation and in several MLL partial duplications found in de novo leukemia with a normal karyotype map within Alu repeats on both partner chromosomes.6,22,26,31-33 Previously, we mapped the location of patient breakpoints within the BCR and showed that 75% of de novo patient breakpoints mapped in the centromeric half of the BCR between two scaffold-associated regions (SARs), whereas 75% of the t-AML patient breakpoints mapped to the telomeric half of the BCR within a strong SAR, which colocalized with 6 of 7 topo II consensus cleavage sites (see Fig 1 in Strissel Broeker et al30). We proposed that the chromatin structure of this 8.3-kb BCR may influence the location of breaks in MLL. Recently, a study of patients with t(4;11) leukemia reported that most de novo adults had a break in the centromeric half of the BCR, whereas most infants (de novo) and all t-AML breaks occurred in the telomeric half.34 These data raise the possibility that a similar mechanism may be involved in breakpoints in both t-AML/t-ALL and in de novo infant leukemia.
In mammalian cells, topo II has both enzymatic and structural functions. There are two genetically and biochemically distinct topo II isoforms, topo II α and topo II β.35,36 Studies show that topo II α localizes in the nucleus, with its peak level of expression occurring at the G2/M boundary in the cell cycle.35 Topo II β localizes in the nucleolus and shows constant levels during the cell cycle.36 As a structural protein, topo II is needed for chromosome condensation, in which it colocalizes to the metaphase scaffold of native chromsomes, where it is thought to bind to SARs.37,38 Topo II has been implicated in recombination events, particularly at the mouse Ig κ light chain gene intronic SAR, and also at the MLLBCR.21,30 39-41
In general, DNase I hypersensitive sites (HS) represent nucleosomal DNA that have become conformationally changed due to the binding of specific proteins to target DNA sequences. They are identified in the genome by their susceptibility to DNase I cleavage. For example, DNase I HS are associated with the enhancers of transcriptionally active genes,42-45 and they map to the boundaries of genes in locus control regions.43,46 DNase I HS also associate with SARs,46,47 where some SARs also show in vitro or in vivo topo II cleavage.39,41,47 In yeast, DNase I sites are hot spots for mitotic and meiotic recombination.48
Because of the association of previous treatment with topo II targeting drugs and MLL rearrangements, we analyzed whether these drugs could induce cleavage in the MLL BCR, particularly in theMLL telomeric SAR.30 49 We have also investigated whether the MLL BCR contains regions that are susceptible to DNase I cleavage.
MATERIALS AND METHODS
Cell lines.
The chronic myelogenous leukemia (CML) BV173 cell line has the phenotype of nondifferentiated stem cells derived from a CML patient in blast crisis.50 The primary clone in this cell line (karyotyped at the University of Chicago, Chicago, IL) contains one normal chromosome 22 and three copies of the Ph chromosome derived from the t(9;22)(q34,q11). The UoC-M1 cell line is derived from a patient with AML.51 This cell line has a complex karyotype (karyotyped at the University of Chicago), with four copies of a germline MLL. Two B-lymphoblastoid cell lines (B-LCL), IB-4 generated from normal cord blood (kindly provided by Dr David Liebowitz, University of Chicago, Chicago, IL) and 9020 generated from a patient with t-AML and a t(9;11) involving MLL(kindly provided by Dr Richard Larson, University of Chicago), were established at the University of Chicago. The ML-2 cell line carrying a t(6;11)(q27;q23), and the Mono Mac 6 (MM6) cell line carrying a t(9;11)(p22;q23) were derived from patients with AML M5 de novo.52,53 The cell lines have MLL fusions with theAF6 and AF9 genes, respectively, which have been characterized in our laboratory.22 54 Additional cell lines studied were YK-M2, HL-60 (human promyelocytic leukemia cells), K562 (erythroleukemia cells derived from a CML patient in blast crisis), and the Burkitt lymphoma cell line, Raji. All cells were cultured in RPMI supplemented with 10% fetal calf serum, 1% HEPES, and sodium bicarbonate (amount adjusted per lot).
In vivo cleavage with DNA topoisomerase II.
Cells grown exponentially in complete media (RPMI 1640, fetal calf serum 10%; GIBCO, Grand Island, NY) were diluted and then treated for 6 or 16 hours with the nonintercalating topo II inhibitors etoposide (VP16 100 μmol/L; Sigma, St Louis, MO), teniposide (VM26 50 μmol/L, 100 μmol/L), or the intercalating topo II inhibitor doxorubicin (10 μmol/L to 50 μmol/L; Sigma) to produce endogenous topo II-cleaved complexes. Cells were then lysed and the DNA was isolated using a previously described method to trap cleaved topo II DNA.55 BV173 cells were also treated with additional damaging agents, including Aclarubicin (0.5 μmol/L to 50 μmol/L; Sigma) and N-Methylformamide (0.5 mol/L; Sigma). To map the approximate location of the drug-induced cleavage site, indirect end labeling was used by hybridizing two probes to Southern blots, an MLLtelomeric probe (bp 7955-8332 of the MLL BCR56) and the 0.74-kb cDNA (exons 5, 6, 7, 9, 10, and 11) polymerase chain reaction (PCR) probe.57
Isolation of nuclei for DNAse I cleavage studies.
For each hematopoietic cell line, approximately 5.0 × 108cells were isolated for nuclei according to Mirkovitch et al,58 with some modifications. All nuclear isolation steps were on ice. Briefly, cells were treated with chilled hypotonic solution I (3.75 mmol/L Tris/HCl, 0.5% thiodiglycol, 0.05 mmol/L spermine, 0.125 mmol/L spermidine, 0.5 mmol/L KOH/EDTA, protease inhibitors [0.1 mmol/L phenylmethysulfonyl fluoride (PMSF), and 0.5% aprotinin], and 20 mmol/L KCl). After two washes, cells were treated with ice-cold solution I + digitonin (0.1%) (solution II). After cellular homogenization, nuclei were centrifuged through a 0.25 mol/L sucrose cushion, washed several times with solution II, optical density (OD) readings were determined, and then the nuclei were frozen in 50% glycerol/solution II at −20°C for up to 4 months.
Treatment of nuclei with the DNase I endonuclease.
For DNase I treatment of nuclei, we used the methods of Kas and Laemmli,47 with modifications. Briefly, a total of 12 OD units of nuclei were washed in a 1× working solution (15 mmol/L Tris/HCl, 0.2 mmol/L spermine, 0.5 mmol/L spermidine, 80 mmol/L KCl, 0.1% digitonin, and the protease inhibitors PMSF [0.2 mmol/L] and aprotinin [1.0%]). Immediately, a range of DNase I enzyme units (0.20 to 20.0 U; Boehringer Mannheim, Indianapolis, IN) were added to each tube containing 1.6 OD units of nuclei and resuspended gently. After an incubation on ice for 5 minutes, all DNase I reactions were stopped by adding 1 μl of 0.5 mol/L EDTA and a 2× buffer containing 100 mmol/L Tris/HCl, 0.5% sodium dodecyl sulfate, and 25 mmol/L EDTA. All samples were diluted into 1× TE containing 250 ng/μL RNase A (Sigma). After 1 hour of incubation at 37°C, proteinase K was added and incubated for an additional 1 hour at 55°C. All DNase I samples were then incubated overnight at 37°C. The following day, each sample was diluted 1:1 with TE, and the DNA was extracted first with phenol and then with phenol/chloroform. The DNA was precipitated with isopropanol in the presence of 0.3 mol/L Na acetate or 0.7 mol/L ammonium acetate.
Southern blot and DNA probe hybridizations.
After digestion of the DNase I samples with restriction enzymes, approximately 15 to 20 μg of DNA (determined by OD readings) was electrophoresed on 0.8% or 1.0% agarose gels. Using standard conditions for Southern blotting (without acid depurination), the DNA was transferred by electroblotting in a 12 mmol/L Tris, 6 mmol/L Na acetate, and 0.3 mmol/L EDTA (pH 7.5)-containing buffer to GeneScreen or Hybond (Amersham, Arlington Heights, IL) positively charged nylon membranes.59 Using indirect end labeling,42 hybridization of DNA probes to Southern blots was performed according to standard protocols, with 50% formamide at 42°C. With certain probes, Cot I DNA (100 μg; GIBCO-BRL, Gaithersburg, MD) was used in the prehybridization and hybridization steps to block repetitive elements. For topo II analysis, high molecular weight DNA was digested withBamHI and analyzed by Southern blot using standard conditions as previously described.12
DNA probe isolation.
Cloned DNA fragments or PCR-amplified products were purified from agarose gels after gel electrophoresis and were used as probes.60 Figure 1 shows a restriction map of the MLL BCR and the location of all theMLL probes used for this study. Primers chosen for MLLamplification corresponded to non-Alu regions of the 8.3-kbBamHI BCR. The following are the DNA fragments listed in the centromeric to telomeric orientation in the MLL gene: a cloned 0.32-kb Rsa I DNA fragment, which maps centromeric to the 8.3-kb MLL BCR (kindly provided by Dr Peter Domer, University of Chicago); a 0.48-kb PCR DNA fragment, the cen probe, which maps adjacent to the centromeric BamHI site and within theMLL BCR (top primer 5′ GGATCCTGCCCCAAAGAAAAGCAGTAGTGAGCC 3′, and bottom primer 5′ AGGCTTCGAACAGGAAATTAAAACAATACCTCC 3′); a 1.2-kbBgl II/Sac I PCR DNA fragment, which maps to the middle of the MLL BCR30; a 0.8-kb NcoI/BamHI cloned DNA fragment, which maps to the telomeric region of the MLL BCR; a 0.385-kb PCR DNA fragment, tel probe, which maps adjacent to the telomeric BamHI site within theMLL BCR (top primer 5′ TTTTCTTACAGCAGCTGCTGGAGTGTAAT 3′, and bottom primer 5′ AGCTCTTACAGCGAACACACTTGGTACAGATC 3′); the 0.74-kb complementary DNA (cDNA) (MLL exons 5, 6, 7, 9, 10, and 11) PCR fragment57; and two DNA fragments that map adjacent and telomeric to the 8.3-kb BamHI BCR—a 0.6-kbPstI DNA fragment isolated from the λ phage clone 14p, a 14-kb telomeric BamHI fragment (0.6-kb P), and a 0.9-kb H/E DNA fragment isolated from the MM6 der(9) EcoRI clone (0.9-kb H/E; Fig 1).22 The AF9MM6 probe was PCR amplified from the AF9 C48 cosmid DNA.22 This AF9MM6 DNA fragment maps centromeric and adjacent to the MM6 breakpoint junction and thus identifies both the AF9 germline and the der(9)EcoRI DNA fragments (top primer 5′ ATATTATGTACAAGAATAAGTTATGCTCTA 3′, bottom primer 5′ AATAGAATTAGAATACTGGAGCTC 3′).
Restriction map of the MLL BCR showing the location of the MLL probes used in this study, the in vivo topo II cleavage site, and the DNA damaging agent cleavage site (large black arrow above the map). Above the map and indicated as grey hatched boxes are the genomic probes from left to right: the 0.32-kb Rsa I DNA fragment, the 0.8-kb Nco I/BamHI DNA fragment, the 0.6-kb P DNA fragment, and the 0.9-kb H/E DNA fragment. Below the map are the PCR probes: the cen 0.48-kb DNA fragment, the 1.2-kbBgl II/Sac I DNA fragment, the 0.74-kb cDNA, and the tel 0.385-kb DNA fragment. Restriction enzyme sites are indicated along the black line showing the MLL BCR and regions centromeric and telomeric to the BCR. BamHI (B) DNA fragments covering 42 kb (black lines indicated above the map), including the centromeric 24-kb fragment, the 8.3-kb BCR DNA fragment, and the 14-kb telomeric DNA fragment, were analyzed for in vivo cleavage of topo II by hybridizing the 0.32-kb Rsa I, the 0.74-kb cDNA, the tel, and the 0.6-kb P probes to BamHI digestions of etoposide-treated cells. Only the 8.3-kb BCR DNA fragment in BV173 cells was analyzed for cleavage using the DNA damaging agents aclarubicin and N-Methylformamide. Black bars below the map are the weak centromeric and strong telomeric SARs.30 Numbered exons are represented as grey hatched rectangles on the map. The restriction map is not drawn to scale centromeric or telomeric to the double thick black diagonal lines (//). Restriction enzymes are noted on the map as follows: B, BamHI; S, Sac I; H,HindIII; E, EcoRI; Bg, Bgl II; X, XbaI. Note that the Bg* enzyme restriction site is polymorphic in BV173 cells.
Restriction map of the MLL BCR showing the location of the MLL probes used in this study, the in vivo topo II cleavage site, and the DNA damaging agent cleavage site (large black arrow above the map). Above the map and indicated as grey hatched boxes are the genomic probes from left to right: the 0.32-kb Rsa I DNA fragment, the 0.8-kb Nco I/BamHI DNA fragment, the 0.6-kb P DNA fragment, and the 0.9-kb H/E DNA fragment. Below the map are the PCR probes: the cen 0.48-kb DNA fragment, the 1.2-kbBgl II/Sac I DNA fragment, the 0.74-kb cDNA, and the tel 0.385-kb DNA fragment. Restriction enzyme sites are indicated along the black line showing the MLL BCR and regions centromeric and telomeric to the BCR. BamHI (B) DNA fragments covering 42 kb (black lines indicated above the map), including the centromeric 24-kb fragment, the 8.3-kb BCR DNA fragment, and the 14-kb telomeric DNA fragment, were analyzed for in vivo cleavage of topo II by hybridizing the 0.32-kb Rsa I, the 0.74-kb cDNA, the tel, and the 0.6-kb P probes to BamHI digestions of etoposide-treated cells. Only the 8.3-kb BCR DNA fragment in BV173 cells was analyzed for cleavage using the DNA damaging agents aclarubicin and N-Methylformamide. Black bars below the map are the weak centromeric and strong telomeric SARs.30 Numbered exons are represented as grey hatched rectangles on the map. The restriction map is not drawn to scale centromeric or telomeric to the double thick black diagonal lines (//). Restriction enzymes are noted on the map as follows: B, BamHI; S, Sac I; H,HindIII; E, EcoRI; Bg, Bgl II; X, XbaI. Note that the Bg* enzyme restriction site is polymorphic in BV173 cells.
We also used DNA probes for other gene regions. The AML1 exon 6 probe was PCR amplified from the AML1B cDNA clone (kindly provided by Dr Guiseppina Nuicifora, Loyola University, Chicago IL; top primer 5′ GACATCGGCAGAAACGAGATGATCAGACC 3′, bottom primer 5′ GCATCTGACTCTGAGGCTGAGGGTTAAAG 3′). BCR gene probes (on chromosome 22) included the 2.3-kb EcoRI/BamHI fragment isolated from the ALL SUPB13 breakpoint junction61 and the 5′ and 3′ BCR DNA fragments isolated from the 5.8-kb major BCR (MBCR).62 The 5′ MBCR BCR DNA probe detects a known HS site approximately 8 kb 5′ to the MBCR.63 The β-globin IVS2 probe (kindly provided by Dr Owen Witte, UCLA, Los Angeles, CA) was used as a negative control for DNase I HS sites in cell lines that do not express the β-globin gene.
RESULTS
In this study, we describe structural elements that map to the same region within the MLL BCR: an in vivo topo II cleavage site, a strong DNase I HS site, and a cleavage site induced with DNA damaging agents. These DNA structural elements may contribute to the location and to the mechanism of breakage leading to leukemia.
An in vivo topo II cleavage site maps near exon 9.
After treatment of BV173 cells with either etoposide, teniposide, or doxorubicin, we identified one additional DNA band within theMLL BCR using BamHI-digested DNA and a probe located at the telomeric end of the MLL BCR (Figs 1 and2A). Two additional DNA bands (approximately 6.9 to 7.0 kb and 1.5 to 1.6 kb) were also observed using the 0.74-kb cDNA probe after treatment with etoposide, doxorubicin, aclarubicin, and N-Methylformamide (Figs 1 and2B and data not shown). In general, treatment with etoposides showed a much stronger DNA cleavage than treatment of cells with aclarubicin or with N-Methylformamide (data not shown). The cleavage site mapped approximately 1.5 to 1.6 kb from the telomeric BamHI site in the MLL BCR, which is near to exon 9 within the telomeric SAR (Fig 1).30 We detected only one in vivo topo II cleavage site mapping within this region, regardless of which topo II–targeting drug was used and with a range of drug concentrations. We identified this same topo II cleavage site in two additional hematopoietic cell lines tested: UoC-M1 and YK-M2 (data not shown). However, we were unable to detect cleavage using similar conditions in some other cell lines, including HL60, K562, Raji, and several B-LCLs (9020 and IB-4; data not shown). In the case of BV173, in which we noted topo II cleavage, we hybridized the same blots with MLL probes (0.32-kbRsa I and 0.6-kb Pst I) that identify the MLL24-kb centromeric and 14-kb telomeric BamHI fragments, respectively, and did not detect any new size fragments (Fig 1 and data not shown). Hybridization of these blots with the β-globinprobe also did not identify any other regions with drug-induced cleavage sites (data not shown). Therefore, this topo II cleavage site was a specific event.
Southern blots of BV173 DNA showing drug-induced cleavage of the MLL BCR. (A) The lanes from left to right show the marker (M), a negative control [(−) where no drugs were added to BV173 cells], VP16 cell treatment (100 μmol/L, 200 μmol/L), VM26 cell treatment (50 μmol/L, 100 μmol/L), and doxorubicin (Dox) cell treatment (10 μmol/L). An increasing amount of drug added is indicated by a triangle above the lanes. The PCR genomic probe (tel; see Fig 1) was hybridized to the Southern blot. The 8.3-kb germline DNA fragment and the new 1.5- to 1.6-kb drug-induced DNA fragment are indicated. (B) The lanes from left to right show DNase I–treated nuclei (0 to 20 U as indicated), a 16-hour drug treatment of BV173 cells, including a dimethyl sulfoxide control (C), VP16 (100 μm—two lanes), and N-Methylformamide (NMF; 0.5 mol/L). The PCR cDNA probe indicated above (0.74 kb) was hybridized toBamHI-digested DNA. The 8.3-kb germline and two new DNase I and drug-induced DNA fragments (1.5 kb and 6.9 to 7.0 kb) are indicated to the right. The DNase I, topo II, and DNA damaging agent cleavage sites either map within exon 9 or just telomeric of exon 9. Note that the new 6.9- to 7.0-kb DNA fragment is strongly hybridizing with exons 5, 6, and 7 and either all or half of exon 9. In contrast, the 1.5-kb DNA fragment is hybridizing more weakly with either none or half of exon 9, plus exon 10 and half of exon 11 (133 bp or 206 bp, respectively). The 1-kb marker (M) indicated to the left identifies a few of the molecular weight bands that cross-hybridize with this probe.
Southern blots of BV173 DNA showing drug-induced cleavage of the MLL BCR. (A) The lanes from left to right show the marker (M), a negative control [(−) where no drugs were added to BV173 cells], VP16 cell treatment (100 μmol/L, 200 μmol/L), VM26 cell treatment (50 μmol/L, 100 μmol/L), and doxorubicin (Dox) cell treatment (10 μmol/L). An increasing amount of drug added is indicated by a triangle above the lanes. The PCR genomic probe (tel; see Fig 1) was hybridized to the Southern blot. The 8.3-kb germline DNA fragment and the new 1.5- to 1.6-kb drug-induced DNA fragment are indicated. (B) The lanes from left to right show DNase I–treated nuclei (0 to 20 U as indicated), a 16-hour drug treatment of BV173 cells, including a dimethyl sulfoxide control (C), VP16 (100 μm—two lanes), and N-Methylformamide (NMF; 0.5 mol/L). The PCR cDNA probe indicated above (0.74 kb) was hybridized toBamHI-digested DNA. The 8.3-kb germline and two new DNase I and drug-induced DNA fragments (1.5 kb and 6.9 to 7.0 kb) are indicated to the right. The DNase I, topo II, and DNA damaging agent cleavage sites either map within exon 9 or just telomeric of exon 9. Note that the new 6.9- to 7.0-kb DNA fragment is strongly hybridizing with exons 5, 6, and 7 and either all or half of exon 9. In contrast, the 1.5-kb DNA fragment is hybridizing more weakly with either none or half of exon 9, plus exon 10 and half of exon 11 (133 bp or 206 bp, respectively). The 1-kb marker (M) indicated to the left identifies a few of the molecular weight bands that cross-hybridize with this probe.
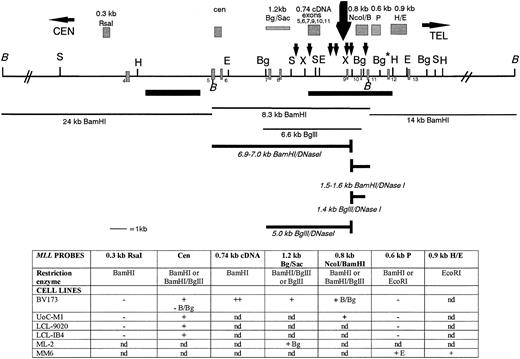
A strong DNase I site maps to the telomeric region of the MLL BCR near exon 9.
We analyzed the DNA regions within and outside the MLL BCR for the presence of DNase I HS. Nuclei from six cell lines were treated with increasing concentrations of DNase I and were then hybridized with various MLL probes. Figure 3 shows a summary of our MLL DNase I mapping results. For all six cell lines tested, we mapped a single strong DNase I HS site near exon 9. In contrast, no DNase I HS mapped in the centromeric half of the BCR or in 24 kb centromeric or 14 kb telomeric to the BCR in the cell lines studied. For example, in BV173 cells, the 1.2-kb BglII/Sac I DNA fragment hybridizes to a 6.6-kb Bgl II germline fragment and a new 5.0-kb Bgl II DNase I fragment (Figs 3 and 4A). In the UoC-M1 cell line, the MLL cen probe hybridizes to the 8.3-kb BamHI germline fragment and a new 6.9- to 7.0-kb BamHI DNase I fragment (Fig 3 and data not shown). No DNase I fragment was observed in the centromeric region of the BCR (Figs 3 and 4B). Because both the 1.2-kb Bgl II/Sac I probe and the MLL cen probe map at the centromeric ends of the digested MLL DNA, we were able to map the DNase I HS near exon 9 (Fig 3).
A DNase I HS site maps near exon 9 in the MLLBCR. Restriction map of the MLL BCR and adjacent regions are represented. Exons, SARs, and restriction enzymes are the same as in Fig 1. The restriction map is not drawn to scale centromeric or telomeric to the double thick black diagonal lines (//). The large black arrow above the map represents the strong DNase I cleavage site near exon 9. Probes are represented as grey hatched boxes above the map, and the 7 small black arrows above the map represent potential topo II in vivo cleavage sites identified by computer analysis.30 Thin black lines below the map identify germline fragments that hybridize with the probes above. Thick lines below the map represent the new DNase I fragments that are only observed after DNase I digestion. Note that only BamHI andBgl II germline (thin lines) and DNase I (thick lines) fragments are represented below the restriction digest map to show examples of our results. The table below the map represents a summary of the DNA restriction digests, the gene probes, and DNase I HS results correlated with the MLL map. The two top rows for each set of results correspond to the probes hybridized (in bold) to DNA isolated from DNase I–treated nuclei and digested with particular enzymes (second row, or otherwise indicated). Restriction enzymes:BamHI (B), Bgl II (Bg), EcoRI (E), NcoI, and Pst I. Results are indicated as + for the presence of DNase I HS, − for negative for DNase I HS, and nd for not determined.
A DNase I HS site maps near exon 9 in the MLLBCR. Restriction map of the MLL BCR and adjacent regions are represented. Exons, SARs, and restriction enzymes are the same as in Fig 1. The restriction map is not drawn to scale centromeric or telomeric to the double thick black diagonal lines (//). The large black arrow above the map represents the strong DNase I cleavage site near exon 9. Probes are represented as grey hatched boxes above the map, and the 7 small black arrows above the map represent potential topo II in vivo cleavage sites identified by computer analysis.30 Thin black lines below the map identify germline fragments that hybridize with the probes above. Thick lines below the map represent the new DNase I fragments that are only observed after DNase I digestion. Note that only BamHI andBgl II germline (thin lines) and DNase I (thick lines) fragments are represented below the restriction digest map to show examples of our results. The table below the map represents a summary of the DNA restriction digests, the gene probes, and DNase I HS results correlated with the MLL map. The two top rows for each set of results correspond to the probes hybridized (in bold) to DNA isolated from DNase I–treated nuclei and digested with particular enzymes (second row, or otherwise indicated). Restriction enzymes:BamHI (B), Bgl II (Bg), EcoRI (E), NcoI, and Pst I. Results are indicated as + for the presence of DNase I HS, − for negative for DNase I HS, and nd for not determined.
BV173 Southern blot showing the MLL BCR DNase I HS site. A Southern blot representing BamHI/BglII-digested DNA from DNase I–treated (DNase I units directly above each lane) and BV173 whole nuclei was hybridized independently withMLL BCR DNA probes (indicated above each panel). λ PhageHindIII/EcoRI-digested marker (left of [A]) correlates with all three hybridizations. (A) Top germline 6.6-kbBgl II DNA fragment hybridizing with the 1.2-kb BglII/Sac I DNA probe is seen in all lanes. A new 5.0-kbBgl II/DNase I DNA fragment (arrow) is not observed in the absence of DNase I, but increases in intensity at higher DNase I concentrations. (B) A germline 2.2-kb DNA fragment is observed in all DNA lanes; thus, the centromeric portion of the MLL BCR is negative for DNase I HS sites. (C) The germline 6.0-kb Bgl II DNA fragment hybridizing with the 0.8-kb Nco I-BamHI DNA probe is seen in all of the DNA lanes. A new 1.4-kb Bgl II DNA fragment is not observed in the absence of DNase I, but increases in intensity at higher concentrations of DNase I (arrow).
BV173 Southern blot showing the MLL BCR DNase I HS site. A Southern blot representing BamHI/BglII-digested DNA from DNase I–treated (DNase I units directly above each lane) and BV173 whole nuclei was hybridized independently withMLL BCR DNA probes (indicated above each panel). λ PhageHindIII/EcoRI-digested marker (left of [A]) correlates with all three hybridizations. (A) Top germline 6.6-kbBgl II DNA fragment hybridizing with the 1.2-kb BglII/Sac I DNA probe is seen in all lanes. A new 5.0-kbBgl II/DNase I DNA fragment (arrow) is not observed in the absence of DNase I, but increases in intensity at higher DNase I concentrations. (B) A germline 2.2-kb DNA fragment is observed in all DNA lanes; thus, the centromeric portion of the MLL BCR is negative for DNase I HS sites. (C) The germline 6.0-kb Bgl II DNA fragment hybridizing with the 0.8-kb Nco I-BamHI DNA probe is seen in all of the DNA lanes. A new 1.4-kb Bgl II DNA fragment is not observed in the absence of DNase I, but increases in intensity at higher concentrations of DNase I (arrow).
To define the location of the DNase I HS on smaller DNA fragments, we hybridized the 0.8-kb Nco I/BamHI MLL telomeric DNA fragment to BamHI/Bgl II- andBamHI-digested DNA from DNase I–treated BV173 and UoC-M1 nuclei (Figs 3 and 4C and data not shown). In addition to MLLgermline fragments, results showed two new DNase I HS DNA fragments, a 1.4-kb Bgl II fragment and a 1.5- to 1.6-kb BamHI fragment, which increased in intensity with higher DNase I concentrations (Fig 4C and data not shown). These results confirm that the HS maps within the previously mapped strong SAR near to exon 9 and, interestingly, maps to the same region as the in vivo topo II cleavage site as well as the cleavage site induced with DNA damaging agents (Figs 2B and 3).30
We studied four other cell lines, including 9020, IB-4, and two leukemia cell lines, the ML-2 [t(6;11)] and MM6 [t(9;11)], in which the MLL gene is rearranged as a consequence of a translocation. Our results showed a strong DNase I HS site mapping to the same region as BV173 and UoC-M1 cells in all four cell lines (Fig 3). Cytogenetic analysis of the ML-2 cell line shows a complex tetraploid karyotype, including two copies of the der(6) and der(11) chromosomes that result from the balanced t(6;11) and two copies of a deleted chromosome 6 and a second abnormal chromosome 11 with an interstitial deletion fromMLL intron 6 through the ETS1 probe at 11q25.52,54 The ML-2 cell line contains rearrangements of all MLL alleles, with the genomic breakpoint in all alleles mapping in the first half of the BCR between exons 6 and 7.54 In this cell line, the DNase I HS site is translocated to the derivative 6 chromosome mapping approximately 5.3 kb telomeric to the MLL/AF6 fusion point (Fig 3 and data not shown).
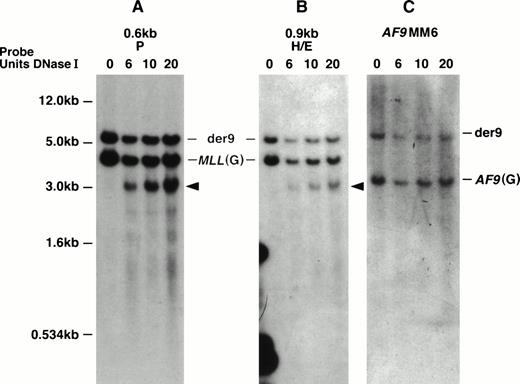
With the MM6 cell line with a t(9;11), we defined the location of the DNase I HS more precisely. This cell line is hypotetraploid and has a complex karyotype including two copies of the normal chromosomes 9 and 11 and two copies of the derivative chromosomes 9 and 11.22,53 Both MLL and AF9 show staggered breaks that result in deletions.22 With more precise mapping, the MLL deletion, which we initially determined to be 499 bp, has been determined to be 507 bp ofMLL (from nucleotides 6577 to 7084 in the MLL BCR). The deletion of AF9 is 165 bp.22 Interestingly, the DNase I HS region appears to map within the region of MLL that is deleted in both derivative chromosomes 9 and 11. Thus, in MM6 cells, we predict that the DNase I HS site maps only on the two normal 11 chromosomes. Hybridizing two MLL probes that map telomeric to the MM6 MLL breakpoint, the 0.6-kb P and the 0.9-kb H/E DNA fragments to EcoRI-digested DNA, we observed a 4.3-kb MLL germline DNA fragment, a 6.0-kb der (9) DNA fragment, and a new 3.2-kb DNase I HS fragment (Figs 3 and 5A and B). Hybridizing the AF9MM6 probe to the same blot only showed a germline AF9 3.4-kb DNA fragment and the der (9) 6.0-kb rearranged DNA fragment (Fig 5C). Because we did not detect the 3.2-kb DNase I HS fragment with the AF9MM6 probe, which we did with both MLL probes (0.6-kb P and 0.9-kb H/E), we conclude that the MLL DNase I HS region did not translocate to AF9 and therefore does not map telomeric to nucleotide (nt) 7087 in the MLL BCR.
MM6 DNA Southern blot showing the MLL DNase I HS site on the normal 11 chromosomes. Southern blot representingEcoRI-digested DNA from DNase I–treated (DNase I units used indicated directly above panels) MM6 whole nuclei was hybridized independently with three DNA probes (indicated above each panel). The 1-kb plasmid marker is shown to the left of (A) and also correlates with hybridizations in (B) and (C). (A) The top 6.0-kb DNA band hybridizing with the MLL 0.6-kb P probe represents the der (9) and is observed in all DNA lanes. The middle MLL 4.5-kb germline (G) DNA band is seen in all DNA lanes. A new 3.2-kbEcoRI/DNase I DNA fragment is only observed in DNase I–treated whole nuclei (arrow). (B) The top 6.0-kb DNA band hybridizing with theMLL 0.9-kb HindIII/EcoRI DNA probe represents the der(9) and is observed in all DNA lanes. The middle MLL4.5-kb germline (G) DNA band is seen in all DNA lanes. A new 3.2-kbEcoRI/DNase I DNA fragment is only observed in DNase I–treated whole nuclei (arrow). This is the same DNA fragment to which the 0.6-kb P probe hybridizes. (C) The top 6.0-kb DNA band hybridizing with theAF9MM6 probe represents the der (9) and is observed in all DNA lanes. The 3.4-kb DNA band represents the AF9 germline (G) region containing the MM6 deletion breakpoint region and is also observed in all DNA lanes.
MM6 DNA Southern blot showing the MLL DNase I HS site on the normal 11 chromosomes. Southern blot representingEcoRI-digested DNA from DNase I–treated (DNase I units used indicated directly above panels) MM6 whole nuclei was hybridized independently with three DNA probes (indicated above each panel). The 1-kb plasmid marker is shown to the left of (A) and also correlates with hybridizations in (B) and (C). (A) The top 6.0-kb DNA band hybridizing with the MLL 0.6-kb P probe represents the der (9) and is observed in all DNA lanes. The middle MLL 4.5-kb germline (G) DNA band is seen in all DNA lanes. A new 3.2-kbEcoRI/DNase I DNA fragment is only observed in DNase I–treated whole nuclei (arrow). (B) The top 6.0-kb DNA band hybridizing with theMLL 0.9-kb HindIII/EcoRI DNA probe represents the der(9) and is observed in all DNA lanes. The middle MLL4.5-kb germline (G) DNA band is seen in all DNA lanes. A new 3.2-kbEcoRI/DNase I DNA fragment is only observed in DNase I–treated whole nuclei (arrow). This is the same DNA fragment to which the 0.6-kb P probe hybridizes. (C) The top 6.0-kb DNA band hybridizing with theAF9MM6 probe represents the der (9) and is observed in all DNA lanes. The 3.4-kb DNA band represents the AF9 germline (G) region containing the MM6 deletion breakpoint region and is also observed in all DNA lanes.
In summary, based on our Southern blot analysis of MM6 cells together with the five other cell lines studied, particularly our hybridizations with the 0.8-kb Nco I/BamHI probe, we predict that a strong DNase I HS site maps to the 387-bp region between nucleotides 6700 and 7087 (containing exon 9) in the MLL BCR. In contrast, no DNase I HS sites mapped in the centromeric half of the BCR or outside the MLL BCR for a total of 42 kb.
Additional gene regions studied for DNase I HS.
Three other gene regions, one of which is involved in topo II-associated t-AML, were also tested for DNase I hypersensitivity. TheAML1 gene on chromosome 21 is involved in the t(8;21)(q22;q22) in both de novo leukemia and in t-AML after therapy with drugs that target DNA topo II.19,64 We examined a 23.9-kbBamHI DNA fragment that contains exon 6 with 1.7 kb of intron 5 and 22.2 kb of intron 6, the location of many of the translocation breakpoints.64 We also examined two BamHI andBgl II DNA regions from the BCR gene on chromosome 22 involved in the t(9;22): the ALL BCR and the CML MBCR; and a 14-kbBamHI DNA region from the β-globin gene that is not involved in translocations.61-63 In the AML1 23-kbBamHI gene region containing intron 6, in both the BV173 and UoC-M1 cell lines, we detected no DNase I HS sites (up to 20 U of DNase I enzyme; data not shown). In the BCR gene, we observed possibly three DNase I HS sites mapping to the ALL BCR in BV173 cells, whereas no DNase I HS sites mapped to this same region in three other cell lines tested (UoC-M1, 9020, and IB-4; P.L. Strissel, unpublished data). We detected a strong DNase I HS site approximately 8 kb 5′ to the CML MBCR in the BCR gene in all cell lines tested (BV173, UoC-M1, 9020, IB4, and MM6). This DNase I HS site had been previously mapped in the K562 cell line.63 In addition to the DNase I HS site 5′ to the CML MBCR, we observed a strong HS site that maps within the 5.8-kb MBCR in the BV173, the UoC-M1, and the ML-2 cell lines but not in the B-LCLs (9020 and IB4; P.L. Strissel, unpublished data). The β-globin gene (the BamHI DNA region including intron 2, exon 3, and the enhancer element) lacked any hypersensitive sites, indicating that it is in a closed chromatin configuration in each of these cell lines (data not shown).
DISCUSSION
The MLL BCR is a unique region to study mechanisms of illegitimate recombination, because virtually all of de novo and therapy-related leukemia patient breakpoints involving MLL map within the 8.3-kb BamHI fragment. As an approach to try to understand MLL BCR illegitimate recombination events, we have studied various DNA structural elements. We mapped an in vivo topo II cleavage site, a strong DNase I HS, and a DNA damaging cleavage site to the same region near exon 9. In contrast, no topo II cleavage sites or DNase I HS sites were found in the centromeric half of the MLLBCR or outside of the BCR for a total of 42 kb.
Our observations confirm our preliminary results49 and those of others40,41 who have detected a single in vivo topo II cleavage site near exon 9 in the MLL BCR and who also noted that no other topo II cleavage sites mapped centromeric in the BCR or centromeric and telomeric in the adjacent BamHI fragments.40 Domer et al21 also observed in vivo topo II cleavage in the MLL BCR of HL-60 cells. Although they did not use indirect or direct end labeling experiments, they observed two in vivo topo II cleavage sites, one of which most likely colocalizes with the cleavage site near to exon 9.21Despite the fact that different MLL BCR probes (representing the centromeric, the middle, and telomeric regions) and various restriction enzyme–digested genomic DNA were used in these investigations, the majority of the studies identified a single in vivo topo II cleavage site on Southern blots, thus confirming that the location of the cleavage site is specific to the MLL BCR region (this report and previous literature21,40 41).
The observations of various groups demonstrate differences in topo II cleavage susceptibility between various cell types and also show DNA cleavage with DNA damaging agents or cell starvation. In these investigations, cells were cultured using various apoptosis-inducing drugs that target topo II (etoposide, tenoposide, and doxorubicin) or genotoxic chemotherapeutic agents or culture conditions (fetal calf serum starvation) that do not target topo II (this report and previous literature21,40,41). We observed drug-induced cleavage in undifferentiated blast cells (BV173) using both topo II targeting drugs and additional DNA damaging agents (Figs 1 and 2A and B). In two myeloid cell lines (UoC-M1 and YK-M2), we observed topo II cleavage (Fig 1). However, we did not observe any topo II cleavage in HL-60 myeloid cells, in K562 erythroleukemia cells, or in the Burkitt lymphoma (Raji) or the two B-LCLs tested (IB-4 and 9020). Thus, we detected differences in topo II cleavage susceptibility between myeloid cell lines; however, we did not observe cleavage in the lymphoblastic cell lines. Domer et al21 observed in vivo topo II cleavage sites in the MLL BCR in the HL-60 cell line, whereas Aplan et al40 observed topo II cleavage only in one (ML-1 cells) of six (HL-60, KG-1, K562, U937, HEL) myeloid cell lines; in contrast to our results, they observed cleavage in Raji cells. Aplan et al40 also observed that topo II cleaved the MLL BCR in normal peripheral blood cells (±phytohemagglutinin), in T-ALL cells at diagnosis, in 4 of 6 T- and 3 of 4 B-cell lines, and in one small cell lung carcinoma cell line. They did not observe cleavage in HeLa or in cell lines from a neuroblastoma, fibrosarcoma, or a bladder cell line. Possible explanations for these conflicting results between our data and those of others21,40 with regard to HL60 and Raji cells could be that cell lines after a different number of passages may have become resistant to etoposide (possibly through mutations in the topo II gene) or may have acquired mutations in the multidrug resistant (MDR) gene65or that the cells could have reduced their topo II enzymatic activity.66 One example supporting resistance to etoposide is a variant HL-60 cell line that was selected for resistance to doxorubicin.67 This HL-60 cell line demonstrates a 10× and 20× resistance to doxorubicin and etoposide, respectively, when compared with the parental HL-60 cells.
The MLL DNase I HS site was present in all six hematopoietic cell lines we tested, including undifferentiated blast cells (BV173), three myeloid cell lines (UoC-M1, ML-2, and MM6), and two B-LCLs (IB-4 and 9020; Fig 3). Thus, in our study, the DNase I HS and the in vivo topo II cleavage site both occur in the same MLL region in BV173 and in UoC-M1 cells. The BV173 cleavage site induced with additional DNA damaging agents also maps to the region where DNase I and topo II cleaves. In contrast, DNase I hypersensitivity but not topo II cleavage was observed in the B-LCLs. In all of these cell lines, the DNase I cleavage was strong, appearing first at 2.0 U DNase I and at 4°C (Figs 4A and C and 5A and B). In the ML-2 cell line, the DNase I HS was located on the derivative 6 chromosome as a result of the t(6;11). In the MM6 t(9;11) cell line, we mapped the DNase I HS cleavage site in the BCR more precisely to a 387-bp region between nucleotides 6700 and 7087 (containing exon 9 on two normal 11 chromosomes). We did not test for DNase I cleavage in any T-cell lines or in other tissues; therefore, at present we do not know whether this site shows tissue specificity. As noted earlier, Aplan et al40 detected the topo II cleavage site in T cells.
In addition to MLL, we examined regions of other genes for DNase I HS sites and topo II cleavage. No HS sites were found in a 23.9-kb region of AML1. This region consists of 1.7 kb of intron 5 and 22.2 kb of intron 6 and is a region where some t(8;21) de novo and t-AML leukemia breakpoints occur. Stannula et al68 observed a weak topo II cleavage site in this same region in two T-cell lines and in one pre-B–cell line. Even at the highest levels of DNase I concentration, we did not observe DNase I HS in BV173 and UoC-M1 cells. Similar to our DNase I results, Stannula et al68 did not observe in vivo topo II cleavage in theAML1 gene in any of the five myeloid cell lines studied. In contrast, the BCR gene on chromosome 22 showed several DNase I HS sites. For example, in BV173 cells, we observed possibly three DNase I HS mapping in the ALL BCR in intron 1.61,69 In contrast, these same sites were not present in the myeloid UoC-M1 cell line or in the lymphoid cell lines 9020 and IB-4. At the CML MBCR located in the second half of the BCR gene,62 we observed a single strong DNase I HS site mapping within the MBCR in BV173, UoC-M1, ML-2, and MM6 cells, but it was not present in the B-cell lines 9020 and IB-4. As a negative control, the β-globin gene region was devoid of DNase I HS sites and in vivo topo II cleavage sites; thus, in these cell lines, this region is in a closed chromatin conformation as expected and does not demonstrate sensitivity to topo II-targeting drugs.
We have previously proposed that the chromatin structure of theMLL BCR may be involved in the mechanism of recombination for both de novo acute leukemias and t-AML.30 In our initial studies, we found differences in the location of MLLbreakpoints in de novo and t-AML patients, that we could correlate with DNA structural features. Thus, 75% of the de novo leukemia breakpoints mapped in the centromeric half of the BCR (a non-SAR DNA loop), compared with 75% of t-AML breakpoints that mapped to the telomeric half of the BCR localizing within the strong SAR.30 Our findings have recently been confirmed by Cimino et al,34who found that 67% of MLL breakpoints in children and adults with de novo leukemia and a t(4;11) mapped in the centromeric half of the BCR, whereas all five of their t-AML patient breakpoints mapped to the telomeric half. Of particular interest is the fact that these investigators showed that 71% (20/28) of breakpoints in infant t(4;11) ALL mapped to the telomeric half of the MLL BCR. It has been suggested that infant ALL may, in part, be caused by the exposure of expectant mothers to pesticides or an excess of natural topo II inhibitors such as flavonoids.70,71 Cimino et al34 speculate that the leukemias in both infants and in t-AML patients may share a common mechanism for breakage in the BCR that may be different from the mechanism involving breaks in the de novo leukemias in older patients.
One proposed mechanism for MLL BCR breakage and recombination is that the topo II cleavage site and the telomeric SAR are initiation sites during early events in apoptosis.41 Stanulla et al41 observed that the same DNA cleavage site mapping near to exon 9 was detected in cells exposed to etoposide treatment as well as in those exposed to other agents (antimetabolites, genotoxic drugs, and cell starvation) that do not affect topo II. This suggests that this is a unique structural region of DNA. Using rapidly growing cells, we observed that this same region is cleaved with DNase I. Thus, another possibility for recombination may be an initial open region (the DNase I HS site/topo II cleavage site) where cleavage occurs more frequently and then promotes illegitimate recombination that is subject to selective pressures. Alternatively, other investigators have proposed that breakage and repair may be involved in MLLrecombination events; after initial breakage, exonuclease degradation and DNA repair is attempted, and this could result in the observed location of the breakpoints mapping at some distances from the site of initial breakage.72
Almost all DNase I HS sites are associated with the binding of a protein to a specific region of DNA.42,73,74 Upon protein/DNA binding, the surrounding chromatin becomes structurally altered, making the DNA more accessible, and this change can be detected using enzymes such as DNase I and S1 nuclease. We previously hypothesized that the MLL telomeric SAR is a protein-enriched region that may reflect SAR function.30 During mitosis, SARs have been shown to be condensation points along the chromosomal axis and appear to be responsible for the remodeling and maintenance of chromatin as well as sites for association with topo II and other nonhistone proteins.75 Thus, the DNase I HS in the telomeric MLL SAR may be a region to which a protein or a protein complex (either topo II or other proteins) binds, perhaps specifically. In the presence of topo II-targeting drugs, such as etoposide in the treatment of cancer patients, or possibly exposure to flavonoids or pesticides in utero as in the infant cases, topo II would lead to cleavage at the DNase I HS. Topo II cleavage occurring at the DNase I HS followed by repair or illegitimate recombination (translocation) could be one explanation for t-AML breakpoints mapping within the telomeric half of the MLL BCR. Elucidation of how DNA breaks occur in the MLL BCR in t-AML patients and in infant leukemia patients will provide critical insights into the mechanism(s) related to translocations, which is likely to be applicable to at least some translocations that occur de novo. Resolution of this question could lead to safer chemotherapeutic drugs as well as, potentially, to a reduced incidence of infant leukemia.
ACKNOWLEDGMENT
The authors thank Dr Ulrich Laemmli and Dr Craig Hart at the University of Geneva for their advice and help in initiating the DNase I studies in their laboratory. The authors also thank Alanna Harden for expert technical assistance.
Supported in part by the International Cancer Research Technology Transfer (ICRETT) Grant No. 246 (Geneva, Switzerland), as a travel grant to (P.L.S.), by the National Cancer Institute (CA 400046; J.D.R. and N.J.Z.-L.) and (CA 42557; J.D.R.), the Spastic Paralysis Foundation of the Illinois Eastern-Iowa District of Kiwanis International (J.D.R.) and by the ILL division ACS (95-42; N.J.Z.-L.). P.L.S. was a fellow supported by an Environmental Carcinogenesis Training Grant (5T32CA09273-19).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Pamela L. Strissel, PhD, 5841 S Maryland Ave, MC2115, Chicago, IL 60637.

![Fig. 2. Southern blots of BV173 DNA showing drug-induced cleavage of the MLL BCR. (A) The lanes from left to right show the marker (M), a negative control [(−) where no drugs were added to BV173 cells], VP16 cell treatment (100 μmol/L, 200 μmol/L), VM26 cell treatment (50 μmol/L, 100 μmol/L), and doxorubicin (Dox) cell treatment (10 μmol/L). An increasing amount of drug added is indicated by a triangle above the lanes. The PCR genomic probe (tel; see Fig 1) was hybridized to the Southern blot. The 8.3-kb germline DNA fragment and the new 1.5- to 1.6-kb drug-induced DNA fragment are indicated. (B) The lanes from left to right show DNase I–treated nuclei (0 to 20 U as indicated), a 16-hour drug treatment of BV173 cells, including a dimethyl sulfoxide control (C), VP16 (100 μm—two lanes), and N-Methylformamide (NMF; 0.5 mol/L). The PCR cDNA probe indicated above (0.74 kb) was hybridized toBamHI-digested DNA. The 8.3-kb germline and two new DNase I and drug-induced DNA fragments (1.5 kb and 6.9 to 7.0 kb) are indicated to the right. The DNase I, topo II, and DNA damaging agent cleavage sites either map within exon 9 or just telomeric of exon 9. Note that the new 6.9- to 7.0-kb DNA fragment is strongly hybridizing with exons 5, 6, and 7 and either all or half of exon 9. In contrast, the 1.5-kb DNA fragment is hybridizing more weakly with either none or half of exon 9, plus exon 10 and half of exon 11 (133 bp or 206 bp, respectively). The 1-kb marker (M) indicated to the left identifies a few of the molecular weight bands that cross-hybridize with this probe.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/10/10.1182_blood.v92.10.3793/4/m_blod42224002aw.jpeg?Expires=1767702227&Signature=NGpbs-6bk5CrfmbtMiYb692OoYR3w5Way1YkKjcs4rB5-BkRYxRmJ-E-AI9b9AFMpo4kk50XUQXx~1f1i-VvbSSZJktPfZvlPGFoW4ROh9U1NmguMB1N4sVvmFg0pTsk87wjBXcc9pvOYh3oVp94dGxN878BMzr8CRUIBk2Xrg5MQD1Doaui~Xc7zgPDmRVp2EDJvPTvP7tnBlZ09fEBJCU0MuEGSqa1xoKbrjxxB8gknEHrTbJaAMeQNQEa2uLRvrSREoe1PRTDdTEAvQoRY6GkPfMKecErKGUvTAv2t8SFermMtsE5Ob6Ep95-qIjxM4qlSy~E8qQHqOeyR8aLKQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Southern blots of BV173 DNA showing drug-induced cleavage of the MLL BCR. (A) The lanes from left to right show the marker (M), a negative control [(−) where no drugs were added to BV173 cells], VP16 cell treatment (100 μmol/L, 200 μmol/L), VM26 cell treatment (50 μmol/L, 100 μmol/L), and doxorubicin (Dox) cell treatment (10 μmol/L). An increasing amount of drug added is indicated by a triangle above the lanes. The PCR genomic probe (tel; see Fig 1) was hybridized to the Southern blot. The 8.3-kb germline DNA fragment and the new 1.5- to 1.6-kb drug-induced DNA fragment are indicated. (B) The lanes from left to right show DNase I–treated nuclei (0 to 20 U as indicated), a 16-hour drug treatment of BV173 cells, including a dimethyl sulfoxide control (C), VP16 (100 μm—two lanes), and N-Methylformamide (NMF; 0.5 mol/L). The PCR cDNA probe indicated above (0.74 kb) was hybridized toBamHI-digested DNA. The 8.3-kb germline and two new DNase I and drug-induced DNA fragments (1.5 kb and 6.9 to 7.0 kb) are indicated to the right. The DNase I, topo II, and DNA damaging agent cleavage sites either map within exon 9 or just telomeric of exon 9. Note that the new 6.9- to 7.0-kb DNA fragment is strongly hybridizing with exons 5, 6, and 7 and either all or half of exon 9. In contrast, the 1.5-kb DNA fragment is hybridizing more weakly with either none or half of exon 9, plus exon 10 and half of exon 11 (133 bp or 206 bp, respectively). The 1-kb marker (M) indicated to the left identifies a few of the molecular weight bands that cross-hybridize with this probe.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/10/10.1182_blood.v92.10.3793/4/m_blod42224002bw.jpeg?Expires=1767702227&Signature=AzISw~BawMdKPbSBbGk3x3xASHRf-VRNuUJdDUHfYBr~0u9PZorvZROPpmcngbx7z9kGrJdzO-CG4fr3G1zk6aZnCK9XkG5P8rVyzyxPrbzRe3f1NBv-RkvkJdtnHNN7TZDt4EYUe3TkRqWAOruXqJxNpUHWrfDJmqbEGv0sZgPMHdQ8VoJffksInYHafINqLwlawD~ZMq~8A0TX9O9TADi2xJ~uH-l77lQOAjr6NmM13RW1h29H~0Ho3fYzerucJyFDAnH~-FWsS3~qs4fySgK8QHYtmhQcOeNE~oYl4bhcuhnoaUpWdPkr84pDi8CsgGHzdTRR94PUCMNCH-A5uw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. BV173 Southern blot showing the MLL BCR DNase I HS site. A Southern blot representing BamHI/BglII-digested DNA from DNase I–treated (DNase I units directly above each lane) and BV173 whole nuclei was hybridized independently withMLL BCR DNA probes (indicated above each panel). λ PhageHindIII/EcoRI-digested marker (left of [A]) correlates with all three hybridizations. (A) Top germline 6.6-kbBgl II DNA fragment hybridizing with the 1.2-kb BglII/Sac I DNA probe is seen in all lanes. A new 5.0-kbBgl II/DNase I DNA fragment (arrow) is not observed in the absence of DNase I, but increases in intensity at higher DNase I concentrations. (B) A germline 2.2-kb DNA fragment is observed in all DNA lanes; thus, the centromeric portion of the MLL BCR is negative for DNase I HS sites. (C) The germline 6.0-kb Bgl II DNA fragment hybridizing with the 0.8-kb Nco I-BamHI DNA probe is seen in all of the DNA lanes. A new 1.4-kb Bgl II DNA fragment is not observed in the absence of DNase I, but increases in intensity at higher concentrations of DNase I (arrow).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/10/10.1182_blood.v92.10.3793/4/m_blod42224004w.jpeg?Expires=1767702228&Signature=kYlegZ2Y1DlL3Q1s~xezxyRtmm4OTL2TzyApf~gBF9VwSYYVTeFHXiLNfMaHdy4e0Bb6zeZuzAsMDpBjLAmgBuS2mwxa3A~95sQAJhZflQ4mz~xIx8yl3TKjFJk4jbdzIADtWPWbzo81dwWvLsNFQmYg2dqkLzftfUcL3Dz-5Va-XalaqX3ByIDuWho2fF9jNgXUFLephS8wCKJexTvAsICtul5ONnkhBuKAJUyMzZloD1RkJjUcTkdD7X47-F0OWFdqadVPdeo5bO4Tc0U1uval1e0Vm84aU1pbLf3EVcEg~P4lyzE7UWU54ha6t087NgUpn2UwtTNcry92TTWacQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal