Abstract
T lymphocytes are implicated in the pathogenesis of systemic vasculitis such as Wegener’s granulomatosis (WG) and polyarteritis nodosa (PAN). In the present study, we have characterized in detail the T-cell receptor (TCR) of peripheral blood T cells from eight vasculitis patients of known HLA class II genotypes. We used flow cytometry to outline the exact TCR V gene expression, complementarity determining region 3 (CDR3) fragment analysis to estimate the degree of clonality and cDNA sequencing to define the exact TCR or β chain sequences. The TCR CDR3 region interacts with antigenic peptides presented by HLA molecules, and it is normally immensely diverse. It was therefore of particular interest to identify a common dominating TCR BV8-F/L-G-G-A/Q-G-J2S3 β chain sequence in the CD4+T cells of four unrelated vasculitis patients. Furthermore, this BV8-associated CDR3 motif was linked to the HLA-DRB1*0401 allele, as well as to active disease and/or an established BV8+ CD4+ T-cell expansion. In contrast, age- and HLA-matched patients with rheumatoid arthritis did not harbor the described BV8 motif. These results strongly suggest that BV8+ CD4+ T cells with the described CDR3 motif recognize a specific antigen presented by DR4 molecules, indicating the existence of a common vasculitis-associated antigen.
WEGENER’S GRANULOMATOSIS (WG) and polyarteritis nodosa (PAN) are both systemic necrotizing vasculitis of small and medium vessels of unknown etiologies.1 T lymphocytes have been suggested to be important in the pathogenesis, eg, they are found infiltrating vascular lesions. The majority of T lymphocytes use the αβ T-cell receptor (TCR) to specifically recognize antigenic peptides in the context of major histocompatability complex (MHC) class I or class II molecules.2 Vasculitis patients often display a disturbed peripheral blood T-cell repertoire, with a high frequency of T-cell expansions, ie, large T-cell populations using a particular TCR V gene segment.3 4 In contrast to healthy individuals, such T-cell expansions are frequently found in the CD4+ subset, and phenotypic as well as functional studies indicate their activated status.4,4a.
In human diseases of unknown etiology and thought to involve T lymphocytes, detailed analysis of the TCR may indicate the nature of postulated T-cell antigens. We thus decided to closer characterize the TCR of T-cell populations from HLA typed patients with WG/PAN. Complementarity determining region 3 (CDR3) fragment analysis and sequencing were used for an exact determination of the α and/or β TCR chains. A strict correlation between the presence of a common, dominating TCR β chain CDR3-motif in several unrelated systemic vasculitis patients and a certain HLA-DR allele is described here. In addition, a remarkably high degree of clonality in all cases of T-cell expansions in these patients was noted. The described TCR motif was not found in age- and HLA-matched controls, and our findings therefore suggest the existence of a specific vasculitis-associated antigen.
MATERIALS AND METHODS
Subjects.
Eight patients (five men, three women, age 45 to 70 years) with necrotizing vasculitis treated at the Departments of Medicine at Karolinska Hospital, Södertälje Sjukhus or Västerviks Lasarett were included in the study. Five of these patients (patients 1 through 5) were chosen for this study because of their dramatic T-cell expansions.3 Patients 6 through 8 were selected because they had signs of BV8+ CD4+ T-cell expansions or because they were HLA-DR4 positive, and here only BV8+CD4+ T cells were analyzed. For details on clinical information, see Table 1.
Four patients with rheumatoid arthritis (RA) served as controls. They were three men and one woman, age 62 to 78 years. The disease duration was 5 to 20 years. They were all treated with nonsteroidal antiinflammatory drugs and one with methotrexate and one with gold. Three RA patients had active disease.
Cell separation.
Peripheral blood mononuclear cells (PBMC) were isolated from heparinized venous blood by Ficoll-Paque gradient centrifugation (Pharmacia, Uppsala, Sweden). For analysis of clonality, CD4+ and CD8+ T cells were separated by magnetic beads (Dynabeads, Oslo, Norway) according to the manufacturer’s description. Briefly, 107 PBMC were incubated end over end for 30 minutes at +4°C with the beads, removed with a magnetic separator, and washed three times. This procedure resulted in >95% enriched CD4+ or CD8+ cell populations.
Immunofluorescence and monoclonal antibodies (MoAb).
For the identification of T-cell expansions (as defined in Grunewald and Wigzell4), the following TCR V specific MoAb were included; BV2 from Immunotech S.A. (Marseille, France), AV12S1, BV3, and BV8 from T Cell Sciences Inc (Cambridge, MA). MoAb specific for CD3, CD4, and CD8 were purchased from Becton Dickinson (BD) (Mountain View, CA). Fluorescein isothiocyanate (FITC)-conjugated F(ab’)2 fragments of rabbit antimouse Ig were purchased from Dakopatts A/S (Glostrup, Denmark). Normal mouse serum (NMS), produced from BALB/c mice was used for negative control at a dilution of 1:500 in phosphate-buffered saline (PBS) containing 0.2% bovine serum albumin (BSA) and 0.01% sodium azide, in all cases staining < 0.5% of the cells. Lymphocyte subset analysis by flow cytometry was performed as described.3
RNA preparation and polymerase chain reaction (PCR) amplifications.
Enriched CD4+ or CD8+ cells were lysed and RNA was extracted using the RNA isolation kit (Stratagene, La Jolla, CA) or RNAzol (Cinna/Biotek Laboratories Inc, Houston, TX) according to the manufacturer’s recommendations. First strand cDNA was generated using random hexanucleotides (Pharmacia) with reverse transcriptase (GIBCO BRL Life Technologies Inc, Gaithersburg, MD).5Amplifications of cDNA were performed in 20, 50, or 100 μL final mixture volumes consisting of: 1x PCR buffer (10 mmol/L Tris-Cl pH 8.3, 50 mmol/L KCl, 0.1% gelatin), 0.2 mmol/L deoxynucleosidetriphosphate (dNTP), 1.5 mmol/L MgCl2, 0.5 μmol/L 5′V specific primers and 0.5 μmol/L 3′constant TCR α or β primer, and 2.5 U of Ampli Taq-DNA polymerase (Perkin-Elmer, Roche Molecular Systems Inc, Branchburg, NJ). TCR AV and -C specific primers were from Genevee et al6 and Oksenberg et al,7while TCR BV and -C specific primers were from Choi et al.8The PCR profile for the TCR α chain was as follows: denaturation at 94°C for 30 seconds, annealing at 61°C for 30 seconds, and extension at 72°C for 30 seconds for 35 cycles with a final extension of 5 minutes at 72°C.9 The PCR profile for the TCR β chain was as follows: denaturation at 94°C for 1 minute, annealing at 55°C for 1 minute, and extension at 72°C for 1 minute for 35 cycles with a final extension of 9 minutes at 72°C.5 An aliquot of each amplification reaction was loaded in ethidium bromide-stained 2% agarose gel to confirm the expected size of the amplified fragment.
CDR3 fragment analysis.
The size distribution of different TCR fragments was studied by using a CDR3 length analysis.10 Briefly, TCR cDNA was amplified in a PCR reaction with a 5′BV or AV specific primer in conjuction with a 3′constant α or β primer labelled with FITC. A fraction of the PCR-amplified product was subsequently denatured (100% formamide, 3 minutes at 90°C) and loaded on a 6.75 % acrylamide gel (Readymix; Pharmacia) and run for 400 minutes at 42°C on an automated DNA sequencer, ALF (Pharmacia). CD4 or CD8 separated PBMC from healthy individuals were run in parallel to show normal CDR3 fragment analysis patterns. The CDR3 length distribution was analyzed with ALF Fragment Manager software version 1.1 (Pharmacia).
Cloning and sequencing.
PCR-amplified TCR V gene products were cloned and sequenced according to standard procedures. Briefly, the PCR product was purified on a spin column (Qiagen, Düsseldorf, Gemany). After phosphorylation with T4 polynucleotide kinase (Pharmacia), the product was ligated to aSmaI cleaved pUC18 vector that was subsequently used to transform competent bacteria. Single colonies were used as templates for solid phase sequencing11 using an Autoread Sequencing Kit (Pharmacia) and analyzed with the ALF (Pharmacia). Alternatively, PCR was performed with a biotinylated C α or β primer, using the same conditions for PCR as described above, and the PCR products were used for “direct” solid phase sequencing with a nonlabelled AV or BV specific primer and Fluoro-dATP (Pharmacia) according to the recommendation of the manufacturer. We defined the CDR3 region and calculated CDR3 lengths using the formula by Rock et al,12ie, the number of amino acids between the conserved GXG triplet in the J region and the nearest preceding C in the V region, minus 4 amino acids.
HLA typing.
HLA genotyping was performed by PCR-based techniques. Polymorphic second exon of the DQA1 and DQB1 genes were amplified. The conditions for amplification and the probes used were as described.13The PCR-amplified products were manually dot blotted onto nylon membranes and 3′ end-labelling of the synthetic oligonucleotide probes with 32P, hybridization, and stringency washes were performed as described.13 HLA typing for DR was performed by using allele specific primer pairs (PCR-SSP technique)14and the subtyping for the DR4 was performed as described.15
Approval was obtained from the Institutional Review Board for these studies. Informed consent was provided according to the Declaration of Helsinki.
RESULTS
T-cell expansions.
Clinical information on the patients as well as data on the T-cell populations analyzed and their respective percentage at the time for sequence analysis is shown in Table 1. Three of the patients showed signs of disease activity at the time of the first analysis (patients 1, 4, and 5) and a fourth patient (patient 3) had active disease at the second analysis. Three T-cell expansions were also analyzed on a second occasion, where they in all cases still constituted major T-cell expansions (Table 1).
General Clinical Data, All of the Analyzed T-Cell Populations and HLA-Haplotypes of the Patients Studied
| Patient No. . | Sex/ Age . | Diagnosis . | ANCA* . | Second Analysis . | Disease Activity† . | Treatment . | Subset‡ . | Percent . | HLA-DR . | DR4 Subtype . | HLA-DQ . | DQA1*-DQB1* . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M/70 | PAN | 25 | Active | Steroids | AV12CD4 | 49 | 4, 4 | 0401-0404 | 7, 8 | 0301-0301/0301-0302 | |
| BV3CD8 | 33 | |||||||||||
| BV8CD4 | 4.3 | |||||||||||
| 2 | M/46 | WG | <10 | Remission (since 8 months) | Steroids + mtx | BV8CD8 BV8CD4 | 42 29 | 4, 4 | 0401-0413 | 7, 7 | 0301-0301/0301-0301 | |
| 3 | M/68 | WG | 18 | (1) Remission | (1) None | BV8CD4 | (1) 15 | 4, 13 | 0401 | 8, 6 | 0301-0302/0103-0603 | |
| 20 | +10 months | (2) Active | (2) Steroids + cph | (2) 29 | ||||||||
| 4 | F/45 | WG | 99 | (1) Active | (1) None | BV2CD4 | (1) 50 | 14, 13 | 5, 6 | 0101-0503/0103-0603 | ||
| 15 | +4 months | (2) Active | (2) Steroids + cph | (2) 60 | ||||||||
| 5 | M/62 | WG | 23 | (1) Active | (1) Steroids + mtx | AV12CD8 | (1) 41 | 4, 4 | 0403-0404 | 7, 7 | 0301-0301/0301-0301 | |
| 26 | +2 months | (2) Active | (2) Steroids + mtx | (2) 35 | ||||||||
| BV8CD4 | (1) 9.9 | |||||||||||
| (2) 5.4 | ||||||||||||
| 61-153 | F/58 | WG | <10 | Remission (2 years) | Steroids + tms | BV8CD4 | (14.7)1-155 8.1 | 1, 3 | 5, 2 | 0101-0501/0501-0201 | ||
| 71-153 | M/66 | WG | <10 | Remission (3 years) | Steroids | BV8CD4 | (17.8)1-15510.0 | 1, 4 | 0401 | 5, 8 | 0101-0501/0301-0302 | |
| 81-153 | F/69 | WG | 17 | Remission (5.5 years) | None | BV8CD4 | (3.0)1-155 5.0 | 4 | 0401 | 7, 7 | 0301-0301/0301-0301 |
| Patient No. . | Sex/ Age . | Diagnosis . | ANCA* . | Second Analysis . | Disease Activity† . | Treatment . | Subset‡ . | Percent . | HLA-DR . | DR4 Subtype . | HLA-DQ . | DQA1*-DQB1* . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M/70 | PAN | 25 | Active | Steroids | AV12CD4 | 49 | 4, 4 | 0401-0404 | 7, 8 | 0301-0301/0301-0302 | |
| BV3CD8 | 33 | |||||||||||
| BV8CD4 | 4.3 | |||||||||||
| 2 | M/46 | WG | <10 | Remission (since 8 months) | Steroids + mtx | BV8CD8 BV8CD4 | 42 29 | 4, 4 | 0401-0413 | 7, 7 | 0301-0301/0301-0301 | |
| 3 | M/68 | WG | 18 | (1) Remission | (1) None | BV8CD4 | (1) 15 | 4, 13 | 0401 | 8, 6 | 0301-0302/0103-0603 | |
| 20 | +10 months | (2) Active | (2) Steroids + cph | (2) 29 | ||||||||
| 4 | F/45 | WG | 99 | (1) Active | (1) None | BV2CD4 | (1) 50 | 14, 13 | 5, 6 | 0101-0503/0103-0603 | ||
| 15 | +4 months | (2) Active | (2) Steroids + cph | (2) 60 | ||||||||
| 5 | M/62 | WG | 23 | (1) Active | (1) Steroids + mtx | AV12CD8 | (1) 41 | 4, 4 | 0403-0404 | 7, 7 | 0301-0301/0301-0301 | |
| 26 | +2 months | (2) Active | (2) Steroids + mtx | (2) 35 | ||||||||
| BV8CD4 | (1) 9.9 | |||||||||||
| (2) 5.4 | ||||||||||||
| 61-153 | F/58 | WG | <10 | Remission (2 years) | Steroids + tms | BV8CD4 | (14.7)1-155 8.1 | 1, 3 | 5, 2 | 0101-0501/0501-0201 | ||
| 71-153 | M/66 | WG | <10 | Remission (3 years) | Steroids | BV8CD4 | (17.8)1-15510.0 | 1, 4 | 0401 | 5, 8 | 0101-0501/0301-0302 | |
| 81-153 | F/69 | WG | 17 | Remission (5.5 years) | None | BV8CD4 | (3.0)1-155 5.0 | 4 | 0401 | 7, 7 | 0301-0301/0301-0301 |
Abbreviations: mtx, methotrexate; cph, cyclophosphamide; tms, trimethoprim-sulphamethoxazole.
At the time for CDR3 fragment analysis and/or cDNA sequencing. Values show cANCA in all cases except for patient 1, where it shows pANCA.
First (1) and second (2) analysis when appropriate.
TCR BV8 was analyzed and found to be normally expressed in CD8+ T cells of patients 1, 3, 5, 6, 7, and 8.
Only BV8+ T cells analyzed.
BV8 expression in CD4+ subsets of patients 6, 7, and 8 at 2 years, 2.5 years, and 1 year, respectively, before sequence analysis.
CDR3 fragment analysis.
The CDR3 fragment length analysis can be used to disclose the size distribution of TCR products from T cells expressing a particular V gene segment and thus estimate the degree of clonality within a T-cell population. Polyclonality is usually seen in normal T-cell populations with, in most cases, 6 to 12 peaks distributed in a gaussian pattern, while dominant peaks showing T cells using a particular CDR3 length are found in oligoclonal or clonal T-cell populations.
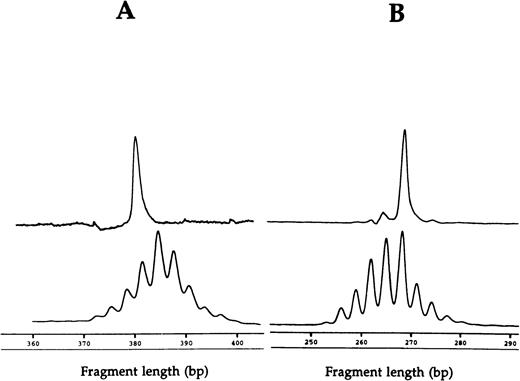
Fragment analyses of the two dramatic T-cell expansions of patient 1 (AV12 CD4 49% and BV3 CD8 33%) each showed a single dominant peak (Fig 1), suggesting that these cells were clonally derived. Similar results were found in the BV8 CD4 and BV8 CD8 T-cell expansions of patient 2 (data not shown). Patient 3 had a BV8 CD4 T-cell expansion that was analyzed on two occasions, 10 months apart (Table 1). BV8 expression increased during this time from 15% to 29%, and in both cases, identical dominant peaks were detected (Fig 2A). A high stability over 4- and 2-month periods, respectively, was also seen in the clones of patients 4 and 5 (Figs 2B and C).
CDR3 fragment analysis of (A) TCR AV 12S1 CD4 T-cell expansion (49%) with AV 12S1 CD4 control and of (B) BV3 CD8 expansion (33%) with BV3 CD8 control of patient 1.
CDR3 fragment analysis of (A) TCR AV 12S1 CD4 T-cell expansion (49%) with AV 12S1 CD4 control and of (B) BV3 CD8 expansion (33%) with BV3 CD8 control of patient 1.
CDR3 fragment analysis of T-cell expansions at separate time points, showing (A) BV8 CD4 T-cell expansions of patient three 10 months apart (15% and 29%) and (B) BV2 CD4 T-cell expansions of patient four 4 months apart (50% and 60%), and (C) AV12S1 CD8 T-cell expansion of patient five 2 months apart (41% and 35%). Relevant controls of healthy subjects are shown at the bottom.
CDR3 fragment analysis of T-cell expansions at separate time points, showing (A) BV8 CD4 T-cell expansions of patient three 10 months apart (15% and 29%) and (B) BV2 CD4 T-cell expansions of patient four 4 months apart (50% and 60%), and (C) AV12S1 CD8 T-cell expansion of patient five 2 months apart (41% and 35%). Relevant controls of healthy subjects are shown at the bottom.
TCR BV8 CD4 sequences.
Almost identical amino acid sequences, including the CDR3 regions as well as the BJ gene segments, were found in BV8+CD4+ T cells of patients 1, 2, 3, and 7 (Table 2). In patients 2, 3, and 7, these clones clearly dominated the BV8+ CD4+sequences, and moreover, the BV8+ T cells made up more or less dramatic peripheral blood CD4+ T-cell expansions either at the time of sequencing or before that (Table 1). In patient 1, who displayed two variants of the BV8-CDR3-motif, no data on BV8 expression before the sequence analysis was available. Additional BV8-sequences of the same dominant CDR3 length were seen in all of these patients, and in patient 3, the same dominating BV8 CD4 clone was identified on two separate occasions, 10 months apart (Table 1). Finally, the nucleotide sequences of the dominating BV8-CDR3-BJ2S3 clones all differed at several locations, ruling out any contamination (Table 3).
Sequences of TCR BV8+ CD4+T-Cell Populations
| Patient BV8 Expression . | V . | . | N ( - D - ) NJ-Segment . | BJ . | Frequency . | CDR3-Length . |
|---|---|---|---|---|---|---|
| Patient 1 | ||||||
| BV8/CD4 4.3% | C A S S - | L G G A G- D T Q Y F | BJ2S3 | 3/12 | 9 | |
| C A S S - | L G G S G- D T Q Y F | BJ2S3 | 1/12 | 9 | ||
| C A S S - | P G P G- N Q P Q H F | BJ1S5 | 1/12 | 9 | ||
| C A S S - | P T E- Y P E N Q Y F | BJ2S5 | 1/12 | 9 | ||
| C A S S - | R Q G F- Y G Y N F | BJ1S2 | 1/12 | 8 | ||
| C A S S - | S P G- P Y E Q Y F | BJ2S7 | 1/12 | 8 | ||
| C A - | T L R G G G- S Y N E Q F F | BJ2S1 | 1/12 | 10 | ||
| C A S - | L P A P S- A K N I Q Y F | BJ2S4 | 1/12 | 10 | ||
| C A S S - | S W D R A- S N Q P Q H F | BJ1S5 | 2/12 | 11 | ||
| Patient 2 | ||||||
| BV8/CD4 29% | C A S S - | F G G A G- D T Q Y F | BJ2S3 | 15/24 | 9 | |
| C A S R - | G L A G V- Y E Q Y F | BJ2S7 | 1/24 | 9 | ||
| C A S S - | P G G- Y Q P Q H F | BJ1S5 | 1/24 | 9 | ||
| C A S R - | F W T E R- N E Q Y F | BJ2S7 | 1/24 | 9 | ||
| C A S S - | V G Q A S- Y G Y T F | BJ1S2 | 1/24 | 9 | ||
| C A S S - | S A K G L V- E A F F | BJ1S1 | 1/24 | 9 | ||
| C A S S - | L D S- N T E A F | BJ1S1 | 1/24 | 7 | ||
| C A S T - | P Y G V R- G Y T F | BJ1S2 | 1/24 | 8 | ||
| C A S S - | L R P S- S T D T Q Y F | BJ2S3 | 1/24 | 10 | ||
| C A S S - | P T G D R I- D I Q Y F | BJ2S4 | 1/24 | 10 | ||
| Patient 3 | ||||||
| BV8/CD4 15%* | C A S S - | F G G Q G- D T Q Y F | BJ2S3 | 4/4 | 9 | |
| BV8/CD4 29%† | C A S S - | F G G Q G- D T Q Y F | BJ2S3 | 13/17 | 9 | |
| C A S S - | L G Q G P- D T Q Y F | BJ2S3 | 1/17 | 9 | ||
| C A S S - | S G Q A- H Q P Q H F | BJ1S5 | 1/17 | 9 | ||
| C A S S - | L A G Q- E Q F F | BJ2S1 | 1/17 | 8 | ||
| C A S S - | P L P G T S V D- S T D T Q Y F | BJ2S3 | 1/17 | 14 | ||
| Patient 5 | ||||||
| BV8/CD4 5.4% | C A S S - | P P Q R P D H- P Y E Q Y F | BJ2S7 | 4/15 | 12 | |
| C A S S - | T G T G- Q F F | BJ2S1 | 1/15 | 6 | ||
| C A S S - | F Q G G G R- I F | BJ2S1 | 1/15 | 7 | ||
| C A S S - | W L- D T E A F F | BJ1S1 | 1/15 | 7 | ||
| C A S R - | M A S- S Y E Q Y F | BJ2S7 | 1/15 | 8 | ||
| C A S S - | L T A G G Y G L H- F | BJ1S2 | 1/15 | 9 | ||
| C A S S - | R T S- G T D T Q Y F | BJ2S3 | 1/15 | 9 | ||
| C A S S - | R T G R G- D T Q Y F | BJ2S3 | 1/15 | 9 | ||
| C A S S - | P S G G A V- G E L F F | BJ2S2 | 1/15 | 10 | ||
| C A S S - | S T M T V- N Y G Y T F | BJ1S2 | 1/15 | 10 | ||
| C A S S - | R S G A G V- A K N I Q Y F | BJ2S4 | 1/15 | 12 | ||
| C A S S - | F R L S T G S L- D E Q F F | BJ2S1 | 1/15 | 12 | ||
| Patient 6 | ||||||
| BV8/CD4 8.1% | C A S S - | R G R- D T Q Y F | BJ2S3 | 1/9 | 7 | |
| C A S S - | L T E- I N T E A F F | BJ1S1 | 1/9 | 9 | ||
| C A S S - | L A G Q G P- Y E Q Y F | BJ2S7 | 2/9 | 10 | ||
| C A S S - | L S S G S- T D T Q Y F | BJ2S3 | 1/9 | 10 | ||
| C A S S - | A Q G P- G A N V L T F | BJ2S6 | 2/9 | 10 | ||
| C A S S - | P P D R N G- P E A F F | BJ1S1 | 1/9 | 10 | ||
| C A S S - | F R P A G G Y S- S T D T Q Y F | BJ2S3 | 1/9 | 14 | ||
| Patient 7 | ||||||
| BV8/CD4 10.0% | C A S S - | L G G A G- D T Q Y F | BJ2S3 | 9/11 | 9 | |
| C A S S - | L G Q G- Y N E Q F F | BJ2S1 | 1/11 | 9 | ||
| C A S S - | R D G- Y Y G Y T F | BJ1S2 | 1/11 | 8 | ||
| Patient 8 | ||||||
| BV8/CD4 5.0% | C A S S - | T- T G E L F F | BJ2S2 | 1/9 | 6 | |
| C A S S - | L R G- E T Q Y F | BJ2S5 | 1/9 | 7 | ||
| C A S S - | T T G- N Q P Q H F | BJ1S5 | 1/9 | 8 | ||
| C A S S - | F G G A- E N Q Y F | BJ2S5 | 1/9 | 8 | ||
| C A S S - | L A V D- S P L H F | BJ1S6 | 1/9 | 8 | ||
| C A S - | R S T G G E- E Q Y F | BJ2S7 | 1/9 | 8 | ||
| C A S S - | A Q G- T N E K L F F | BJ1S4 | 1/9 | 9 | ||
| C A S - | I G G S- S G G N V L T F | BJ2S6 | 1/9 | 10 | ||
| C A S S - | S C G E L- S Y N E Q F F | BJ2S1 | 1/9 | 11 |
| Patient BV8 Expression . | V . | . | N ( - D - ) NJ-Segment . | BJ . | Frequency . | CDR3-Length . |
|---|---|---|---|---|---|---|
| Patient 1 | ||||||
| BV8/CD4 4.3% | C A S S - | L G G A G- D T Q Y F | BJ2S3 | 3/12 | 9 | |
| C A S S - | L G G S G- D T Q Y F | BJ2S3 | 1/12 | 9 | ||
| C A S S - | P G P G- N Q P Q H F | BJ1S5 | 1/12 | 9 | ||
| C A S S - | P T E- Y P E N Q Y F | BJ2S5 | 1/12 | 9 | ||
| C A S S - | R Q G F- Y G Y N F | BJ1S2 | 1/12 | 8 | ||
| C A S S - | S P G- P Y E Q Y F | BJ2S7 | 1/12 | 8 | ||
| C A - | T L R G G G- S Y N E Q F F | BJ2S1 | 1/12 | 10 | ||
| C A S - | L P A P S- A K N I Q Y F | BJ2S4 | 1/12 | 10 | ||
| C A S S - | S W D R A- S N Q P Q H F | BJ1S5 | 2/12 | 11 | ||
| Patient 2 | ||||||
| BV8/CD4 29% | C A S S - | F G G A G- D T Q Y F | BJ2S3 | 15/24 | 9 | |
| C A S R - | G L A G V- Y E Q Y F | BJ2S7 | 1/24 | 9 | ||
| C A S S - | P G G- Y Q P Q H F | BJ1S5 | 1/24 | 9 | ||
| C A S R - | F W T E R- N E Q Y F | BJ2S7 | 1/24 | 9 | ||
| C A S S - | V G Q A S- Y G Y T F | BJ1S2 | 1/24 | 9 | ||
| C A S S - | S A K G L V- E A F F | BJ1S1 | 1/24 | 9 | ||
| C A S S - | L D S- N T E A F | BJ1S1 | 1/24 | 7 | ||
| C A S T - | P Y G V R- G Y T F | BJ1S2 | 1/24 | 8 | ||
| C A S S - | L R P S- S T D T Q Y F | BJ2S3 | 1/24 | 10 | ||
| C A S S - | P T G D R I- D I Q Y F | BJ2S4 | 1/24 | 10 | ||
| Patient 3 | ||||||
| BV8/CD4 15%* | C A S S - | F G G Q G- D T Q Y F | BJ2S3 | 4/4 | 9 | |
| BV8/CD4 29%† | C A S S - | F G G Q G- D T Q Y F | BJ2S3 | 13/17 | 9 | |
| C A S S - | L G Q G P- D T Q Y F | BJ2S3 | 1/17 | 9 | ||
| C A S S - | S G Q A- H Q P Q H F | BJ1S5 | 1/17 | 9 | ||
| C A S S - | L A G Q- E Q F F | BJ2S1 | 1/17 | 8 | ||
| C A S S - | P L P G T S V D- S T D T Q Y F | BJ2S3 | 1/17 | 14 | ||
| Patient 5 | ||||||
| BV8/CD4 5.4% | C A S S - | P P Q R P D H- P Y E Q Y F | BJ2S7 | 4/15 | 12 | |
| C A S S - | T G T G- Q F F | BJ2S1 | 1/15 | 6 | ||
| C A S S - | F Q G G G R- I F | BJ2S1 | 1/15 | 7 | ||
| C A S S - | W L- D T E A F F | BJ1S1 | 1/15 | 7 | ||
| C A S R - | M A S- S Y E Q Y F | BJ2S7 | 1/15 | 8 | ||
| C A S S - | L T A G G Y G L H- F | BJ1S2 | 1/15 | 9 | ||
| C A S S - | R T S- G T D T Q Y F | BJ2S3 | 1/15 | 9 | ||
| C A S S - | R T G R G- D T Q Y F | BJ2S3 | 1/15 | 9 | ||
| C A S S - | P S G G A V- G E L F F | BJ2S2 | 1/15 | 10 | ||
| C A S S - | S T M T V- N Y G Y T F | BJ1S2 | 1/15 | 10 | ||
| C A S S - | R S G A G V- A K N I Q Y F | BJ2S4 | 1/15 | 12 | ||
| C A S S - | F R L S T G S L- D E Q F F | BJ2S1 | 1/15 | 12 | ||
| Patient 6 | ||||||
| BV8/CD4 8.1% | C A S S - | R G R- D T Q Y F | BJ2S3 | 1/9 | 7 | |
| C A S S - | L T E- I N T E A F F | BJ1S1 | 1/9 | 9 | ||
| C A S S - | L A G Q G P- Y E Q Y F | BJ2S7 | 2/9 | 10 | ||
| C A S S - | L S S G S- T D T Q Y F | BJ2S3 | 1/9 | 10 | ||
| C A S S - | A Q G P- G A N V L T F | BJ2S6 | 2/9 | 10 | ||
| C A S S - | P P D R N G- P E A F F | BJ1S1 | 1/9 | 10 | ||
| C A S S - | F R P A G G Y S- S T D T Q Y F | BJ2S3 | 1/9 | 14 | ||
| Patient 7 | ||||||
| BV8/CD4 10.0% | C A S S - | L G G A G- D T Q Y F | BJ2S3 | 9/11 | 9 | |
| C A S S - | L G Q G- Y N E Q F F | BJ2S1 | 1/11 | 9 | ||
| C A S S - | R D G- Y Y G Y T F | BJ1S2 | 1/11 | 8 | ||
| Patient 8 | ||||||
| BV8/CD4 5.0% | C A S S - | T- T G E L F F | BJ2S2 | 1/9 | 6 | |
| C A S S - | L R G- E T Q Y F | BJ2S5 | 1/9 | 7 | ||
| C A S S - | T T G- N Q P Q H F | BJ1S5 | 1/9 | 8 | ||
| C A S S - | F G G A- E N Q Y F | BJ2S5 | 1/9 | 8 | ||
| C A S S - | L A V D- S P L H F | BJ1S6 | 1/9 | 8 | ||
| C A S - | R S T G G E- E Q Y F | BJ2S7 | 1/9 | 8 | ||
| C A S S - | A Q G- T N E K L F F | BJ1S4 | 1/9 | 9 | ||
| C A S - | I G G S- S G G N V L T F | BJ2S6 | 1/9 | 10 | ||
| C A S S - | S C G E L- S Y N E Q F F | BJ2S1 | 1/9 | 11 |
First occasion.
Second occasion.
Nucleotide and Amino Acid Sequences of BV8 CD4-Associated CDR3 Region Motifs From Four Patients, All Being HLA-DRB1*0401 Allele Positive
| Patient No. . | BV8 . | N-D-N . | BJ2S3-Segment . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | C | A | S | S | - | L | G | G | A | G | - | D | T | Q | Y | F |
| TGT | GCC | AGC | AGT | TTA | GGG | GGA | GCG | GGT | GAT | ACG | CAG | TAT | TTT | |||
| 2 | C | A | S | S | - | F | G | G | A | G | - | D | T | Q | Y | F |
| TGT | GCC | AGC | AGT | - | TTC | GGG | GGG | GCC | GGA | GAT | ACG | CAG | TAT | TTT | ||
| 3 | C | A | S | S | - | F | G | G | Q | G | - | D | T | Q | Y | F |
| TGT | GCC | AGC | AGT | - | TTC | GGT | GGA | CAG | GGA | GAT | ACG | CAG | TAT | TTT | ||
| 7 | C | A | S | S | - | L | G | G | A | G | - | D | T | Q | Y | F |
| TGT | GCC | AGC | AGT | TTA | GGC | GGG | GCC | GGT | - | GAT | ACG | CAG | TAT | TTT | ||
| Patient No. . | BV8 . | N-D-N . | BJ2S3-Segment . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | C | A | S | S | - | L | G | G | A | G | - | D | T | Q | Y | F |
| TGT | GCC | AGC | AGT | TTA | GGG | GGA | GCG | GGT | GAT | ACG | CAG | TAT | TTT | |||
| 2 | C | A | S | S | - | F | G | G | A | G | - | D | T | Q | Y | F |
| TGT | GCC | AGC | AGT | - | TTC | GGG | GGG | GCC | GGA | GAT | ACG | CAG | TAT | TTT | ||
| 3 | C | A | S | S | - | F | G | G | Q | G | - | D | T | Q | Y | F |
| TGT | GCC | AGC | AGT | - | TTC | GGT | GGA | CAG | GGA | GAT | ACG | CAG | TAT | TTT | ||
| 7 | C | A | S | S | - | L | G | G | A | G | - | D | T | Q | Y | F |
| TGT | GCC | AGC | AGT | TTA | GGC | GGG | GCC | GGT | - | GAT | ACG | CAG | TAT | TTT | ||
Amino acid sequences are shown in bold.
Patients 5, 6, and 8 did not harbor the same sequence at all (Table 2). These patients had a normal BV8 expression in CD4+ T cells at the time of sequence analysis, although patients 5 and 6 had signs of increased BV8 expression in the CD4+ T-cell subset before that (Table 1). In patient 5, a different BV8-BJ2S7 clone, making up 4 of 15 sequences, was detected.
All control individuals (1 through 4) had a normal TCR BV8 expression in CD4+ peripheral blood cells (5.4%, 4.7 %, 2.8%, and 5.1%, respectively). Altogether 32 clones from these BV8+CD4+ T cells of the four controls were sequenced. Only 3 of 32 sequences included the BJ2S3 gene segment, but in no case did we identify the described sequence motif (data not shown).
TCR AV and BV sequences of T-cell expansions other than BV8.
Sequence analyses of the remaining T-cell expansions, where CDR3 fragment analyses had indicated a high degree of clonality, were in all cases in concordance with the fragment analyses (Table 4). The two AV12-associated α chain sequences shared some CDR3 similarities, although they used different AJ segments and were CD4+ and CD8+, respectively. In addition, the two major clonal BV T-cell expansions in the CD8 subset expressing BV3 (patient 1) or BV8 (patient 2) both used CDR3 regions that were 10 amino acids long, and both had a positively charged amino acid (arginin) at the beginning of the junctional region (third and second position in the CDR3 region, respectively) and a negatively charged amino acid (glutamic acid or aspartic acid) 5 amino acids downstream (counting from the arginin). Thus, there were additional similarities in CDR3 regions of T-cell expansions expressing other TCR AV or BV gene segments from unrelated patients.
Amino Acid Sequences Obtained From T-Cell Expansions
| Patient No./Expansion . | Percent . | BVN ( - D - ) N . | J-Segment . | BJ . | Clonality . | CDR3-Length . |
|---|---|---|---|---|---|---|
| 1/BV3 CD8 | 33 | C A S S - S T R T V G | - Y E Q Y F | BJ2S7 | Direct4-150 | 10 |
| 2/BV8 CD8 | 42 | C A S S - L R P S | - S T D T Q Y F | BJ2S3 | 7/7 | 10 |
| 4/BV2 CD4 | 50-60 | I C S A - Q D K G S | - E T Q Y F | BJ2S5 | Direct4-150 | 8 |
| 1/AV12 CD4 | 49 | Y F C A - L S S | - T N S G Y A L N F | AJ41 | Direct4-150 | 9 |
| 5/AV12 CD8 | 41-35 | Y F C A - L S E R | - D T D K L I F | AJ34 | Direct4-150 | 8 |
| Patient No./Expansion . | Percent . | BVN ( - D - ) N . | J-Segment . | BJ . | Clonality . | CDR3-Length . |
|---|---|---|---|---|---|---|
| 1/BV3 CD8 | 33 | C A S S - S T R T V G | - Y E Q Y F | BJ2S7 | Direct4-150 | 10 |
| 2/BV8 CD8 | 42 | C A S S - L R P S | - S T D T Q Y F | BJ2S3 | 7/7 | 10 |
| 4/BV2 CD4 | 50-60 | I C S A - Q D K G S | - E T Q Y F | BJ2S5 | Direct4-150 | 8 |
| 1/AV12 CD4 | 49 | Y F C A - L S S | - T N S G Y A L N F | AJ41 | Direct4-150 | 9 |
| 5/AV12 CD8 | 41-35 | Y F C A - L S E R | - D T D K L I F | AJ34 | Direct4-150 | 8 |
Show results of direct sequencing.
HLA-DR and –DQ and associations with TCR expression.
All patients were HLA typed for HLA-DR and -DQ (Table 1). Patients 1, 2, 3, 5, 7, and 8 were HLA-DR4 positive, and all except patient 5 had the DRB1*0401 allele. Among the DRB1*0401 allele positive patients (1, 2, 3, 7, and 8), all except patient 8 had the above-described BV8-CDR3-BJ2S3 sequence in their peripheral blood CD4+ T cells (Table 5). Notably, patient 8, who did not have the BV8-CDR3-BJ2S3 clone, was in clinical remission since 5.5 years and had no detectable BV8+ CD4+expansion. In patient 5, who was HLA-DRB1*0403-0404 positive (Table 1), we detected a repeated clone using a different TCR BV8-linked β chain (Table 2). Patient 6 had a BV8+ CD4+ T-cell expansion, but was DRB1*0401 negative and did not have the BV8-motif (Table 5).
Summary of BV8-CDR3-BJ2S3 Sequences and Their Associations With HLA-DRB15-1500401 Alleles and BV8 CD4 T-Cell Expansions
| Patient No. . | BV8-CDR3-J2S3 Motif5-150 . | DRB15-1500401 Allele . | BV8+ CD4 Expansion5-151 . |
|---|---|---|---|
| 1 | Yes (3/12)5-152 | Yes | No5-153 |
| 2 | Yes (15/24) | Yes | Yes |
| 3 | Yes (4/4-13/17) | Yes | Yes |
| 4 | ND | No | No |
| 5 | No (0/15) | No | No |
| 6 | No (0/11) | No | Yes |
| 7 | Yes (9/11) | Yes | Yes |
| 8 | No (0/9) | Yes | No |
| Patient No. . | BV8-CDR3-J2S3 Motif5-150 . | DRB15-1500401 Allele . | BV8+ CD4 Expansion5-151 . |
|---|---|---|---|
| 1 | Yes (3/12)5-152 | Yes | No5-153 |
| 2 | Yes (15/24) | Yes | Yes |
| 3 | Yes (4/4-13/17) | Yes | Yes |
| 4 | ND | No | No |
| 5 | No (0/15) | No | No |
| 6 | No (0/11) | No | Yes |
| 7 | Yes (9/11) | Yes | Yes |
| 8 | No (0/9) | Yes | No |
Abbreviation: nd, not determined.
Presence and frequency of the BV8-F/L-G-G-A/Q-G-J2S3 motif.
BV8 expansion in CD4+ T cells as defined in reference 4 (ie >14.6%) at the time of or before sequencing analysis.
One additional similar sequence.
No data on BV8 CD4 before the sequence analysis was available.
All four control individuals had the DRB1*0401 allele, but lacked completely the described TCR BV8 motif.
DISCUSSION
We have in this study closely characterized the TCR on T-cell peripheral blood subpopulations of HLA-typed patients with necrotizing vasculitis, who are known to have a disturbed T-cell repertoire with a high frequency of T-cell expansions.3 A large number of such T-cell expansions, ie, big populations of T cells using a particular TCR V gene segment, are frequently detected in the CD4+ T-cell subset, an extremely rare finding in healthy individuals.4 Here we describe a common TCR BV8-F/L-G-G-A/Q-G-J2S3 sequence in the CD4+ subsets of four unrelated vasculitis patients, all being HLA-DRB1*0401 allele positive, and moreover corresponding to active disease and/or BV8+ CD4+ T-cell expansions. No such T cells were found in age- and HLA-matched control subjects suffering from another inflammatory disease, RA. These findings strongly suggest that patients with systemic vasculitis have been exposed to and elicited a cellular immune response against a common antigen.
Recent findings of Th1 skewed, activated CD4+ peripheral blood T cells in WG patients,16 as well as previous reports of activated CD4+ T cells in lesions,17,18correlations between soluble interleukin-2 receptor (sIL-2R) and disease activity,17,19,20 and the granuloma formation in the lesions suggest a role for T cells in the pathogenesis of granulomatous vasculitis syndromes. A central role for γ-interferon (IFN) producing T cells, infiltrating the vascular wall in giant cell arteritis (GCA), has been discussed.21 The T lymphocytes may help in the production of the typically WG associated antineutrophil cytoplasmic antibodies (ANCA).22 23
Analyses of the TCR may help in understanding the nature of a presumed antigen responsible for eliciting T-cell responses, subsequently creating T-cell expansions and may open the possibility of a highly selective immunotherapy by targeting disease-mediating T cells.24-26 T cells using a restricted TCR can expand after interaction with certain well characterized antigens, as shown not only in animal models,27 but also in humans. Thus, in the recognition of certain Epstein-Barr virus (EBV) epitopes,28mycobacterial heat shock proteins,29 influenza A virus,30 or house dust mite allergens,31 human T cells were shown to use a highly restricted TCR. Superantigens are known to be able to selectively stimulate polyclonal T cells expressing the appropriate TCR BV segment, regardless of the composition of the rest of the TCR.32 The present finding of a high degree of clonality in all of the investigated T-cell expansions argues strongly against a superantigen as the cause of the expansions in these patients.
What specific antigen, able to elicit strong T-cell responses, could be associated with systemic vasculitides? An infectious cause for vasculitis was implicated by findings in animal models where it has been shown that certain herpes viruses, ie, α-, β-, or γ-herpes viruses, can cause chronic vascular inflammatory conditions.33-35 In humans, vasculitic complications can emerge after chronic viral infections such as hepatitis C infection.36 Interestingly, infections with the human γ-herpes virus EBV has been shown to create dramatic T-cell expansions in humans.37 Disease-associated antigens could be derived from an infectious agent itself, but the infection could also lead to the release of sequestered self-antigens, which may trigger an immune response.38 In line with this are reports of T cells of WG patients being autoreactive and specific for proteinase 3 (PR3), the most common target antigen for the WG-associated autoantibodies (ANCA).39,40 Cross-reactivity between a microbial antigen and PR3 as a possible pathogenetic mechanism in WG was also suggested previously.22
We detected a BV8-BJ2S3 clone with a common amino acid CDR3 motif in four of five unrelated DRB1*0401 allele positive patients, while in all patients lacking the HLA-DRB1*0401 allele, this specific BV8-CDR3 motif was absent. Interestingly, the only DRB1*0401 allele positive patient without this specific clone had inactive disease since 5.5 years, and moreover no signs of any BV8 CD4 T-cell expansion as analyzed 1 year before the sequencing analysis. The four patients with the same T-cell clone in their peripheral blood not only had the identical HLA-DRB1*0401 allele in common, but also had signs of either a BV8+ CD4+ T-cell expansion and/or active disease. Importantly, a BV8+ CD4+ T-cell expansion per se did not generate the specific BV8-motif, as seen in the HLA-DRB1*0401 allele negative patient 6 (and also indicated in patient 5). The randomly generated CDR3 region is under normal circumstances immensely diverse and considered to interact with antigenic peptides presented by HLA molecules.2 41-43 The perfect positive correlation between a dominating TCR β chain sequence and a certain HLA-DR molecule therefore strongly suggest that CD4 positive T cells of these four unrelated patients have encountered the same antigen, presented by the HLA-DR4 molecule and triggering BV8 CD4 positive T cells using the same CDR3 motif. The only differences in amino acids in the BV8-associated CDR3 regions would in this respect most likely not confer any significantly altered capabilities to recognize a presumed antigen. In addition, the BV8-CDR3-BJ2S3 clones in the four patients were coded for by different nucleotide sequences. This finding rules out any contamination and more importantly, strongly supports the suggestion that these cells have been selected for by a specific (and shared) antigen.
A number of clones with identical CDR3 lengths, but using slightly different amino acid sequences compared with the dominating clone, were frequently detected in the T-cell expansions. This phenomenon can be seen after antigen stimulation, where closely related T-cell clones are selected in response to an antigen, as described in mice44and also suggested in vivo in human peripheral blood T cells subsequent to vaccination for hepatitis B.45 Moreover, although two of the major T-cell expansions in the CD8 subset in these patients used different BV genes, they had identical CDR3 lengths and positively and negatively charged amino acids positioned similarly. Because charged amino acids in the CDR3 region have been proven experimentally to be important for antigen recognition,41 these results altogether add to the suggestion that necrotizing vasculitis patients have been exposed to a common antigen.
There is no association of the DRB1*0401 allele and vasculitis, but it has been associated with a more systemic form of RA.46 The DRB1*0401 allele may be associated with presentation of highly selected sets of antigenic peptides, as has been described for the HLA-DR3 molecule.47 Such properties could influence the formation of the TCR repertoire during selection, or could increase the risk for presentation of certain autoantigens.
We previously described the occurrence of dramatic T-cell expansions in patients with another necrotizing vasculitis, GCA.48Interestingly, there was a preference for BV8 T-cell expansions also in that group of patients, and in one case where we were able to characterize the TCR β chain BJ usage, the BV8 CD4 positive T-cell expansion used the BJ2S3 gene segment to an unusually high degree. Moreover, that particular patient was HLA-DR4 positive (the DR4 subtype was not defined). It is therefore possible that the BV8-CDR3-J2S3 T-cell clone may exist in related systemic vasculitides, linked to HLA-DR4 (DRB1*0401 allele) and disease activity/BV8 CD4 T-cell expansions. RA patients, in contrast, did not harbor T cells with this particular TCR motif, and one may accordingly speculate on the existence of a common vasculitis-associated antigen.
Healthy individuals may accommodate (CD8 positive) T-cell expansions,49-52 while in certain patient groups, T-cell expansions seem more frequent and of a higher magnitude.4In particular, patients suffering from systemic vasculitides frequently harbor T-cell expansions, as described in patients with necrotizing vasculitis,3 giant cell arteritis/temporal arteritis,48 Takayasu’s disease,53 Kawasakis disease,54 and microscopic polyarteritis.55 The common occurrence of expansions in the CD4 positive T-cell subset clearly delineates the expansions of systemic vasculitis patients from those found in healthy individuals. Although there was no clear association between disease activity and the T-cell expansions, in one patient (patient 3), the increase of BV8 CD4 cells in time correlated with a relapse of the disease. Clearly, it would be necessary to study a much larger group of patients to find any association of T-cell expansions with disease activity. However, the BV8 motif described here may become useful as a diagnostic marker, or a marker of disease activity. Finally, studies on the specificity of the BV8+ CD4+ T cells might lead to the identification of a systemic vasculitis-associated antigen, which in turn could be used for the development of new immunotherapies.
ACKNOWLEDGMENT
The authors wish to thank Louise Berg for her help with the control subjects. Professor Hans Wigzell, Dr Olle Olerup and Dr Elisabeth Svennungsson are acknowledged for their assistance in this work.
Supported by grants from AFA (Labour Market Insurance Company).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Johan Grunewald, MD, PhD, Microbiology and Tumorbiology Center (MTC), Karolinska Institute, 171 77 Stockholm, Sweden; e-mail: Johan.Grunewald@mtc.ki.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal