Abstract
An increasing frequency of malignant lymphomas occurs among patients infected by human immunodeficiency virus. Because of the close similarities to human malignancies, we used a nonhuman primate model to study the pathogenesis of simian immunodeficiency virus (SIV)–associated malignancies. Specifically, we investigated (1) the presence of the SIV genome in tumor cells, (2) the presence of coinfecting viruses, and (3) the presence of a rearrangement of the immunoglobulin and c-myc genes. We observed 5 cases of non-Hodgkin's lymphomas (4 of B- and 1 of T-cell origin) among 14 SIV-infected cynomolgus monkeys. No c-myc translocation was observed in the tumors, whereas B-cell lymphomas were characterized either by a monoclonal (in 2 of 4) or by an oligoclonal (in 2 of 4) VDJ rearrangements of the immunoglobulin heavy chain gene. Molecular, biological, and immunological analyses did show the presence of infectious SIV in the tumor cells of 1 T-cell and 2 oligoclonal B-cell lymphomas. Neither Simian T-lymphotropic nor Epstein-Barr viruses were detectable, whereas Simian herpes virus Macaca fascicularis-1 was detectable at a very low copy number in 3 of 4 B-cell lymphomas; however, only 1 of these also harbored the SIV genome. These results support the possibility that SIV may be directly involved in the process of B or T lymphomagenesis occurring in simian acquired immunodeficiency syndrome.
THE INCIDENCE of non-Hodgkin's lymphomas (NHLs) in patients infected with the human immunodeficiency virus (HIV) has increased since the first report describing a high frequency of high-grade B-cell lymphomas in HIV-1–infected homosexual men1,2; therefore, the appearance of B-cell lymphomas was included among the diagnostic criteria of the acquired immunodeficiency syndrome (AIDS).3 It is expected that the frequency of lymphomas will further increase among HIV-positive patients as they live longer following antiretroviral therapy.4,5Immunohistological studies have proved that most of these NHLs are high-grade B-cell lymphomas characterized by a heterogeneous morphology and by an unusual distribution in extranodal sites. Epidemiological cofactors have not yet been identified, and the pathogenesis of the AIDS-related lymphomas is not yet completely understood, although the role of Epstein-Barr virus (EBV) as well as of chromosomal translocations involving the c-myc locus has been reported in numerous studies.6
Nonhuman primates experimentally infected by the simian immunodeficiency virus (SIV) have been widely used to study the pathogenesis of AIDS. SIV infection induces in macaques a severe immunosuppression that mimics the course of HIV infection in humans.7 Like in human HIV infection, an increased frequency of B-cell lymphomas has been observed by Feichtinger et al in immunosuppressed monkeys infected with SIV.8 Because of their clinical, morphological, and immunological characteristics, these malignancies closely resemble the EBV-associated lymphomas in HIV-infected patients. EBV-like herpesviruses have been isolated from several nonhuman primate species but, with the exception of few cases of lymphomas in a Macaca mulatta9 and in a baboon,10 they were not usually associated with any known disease of monkeys. However, experimental infection of cottontop tamarins with EBV gives rise to B-cell lymphomas containing multiple EBV genomes, histologically similar to the human posttransplant lymphomas.11 An EBV-like B-lymphotropic Simian herpesvirus termed herpesvirus Macaca fascicularis-1 (HVMF-1) showing variable homology with several regions of the EBV genome has been recently identified12 and found to be associated with lymphomas in SIV-infected monkeys.13 Nevertheless, like HIV in human B-cell lymphomas, the SIV genome has never been found in tumor cells from these monkey lymphomas.
During the course of studies in Macaca fascicularisexperimentally infected with SIV, we observed 5 cases of high-grade non-Hodgkin's lymphoma. Four of them were large B-cell lymphomas of the centroblastic type, and 1 was a peripheral, pleomorfic, small-medium–sized T-cell lymphoma. In the attempt to understand the lymphomagenic processes in the SIV/Macaca model, we have analyzed the tumor for the presence of SIV and for coinfecting viruses such as Simian T-lymphotropic virus type 1 (STLV-1), EBV, HVMF-1; for the clonality of the VDJ rearrangement of the immunoglobulin heavy chain (IgH) gene and for the chromosomal translocation of thec-myc locus.
MATERIALS AND METHODS
Animals.
Adult cynomolgus monkeys (Macaca fascicularis), originated from the breeding colony of the Istituto Superiore di Sanità, were housed individually in stainless steel cages according to the European guidelines (EEC, Directive No. 86-609, November 26, 1986) at a constant room temperature (24 ± 2°C) and humidity level (60 ± 5%) on a 12-hour-light/12-hour-dark cycle. At regular intervals of time and after mild sedation of the monkeys (Ketamine HCl, 10 mg/kg; Parke-Davis, Milan, Italy), weight and rectal temperature were recorded, clinical examinations performed, and blood collected from the femoral vein. All experimental procedures were done according to the institutional guidelines “Care and use of laboratory animals” (D.L. publication No. 116, 27, January 1992, Italian Ministry of Health). Animals whose conditions of life were not acceptable were killed by intracardiac injection of Tanax (0.5 mg/kg; Hoechst, Frankfurt, Germany).
Viruses, virus isolation, and titration.
The 5 monkeys (no. 4, 8503, OD5, 208, and S1) that developed a lymphoproliferative disease were part of a study14 in which 14 Cynomolgus monkeys of either sex were inoculated intravenously with 10 to 20 MID50 of either SIVmac251/32H15 grown in human T cells (provided by M. Cranage and P. Greenway, PHLS Center for Applied Microbiology and Research, Porton Down, UK, through the Program EVA of the EC program on AIDS research directed by H. Holmes) or SIVmac2517 grown in monkey peripheral blood mononuclear cells (PBMC; provided by A.M. Aubertine, Institut National de la Santè et de la Recherche Medicale, Strasbourg, France). Two monkeys (no. 4 and 8503) had a history of immunization with a whole formalin–inactivated SIV. The other animals (no. OD5, 208, and S1) were naive monkeys inoculated with the same strains of SIV. In the present study, both the vaccinated as well the naive-infected monkeys are referred to as infected monkeys.
For virus isolation, Ficoll-Paque purified (Pharmacia, Uppsala, Sweden) PBMC (4 × 106) were cocultured with human CEMX174 cells (1 × 106)16 (obtained through the AIDS Research and reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health; courtesy of Dr Peter Cresswell) for 30 to 40 days in RPMI 1640 containing 10% fetal calf serum (FCS) and antibiotics. Cultures were fed twice a week and scored for syncytia formation; supernatants were tested for the presence of p27 by an antigen capture assay (SIV p27 Core Antigen; Coulter, Hialeah, FL) and for reverse transcriptase activity as described previously.17
Tumor tissues and the spleen of each monkey were mechanically minced with a dounce, resuspended in complete medium, clarified by centrifugation (6,000 rpm at 4°C for 20 minutes) and stored at −152°C. Cell-free supernatants were used to determine the virus titer on C8166 human T cells that form syncytia upon infection with SIV.
Detection of anti-SIV antibodies.
Antibody titers to SIV proteins were determined by end-point plasma dilutions using an HIV-2 enzyme-linked immunosorbent assay (ELISA) (Elavia II, Diagnostic Pasteur, Paris, France) because the coated antigens, derived from HIV-2 infected cells, are recognized by anti-SIV antibodies.
Lymphocyte subsets determination.
Lymphocyte subsets were evaluated by direct immunofluorescence using R-phycoerithryn or fluorescein-labeled monoclonal antibodies (MoAb) directed against human CD2, CD20, CD4, and CD8 cell surface markers (DAKO, Glostrup, Denmark). Citrated whole blood (100 μL) was incubated for 30 minutes at 4°C with the MoAb (10 μL each) and then lysed with lysis buffer (Becton Dickinson, Palo Alto, CA). After being washed twice with phosphate-buffered saline (PBS) containing 2.5% FCS, cells were resuspended and fixed in PBS pH 7.4 containing paraformaldehyde 1% (wt/vol). Ten thousand lymphocytes were gated from leukocytes based on forward and 90° light scatter and analyzed for each sample by using a FACScan cytometer (Becton Dickinson).
Histology and immunohistochemistry.
Autopsies were performed immediately after spontaneous death or sacrifice, and tissue samples were fixed in 10% buffered neutral formalin, embedded in paraffin, sectioned at 5 μ, and stained with hematoxylin-eosin and Giemsa; additional paraffin sections were processed for in situ hybridization analysis. Care was taken to ensure that samples of lymphoma tissues did not contain significant amounts of soft tissues or other structures. Other tissue aliquots were frozen in liquid nitrogen and stored at −80°C. Cryostat sections were immunohistochemically stained using mouse monoclonal primary antibodies, anti-mouse secondary antibody conjugated with biotin, and a final avidin-peroxidase color development stage (PK 4002; Vector Laboratories, Burlingame, CA) as described elsewhere.18Positive and negative controls and isotype-matched antibodies were also included in each experiment. The following antibodies were used to establish the phenotype of the lymphomas according to the manufacturer's specifications: CD20-L26, CD3-UCHT1, CD4-MT310, CD8-DK25, CD68-PGM1 (Dakopatt, Glostrup, Denmark).
Clinical and Immunological Status of Lymphoma-Bearing Monkeys (4, 8503, OD5, 208, S1) Compared With That of Monkeys Which Did Not Develop Lymphomas (C1, 8302, H8, OD3, 203)
| Monkey Code . | Treatment . | SIV Strain* . | Ab Anti-SIV-151 . | VC-152 . | PCR-153 . | Time to Death After Infection (wk) . | SIV Disease . | Immune Status-155 (before death) . | Lymphoma . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD20+ . | CD2+ . | CD4+ . | CD8+ . | |||||||||
| 4 | Vaccinated¶ | SIVmac251 | 4.4 | + | + | 81 | Chronic | 340 | 1,500 | 210 | 1,710 | + |
| 8503 | Vaccinated | SIVmac251/32H | 4.4 | + | + | 139 | Chronic | 500 | 780 | 80 | 790 | + |
| OD5# | Naive infected | SIVmac251 | 4.7 | + | + | 124 | Chronic | 670 | 9,450 | 200 | 10,040 | + |
| 208 | Naive infected | SIVmac251/32H | 5.0 | + | + | 94 | Chronic | 380 | 1,590 | 50 | 440 | + |
| S1 | Naive infected | SIVmac251/32H | <2.0 | + | + | 12 | Acute | 248 | 1,292 | 795 | 745 | + |
| C1 | Vaccinated | SIVmac251/32H | 5.0 | + | + | 106 | Chronic | 380 | 1,140 | 80 | 1,220 | − |
| 8302 | Vaccinated | SIVmac251/32H | 5.0 | + | + | 125 | Chronic | 190 | 650 | 200 | 480 | − |
| OD3 | Naive infected | SIVmac251 | 5.3 | + | + | 71 | Chronic | 540 | 1,560 | 390 | 1,510 | − |
| 203 | Naive infected | SIVmac251/32H | 5.0 | + | + | 66 | Chronic | 680 | 1,020 | 20 | 1,330 | − |
| H8 | Naive infected | SIVmac251/32H | <2.0 | + | + | 16 | Acute | 849 | 3,184 | 1,592 | 1,432 | − |
| Monkey Code . | Treatment . | SIV Strain* . | Ab Anti-SIV-151 . | VC-152 . | PCR-153 . | Time to Death After Infection (wk) . | SIV Disease . | Immune Status-155 (before death) . | Lymphoma . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD20+ . | CD2+ . | CD4+ . | CD8+ . | |||||||||
| 4 | Vaccinated¶ | SIVmac251 | 4.4 | + | + | 81 | Chronic | 340 | 1,500 | 210 | 1,710 | + |
| 8503 | Vaccinated | SIVmac251/32H | 4.4 | + | + | 139 | Chronic | 500 | 780 | 80 | 790 | + |
| OD5# | Naive infected | SIVmac251 | 4.7 | + | + | 124 | Chronic | 670 | 9,450 | 200 | 10,040 | + |
| 208 | Naive infected | SIVmac251/32H | 5.0 | + | + | 94 | Chronic | 380 | 1,590 | 50 | 440 | + |
| S1 | Naive infected | SIVmac251/32H | <2.0 | + | + | 12 | Acute | 248 | 1,292 | 795 | 745 | + |
| C1 | Vaccinated | SIVmac251/32H | 5.0 | + | + | 106 | Chronic | 380 | 1,140 | 80 | 1,220 | − |
| 8302 | Vaccinated | SIVmac251/32H | 5.0 | + | + | 125 | Chronic | 190 | 650 | 200 | 480 | − |
| OD3 | Naive infected | SIVmac251 | 5.3 | + | + | 71 | Chronic | 540 | 1,560 | 390 | 1,510 | − |
| 203 | Naive infected | SIVmac251/32H | 5.0 | + | + | 66 | Chronic | 680 | 1,020 | 20 | 1,330 | − |
| H8 | Naive infected | SIVmac251/32H | <2.0 | + | + | 16 | Acute | 849 | 3,184 | 1,592 | 1,432 | − |
*SIV strain used to infect the monkeys. SIVmac251/32H derived from an in vivo passage of SIVmac251.
End-point anti-SIV antibody titers detected by ELISA.
Viral isolation by coculture with C8166 human T-cell line.
PCR DNA analysis using primers amplifying the gag region of SIV.
Number of CD20+, CD2+, CD4+, and CD8+ cells (×103/mL) as determined from the last bleeding before death.
¶Formalin-inactivated whole SIV derived from HUT-78 chronically infected cells.
#Monkey OD5 was splenectomized 1 year after infection.
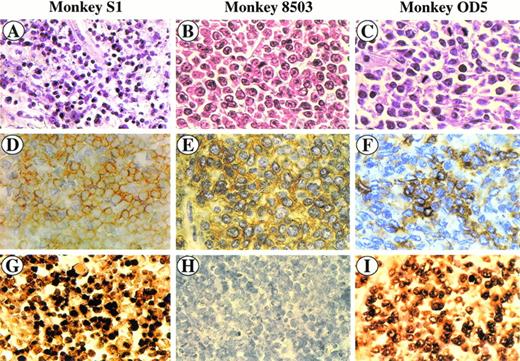
SIV-associated lymphomas. Sections were stained with hematoxylin and eosin (A, B, and C), immunostained with anti-CD3 MoAb (D) and anti-CD20 MoAb (E, F), and hybridized with the SG5 SIV gag oligoprobe (G, H, I). Monkey S1 (A, D, and G) is a peripheral CD3+ T-cell lymphoma (D); CD3+ cells (yellow) show a positive in situ hybridization signal (brown) (G). Monkey 8503 (B, E, and H) is a centroblastic CD20+ B-cell lymphoma (E) showing a negative in situ hybridization signal (H). Monkey OD5 (C, F, and I) is a centroblastic CD20+ B-cell lymphoma (F) showing a positive in situ hybridization signal (I).
SIV-associated lymphomas. Sections were stained with hematoxylin and eosin (A, B, and C), immunostained with anti-CD3 MoAb (D) and anti-CD20 MoAb (E, F), and hybridized with the SG5 SIV gag oligoprobe (G, H, I). Monkey S1 (A, D, and G) is a peripheral CD3+ T-cell lymphoma (D); CD3+ cells (yellow) show a positive in situ hybridization signal (brown) (G). Monkey 8503 (B, E, and H) is a centroblastic CD20+ B-cell lymphoma (E) showing a negative in situ hybridization signal (H). Monkey OD5 (C, F, and I) is a centroblastic CD20+ B-cell lymphoma (F) showing a positive in situ hybridization signal (I).
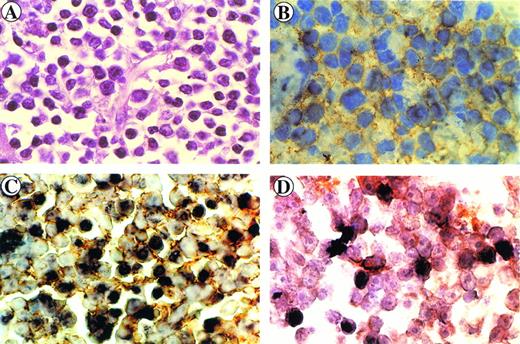
Immunohistology of the retro-orbital centroblastic B-cell lymphoma arising in monkey 4. Sections were stained with hematoxylin-eosin (A) and immunostained with anti-CD20 MoAb (B). Immunohistochemical and in situ hybridization (brown) double-staining of CD20+ cells (yellow ring) (C) and of CD68+ cells (orange ring) (D) displaying intracellular SIV genome signals (dark brown).
Immunohistology of the retro-orbital centroblastic B-cell lymphoma arising in monkey 4. Sections were stained with hematoxylin-eosin (A) and immunostained with anti-CD20 MoAb (B). Immunohistochemical and in situ hybridization (brown) double-staining of CD20+ cells (yellow ring) (C) and of CD68+ cells (orange ring) (D) displaying intracellular SIV genome signals (dark brown).
Features of the SIV-Associated NHL in InfectedMacaca fascicularis
| Monkey Code . | Histology and Anatomic Site . | Immunophenotype . | ||||
|---|---|---|---|---|---|---|
| CD20 . | CD3 . | CD4 . | CD8 . | CD68 . | ||
| 4 | Centroblastic, large cell NHL; retroorbital tissues | +* | − | − | − | ± |
| 8503 | Centroblastic, large cell NHL; mesenteric lymph nodes, stomach and liver | + | − | − | − | ± |
| OD5 | Centroblastic, large cell NHL; submaxillary and mesenteric lymph nodes, skin and heart | + | − | − | − | ± |
| 208 | Centroblastic, large cell NHL; retroorbital tissues | + | − | − | − | ± |
| S1 | Peripheral, pleomorphic, small-medium–sized cell NHL; axillary lymph nodes, spleen and retroorbital tissues | − | + | + | + | ± |
| Monkey Code . | Histology and Anatomic Site . | Immunophenotype . | ||||
|---|---|---|---|---|---|---|
| CD20 . | CD3 . | CD4 . | CD8 . | CD68 . | ||
| 4 | Centroblastic, large cell NHL; retroorbital tissues | +* | − | − | − | ± |
| 8503 | Centroblastic, large cell NHL; mesenteric lymph nodes, stomach and liver | + | − | − | − | ± |
| OD5 | Centroblastic, large cell NHL; submaxillary and mesenteric lymph nodes, skin and heart | + | − | − | − | ± |
| 208 | Centroblastic, large cell NHL; retroorbital tissues | + | − | − | − | ± |
| S1 | Peripheral, pleomorphic, small-medium–sized cell NHL; axillary lymph nodes, spleen and retroorbital tissues | − | + | + | + | ± |
*Sections were examined microscopically and scored semiquantitatively as follows: +, ≥20 positive cells per high power field (×400); ±, ≤5 positive cells per high power field (×400); −, absence of positive cells.
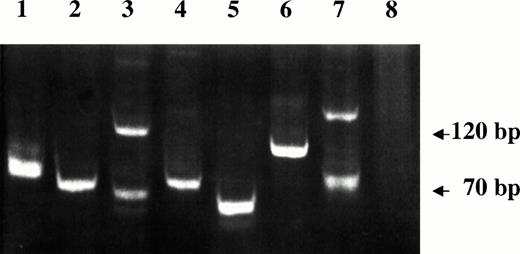
VDJ-IgH PCR analysis of DNA samples from lymphoma tissues of SIV-infected monkeys. Lanes 1 through 4 are from monkeys 4, 208, OD5, and 8503, respectively. A spontaneous nasal non-Hodgkin's B-cell lymphoma from uninfected monkey (lane 5), Rajii cells (lane 6), SL-691, an in vitro established Simian Lymphoblastoid B-cell line (lane 7), and monkey PBMC (lane 8) were also included as controls.
VDJ-IgH PCR analysis of DNA samples from lymphoma tissues of SIV-infected monkeys. Lanes 1 through 4 are from monkeys 4, 208, OD5, and 8503, respectively. A spontaneous nasal non-Hodgkin's B-cell lymphoma from uninfected monkey (lane 5), Rajii cells (lane 6), SL-691, an in vitro established Simian Lymphoblastoid B-cell line (lane 7), and monkey PBMC (lane 8) were also included as controls.
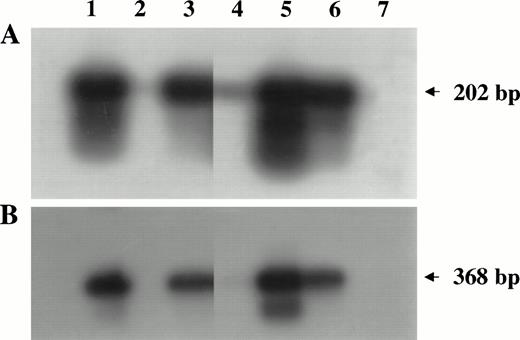
PCR analysis of high-molecular-weight DNA extracted from lymphoma tissues of infected monkeys. Primers and probes amplifyinggag (A) and env (B) regions of the SIV genome were used as described in Materials and Methods. Lanes 1 through 5 are samples of monkeys 4, 208, OD5, 8503, and S1, respectively. DNA samples derived from SIV chronically infected HUT 78 cells (lane 6) or HUT 78 uninfected (lane 7) cells were included as positive or negative control, respectively.
PCR analysis of high-molecular-weight DNA extracted from lymphoma tissues of infected monkeys. Primers and probes amplifyinggag (A) and env (B) regions of the SIV genome were used as described in Materials and Methods. Lanes 1 through 5 are samples of monkeys 4, 208, OD5, 8503, and S1, respectively. DNA samples derived from SIV chronically infected HUT 78 cells (lane 6) or HUT 78 uninfected (lane 7) cells were included as positive or negative control, respectively.
Viral Load in Lymphoma Tissue and Spleen Homogenate Based on Syncytia Induction on C8166 Human T Cells
| Monkey Code . | Tissue . | Grams* . | Volume of Homogenate (mL) . | TCID50/mL† . |
|---|---|---|---|---|
| 4 | Tumor mass | 0.65 | 2 | 1.0 × 105 |
| Spleen | 0.49 | 2 | 2.2 × 103 | |
| 8503 | Tumor mass | 0.73 | 2 | 1.7 × 104 |
| Spleen | 0.98 | 2.5 | 8.9 × 103 | |
| OD5 | Tumor mass | 1.25 | 2 | 3.1 × 103 |
| Spleen | ND‡ | |||
| 208 | Tumor mass | 0.51 | 2 | 3.1 × 103 |
| Spleen | 0.65 | 2 | 2.2 × 103 | |
| S1 | Tumor | 0.19 | 2 | 2.5 × 107 |
| Spleen | 0.80 | 2 | 3.1 × 103 |
| Monkey Code . | Tissue . | Grams* . | Volume of Homogenate (mL) . | TCID50/mL† . |
|---|---|---|---|---|
| 4 | Tumor mass | 0.65 | 2 | 1.0 × 105 |
| Spleen | 0.49 | 2 | 2.2 × 103 | |
| 8503 | Tumor mass | 0.73 | 2 | 1.7 × 104 |
| Spleen | 0.98 | 2.5 | 8.9 × 103 | |
| OD5 | Tumor mass | 1.25 | 2 | 3.1 × 103 |
| Spleen | ND‡ | |||
| 208 | Tumor mass | 0.51 | 2 | 3.1 × 103 |
| Spleen | 0.65 | 2 | 2.2 × 103 | |
| S1 | Tumor | 0.19 | 2 | 2.5 × 107 |
| Spleen | 0.80 | 2 | 3.1 × 103 |
*Grams are refereed to the weight of the tumor mass or the spleen homogenized.
TCID50 is defined by end-point dilution at which virus is able to infect 50% of cell cultures in an in vitro assay. The interpolation to obtain the tissue culture infectious dose 50% (TCID50) has been performed by the Reed and Muench method.
This monkey was splenectomized 1 year after infection.
Abbreviation: ND, not determined.
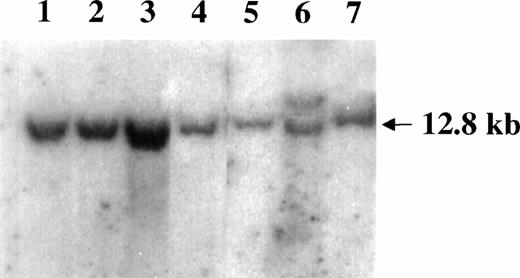
Southern blot analysis of the c-myc locus in SIV-associated lymphomas. DNA samples were digested with EcoRI and hybridized with the pMC41-3DC probe covering the 3rd exon of thec-myc locus. Lanes 1 through 5 are samples of monkeys 4, 208, OD5, 8503, and S1, respectively. The Raji cells (lane 6) carrying the germ line band (12.8 Kd) and a higher rearranged band (<12.8 kd) and monkey PBMC (lane 7) were included as controls.
Southern blot analysis of the c-myc locus in SIV-associated lymphomas. DNA samples were digested with EcoRI and hybridized with the pMC41-3DC probe covering the 3rd exon of thec-myc locus. Lanes 1 through 5 are samples of monkeys 4, 208, OD5, 8503, and S1, respectively. The Raji cells (lane 6) carrying the germ line band (12.8 Kd) and a higher rearranged band (<12.8 kd) and monkey PBMC (lane 7) were included as controls.
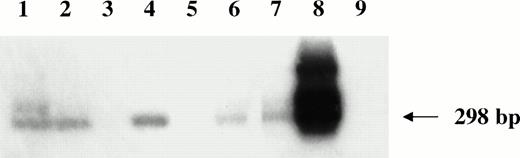
Nested HVMF-1 DNA-PCR analysis of monkey lymphomas. Lanes 1 through 5 are samples of monkeys 4, 208, OD5, 8503, and S1, respectively. Furthermore, samples from a spontaneous nasal non-Hodgkin's B-cell lymphoma from uninfected monkey (lane 6), from SL-P1 and SL-691 cells (in vitro established Simian Lymphoblastoid cell lines; lane 7 and 8, respectively), and from HUT 78 human T-cell line (lane 9) were included as controls.
Nested HVMF-1 DNA-PCR analysis of monkey lymphomas. Lanes 1 through 5 are samples of monkeys 4, 208, OD5, 8503, and S1, respectively. Furthermore, samples from a spontaneous nasal non-Hodgkin's B-cell lymphoma from uninfected monkey (lane 6), from SL-P1 and SL-691 cells (in vitro established Simian Lymphoblastoid cell lines; lane 7 and 8, respectively), and from HUT 78 human T-cell line (lane 9) were included as controls.
DNA-polymerase chain reaction (PCR) analysis of SIV, STLV-1, EBV, and HVMF-1.
Genomic DNA was extracted from tissues following the phenol-chloroform method by precipitation with 3 mol/L sodium acetate and cold ethanol. DNA was then amplified with primers (PCO3/PCO4) specific for a 110-bp sequence of human β-globin gene to assess its competence for PCR.19 To detect the SIV genome, DNA PCR was performed using primers amplifying a 202-bp region of the gag gene (Gagf3, nt 1297-1316, 5′-GGTGCATTCACGCAGAAGAG-3′, Gagr4, nt 1498-1478, 5′-GTTCTCGGGCTTAATGGCAGG-3′) or a 368-bp region of the env gene (EnvLfl, nt 8153-8176, 5′-ATAAAAGGGGTCTTTGTGCTAG-3′; EnvLr2, nt 8519-8496, 5′-TCAACCTTTCGCTCCCACTCTTGC-3′) of the SIVmac251 genome (GenEMBL access number M19499). PCR was performed in 100 μL of reaction mixture containing 1 μg of DNA, 200 μmol/L of each of the four deoxynucleotide triphosphates, 60 pmol/L of each primer, 2.5 U Taq polymerase (Amplitaq, Perkin Elmer, Norwalk, CT), 50 mmol/L of KCl, 10 mmol/L of Tris-HCl (pH 8.3), and 1.5 mmol/L of MgCl2. After a denaturation step at 94°C for 5 minutes, DNA was amplified by using the DNA thermal cycler 9600 (Perkin Elmer Cetus) for 35 cycles, each at 95°C for 90 seconds, at 55°C for 90 seconds, and at 72°C for 2 minutes with a final extension step at 72°C for 10 minutes.
Conserved pol region of the human T-lymphotropic virus type 1 genome, which shares 90% genomic identity with the pol region of STLV, was chosen for PCR amplification using primers and a probe described elsewhere.20 PCR started with 5 minutes denaturation step at 94°C followed by 30 cycles each at 94°C for 90 seconds, at 50°C for 90 seconds, and at 72°C for 2 minutes and by a final extension step at 72°C for 10 minutes.
EBV was analyzed using oligonucleotide primers common for EBNA2 type A and B sequences (EBVf, nt 48170-48189, 5′-AGGCTGCCCACCCTGAGGAT-3′; EBVr, nt 48339-48320, 5′-GCCACCTGGCAGCCCTAAAG-3′; EBVp, nt 48262-48280, 5′-GTTGCCGCCAGGTGGCAGC-3′) derived from human EBV sequences (GenEMBL access number V01555, K03333, and K3332). PCR was performed in a 100-μL reaction mixture with the same composition as it was for the SIV gag PCR, started with a denaturation step at 95°C for 4 minutes followed by 30 cycles each at 94°C for 45 seconds, at 62°C for 45 seconds, at 72°C for 90 seconds with a final extension step at 72°C for 10 minutes.
A nested PCR was used to detect the EBV-like HVMF-1 genome. The outer set of primers (Ws and Was-3) has already been published by Li et al.12 The inner set of primers (Ws-4, 5′-CAGTTATTCTGCTCAGCCCAC-3′; Was-2, 5′-CCAGACTAGACCCCAGGTTCC-3′), and the probe (Wdp, 5′-AAGGTGCAGGCACAACAGCC-3′) were chosen from the nucleotide sequence data reported in the GenEMBL access number X77781HVMFINA5. The first round of PCR was performed by using the same mixture composition and cycling conditions as were for the SIVgag PCR. The final product of the first PCR (10 μL) was subjected to a second round of amplification by using the same buffer composition. PCR started with a 5-minute denaturation step at 94°C, followed by 30 cycles each at 94°C for 30 seconds, at 55°C for 30 seconds, at 72°C for 1 minute, and a final extension step at 72°C for 10 minutes.
Ten microliters of the PCR products were electrophoresed on 1.5% to 2% agarose gel, transferred to filter (Nytran-N; S&S, GmbH, Germany), and hybridized with 5′-end 32P-labeled–specific oligoprobes.
Rearrangement of the c-myc locus and of the VDJ region of IgH gene.
To analyze the c-myc gene, 20 μg of DNA from lymphoma tissues were digested with HindIII and EcoRI restriction enzymes, separated onto agarose gel, transferred to Nytran filters, and hybridized with a 32P-labeled 1.6-kilobase (kb) pMC41-3RC probe21 representing the third exon of thec-myc locus. The c-myc translocation could be identified when bands other than the germ line c-mycconfiguration (12.8 kb) were present.
A seminested DNA-PCR amplification of rearranged VDJ fragments of IgH gene was performed by using primers corresponding to the human complementarity determining region 3 (CDR 3) as described elsewhere.22 Final PCR products (20 μL) were electrophoresed onto a 12% polyacrylamide gel, and then DNA was stained by ethidium bromide and visualized by ultraviolet light.
In situ hybridization and immunostaining.
Paraffin sections were deparaffinized for 20 minutes at room temperature with xilene, rehydrated with ethyl alcohol, and washed with distilled water for 5 minutes and with PBS for 10 minutes. Standard in situ hybridization was done by using a probe homologous to thegag region of the SIV genome (SG5p, nt 1470-1502 5′-AATAGGTGGTAACTATGTCCACCTGCCATTAAG-3′) or a SIV-unrelated oligo probe (SK19, nt 1134-1174 gag, HIV-1; GenEMBL access number K02013) that was used as the negative control. After protease digestion (10 μg/mL for 15 minutes at 37°C), the slides were treated for 5 minutes with cold paraformaldehyde 4% (wt/vol) and then sequentially washed with cold PBS and 20 × SSC (1 × SSC = 0.15 mol/L of NaCl, 15 mmol/L of sodium citrate) for 10 minutes at room temperature. The excess of liquid was soaked out, and 50 μL of the prehybridization solution (4 × SSC, 50% formamide, 1× Denhardt's, 5% dextran sulfate, 0.5 mg/mL salmon sperm) was added and the slides covered with a cover slip. The samples were then incubated for 2 hours at 42°C (Omnigene Temperature Cycler, Hybaid Lt, Middlesex, UK). The probe was labeled with digoxigenin (dig)-dUTP using the 3′-tailing method according to the manufacturer's instructions (3′-labeling DNA kit; Boehringer Mannheim, Indianapolis, IN). The dig-labeled probe was added to the same solution and, after a denaturation step at 90°C for 10 minutes, the hybridization was performed at 42°C overnight. After hybridization and washing, the slides were incubated with a sheep anti-dig MoAb conjugated with alkaline phosphatase (dilution 1:400) for 2 hours at room temperature. The probe/target complex was visualized by incubation with chromogen (NBT/Xphosphate; Boehringer Mannheim). The hybridization signal was evident as a dark brown precipitate. For immunohistochemistry, after the final wash of the in situ hybridization procedure, the slides were overlaid with PBS containing 10% FCS and the immunoreaction was performed as described above.
RESULTS
Clinical and immunological status of SIV-infected monkeys developing lymphomas.
The individual histories of the monkeys with lymphomas (4, 8503, OD5,208, and S1) compared with those of monkeys without lymphomas (C1, 8302, OD3, 203, and H8) are summarized in Table 1. Because the strains of viruses used to infect the animals were almost identical in their pathogenic potential, the monkeys have been matched according to the treatment (4 and 8503 v C1 and 8302; OD5 and 208 v OD3 and 203; S1 v H8). All but 1 monkey (monkey S1) had a mean survival time longer than that observed in a comparable group of 4 monkeys that did not develop lymphomas (109.5 weeks v 92 weeks after infection, respectively). Monkey S1 did not seroconvert although virus was frequently recovered from PBMC. This monkey appeared healthy before SIV inoculation; however, soon after this, a rapidly increasing mass in the right-side ocular bulb was observed.
Monkeys 4, 8503, OD5, and 208 showed a marked depletion of CD4+ T cells and an altered CD4/CD8 ratio similar to that observed in the comparable tumor-free SIV-infected monkeys (C1, 8302, OD3 and 203; Table 1). Immunodeficiency was not observed in monkeys S1 (with lymphoma) and H8 (without lymphoma), likely due to the very short period of time between infection and death. Monkey OD5 had an abnormal level of CD2+ and CD8+ cells, likely due to the splenectomy performed at 1 year after infection or to the presence of cutaneous inflammatory necrotic lesions.
Immunohistology and VDJ-IgH clonality analysis of lymphomas.
The tumor masses histologically diagnosed according to the Kiel classification23 were classified as large-cell NHL centroblastic type (monkeys 8503, OD5, 208, and 4; Fig 1B through C and Fig 2A; monkey 208 not shown) and peripheral, pleomorphic, small-medium–sized cell NHL (monkey S1; Fig 1A). As shown in Table 2, lymphomas displayed a different localization independent of the phenotype. Four out of the 5 tumors (monkeys 8503, OD5, 208, and 4) stained positive for the B-cell marker CD20 and were therefore classified as B-cell lymphomas (Table 2, Fig 1E through F, and Fig 2B; monkey 208 not shown), whereas the tumor tissue from monkey S1 was positive for the CD3 cell-surface marker and was classified as a T-cell lymphoma (Table 2; Fig 1D).
A molecular study was performed on SIV-associated NHLs specimens to determine the clonality of the B-cell lymphomas. Figure 3 shows the single band that represents the monoclonal rearranged VDJ region, which was detected in tumor samples of monkeys 208 and 8503. These were therefore classified as monoclonal lymphomas. Two bands plus an additional band below 70 bp and at least three other bands (size 70 to 120 bp) were observed in the PCR products of amplified DNA of monkeys OD5 and 4, respectively, that were classified as oligoclonal lymphomas.
Detection of SIV in B- and T-cell lymphomas.
The presence of the SIV genome was demonstrated in 3 of 5 lymphomas (monkeys 4, OD5, and S1) by DNA-PCR analysis, using primers amplifying fragments of the gag and env regions of the SIV genome (Fig 4). In one case (monkey 8503), a faint band was observed when thegag gene was analyzed.
Paraffin sections were then analyzed by in situ nucleic acid hybridization for the SIV genome. A clear SIV-specific signal was found in cells of lymphoma tissues of monkeys S1, OD5 (Fig 1G and I, respectively) and 4 (Fig 2C) but not in those of monkeys 8503 (Fig 1H) and 208 (not shown). In the case of the T-cell lymphoma (monkey S1) the SIV-bearing tumor cells were numerous (about 60% to 80%) and homogeneously distributed throughout the section, whereas in the two cases of B-cell lymphomas (monkeys 4 and OD5), they appeared to have a less homogeneous distribution. In addition, the number of SIV-positive areas appeared to be more numerous in the tissue of monkey 4 than in that of monkey OD5.
To verify the phenotype of the SIV-positive cells, an immunohistochemical analysis was performed at the end of the in situ hybridization procedure. SIV-positive cells were stained positively for the CD3 marker in monkey S1 (Fig 1G) or for the CD20 marker in monkeys OD5 and 4 (Fig 1I and Fig 2C, respectively). Double-staining experiments performed on tissue sections of monkey 4 showed the presence of a low number of CD68-positive cells displaying also positivity for the SIV genome (Fig 2D).
The lymphomatous tissues of monkeys 4, OD5, and S1 that resulted SIV-positive by DNA-PCR and by in situ hybridization analyses did reveal a viral load higher than that found in the spleen of the same monkey. In particular, in line with the number of the SIV-positive cells, the titer of the virus present in tumor samples of monkey S1 was about from two to four log higher than that detected in tissues of monkeys 4 and OD5 (Table 3).
C-myc translocation and detection of STLV, EBV, and HVMF-1 genomes in monkey lymphomas.
Genomic DNA extracted from the lymphomatous tissues of all monkeys was analyzed by Southern blot hybridization for the presence of the translocation of the c-myc locus. In all cases, no bands other than the 12.8 kb of the germ line were observed (Fig 5).
We next examined the possibility that coinfecting viruses could be present in the lymphomatous tissues of the monkeys. All monkeys were negative for STLV-1 and EBV-like sequences as determined by DNA-PCR analysis (data not shown), whereas 3 of 4 B-cell lymphomas were positive by nested PCR for HVMF-1 sequences (Fig 6).
DISCUSSION
Lymphoid neoplasms, the most frequent tumors observed in nonhuman primates,24-26 have evoked a great interest because their anatomic and physiological features closely resemble those observed in humans. In this study we have described 5 cases of NHL in 14 monkeys experimentally infected with SIV. The immunodeficiency was a hallmark of monkeys affected by B-cell lymphomas, whereas CD4+T-cell depletion was not observed in the monkey developing the T-cell lymphoma. Interestingly, SIV sequences were present at levels detectable by in situ hybridization within the cells of 2 cases of the B- and in one case of the T-cell lymphoma. To the best of our knowledge this is the first demonstration that B- and T-cell simian lymphomas can harbor SIV sequences in the tumor cells, although SIV was also detected in some infiltrating macrophages. Several attempts have been made to detect HIV genome in AIDS-associated B-cell malignancies, but the results were not conclusive.27,28 Furthermore, the phenotype of the “infected cells” was not determined. Similarly, in neoplasms from SIV-infected monkeys, some non-neoplastic cells, mainly macrophages and multinucleated cells, were occasionally found positive for the presence of p27 SIV protein. Moreover, even in in vitro cell lines derived from monkey lymphomas, no reverse transcriptase activity has been detected.8 In addition, we observed that the SIV titer in lymphomatous tissues was higher than that found in the spleen, a macrophage-rich tissue known to be a relevant source of the virus. However, this does not exclude the contribution of infected cells, other than “transformed” B cells or T cells, in the SIV viral load of tumor tissues.
It has been suggested that monoclonal or oligoclonal NHLS in human29 and simian AIDS30 as well as in murine retrovirus-induced immunodeficiency syndrome lymphomas31progress from an initial polyclonal expansion of activated B cells.32 Although lymphomagenesis in vivo might depend on a complex cascade of events also involving a complex network of cellular interactions,33-38 it is likely that “transformation” in some cases can be initiated through chronic antigen stimulation, heightening the probability of chromosomal alterations or genetic lesions.39 Indeed, in vitro studies on human B lymphocytes have suggested that HIV or its transactivating protein Tat may have transforming properties because they may mediate c-mycexpression that, in some cases, seems to be closely linked to the development of B-cell neoplasia.40-43 Nevertheless, as already reported for a group of human AIDS-associated lymphomas,44c-myc rearrangement was not detected in the lymphomas we have observed in infected monkeys, including those harboring the SIV genome. Thus, it seems that alternative molecular pathways may be involved in the transformation processes.
Interestingly, we have observed that only B-cell lymphomas positive for the presence of the SIV genome had an oligoclonal rearrangement of the VDJ region of the IgH gene. Taking into consideration the oligoclonal expansion of the lymphocytes, one could suppose that a B cell at one time became susceptible to SIV infection45-47 with resultant further proliferation. In line with this hypothesis, we observed that only some B cells were infected by SIV. The presence of the SIV genome in simian tumor cells might suggest a possible transforming potential of SIV through insertional mutagenesis, as already described in AIDS-related T-cell lymphoma48,49 and in some virus-infected T-cell neoplasms in rhesus.50,51Transformation may occur also by a fusion process between lymphocytes and infected macrophages as shown by the CD3 and CD14 double immunostaining of tumor cells of AIDS-related lymphoma.52Although such analysis was not performed on our samples, we observed giant cells doubly positive for CD3/SIV, CD20/SIV, and CD68/SIV in the case of T-cell lymphoma and in two cases of B-cell lymphomas, respectively.
Because lymphomagenesis is thought to be a multistep process, we analyzed the tumors for the presence of coinfecting viruses that have been reported to be present in human or in simian malignancies.53-55 All lymphomas were negative for STLV and EBV, but three of them (all of B-cell origin) were associated with HVMF-1 infection. However, HVMF-1 sequences were not detected in one (monkey OD5) of the two cases of SIV-positive lymphomas. Its detection required a nested PCR analysis, thus indicating a low HVMF-1 viral load as compared with the SIV load that was detected by a single run of PCR and in situ analysis.
In the same species of monkeys (Cynomolgus), Feichtinger et al originally reported a high incidence of lymphomas in a group of monkeys infected by SIVsm but not in a comparable group of monkeys infected by HIV-2.8 Whether this observation means that lymphoproliferative diseases depend on the pathogenic potential of the virus strain used for infection7,56 remains to be shown. In the same model HVMF-1 has been associated with an high incidence of B-cell lymphomas in SIVsm-infected monkeys. However, HVMF-1 was detected not only in tumor-bearing infected monkeys but also in HIV-2–infected and noninfected monkeys that did not develop lymphomas.57 Thus, HVMF-1 infection per se may not be fully responsible for the development of lymphomas and may require the presence of an immunodeficient state. Consistent with this observation, human EBV-like Rhesus lymphocryptic virus (LCV) can reproduce in monkeys the key aspects of the EBV infection in humans but was unable to induce tumors in naive pathogen-free animals up to 24 months after infection.58 On the other hand, AIDS-associated polyclonal B-cell lymphomas with no evidence of c-myc rearrangement and negative for EBV have been reported.43 Thus, according to our data, neoplasia may develop in the absence of a concomitant HVMF-1 infection in immunosuppressed monkeys. Nevertheless, we cannot provide further evidence against or in favor of a direct involvement of HVMF-1 in lymphomagenesis because our data are based on one only case of SIV-positive, HVMF-negative simian B-cell lymphoma. A serological and molecular survey to asses the spread of HVMF-1 infection in our Cynomolgus colony and the incidence of neoplasia is currently in progress. In this framework, we have already observed some cases of lymphomas in SIVmac-infected monkeys that were certified to be serologically negative for STLV, type-D retroviruses, cytomegalovirus, and EBV nuclear antigen (Shamrock Limited, England; personal communication and unpublished results, May 1995). It is quite clear that both simian models of SIV-associated neoplasia, although different in some aspects, may represent the different subtypes of AIDS-associated malignancies.59 Although from our data we cannot definitely prove the transforming potential of SIV, it seems reasonable to suggest that, under certain circumstances, SIV may play a role in lymphomagenesis. Thus, monkey lymphoma models might be useful to study the pathogenesis and treatment of immunodeficiency-associated tumors.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Dr B. Ensoli for helpful discussion and comments on the manuscript. We also thank S. Mochi for the synthesis of primers and probes, to A. Carè for providing the pMC41-3RC used to detect the c-myc translocation, and to A. Lippa and F.M. Regini for the excellent editorial assistance. We also thank F. Varano, A. Cesolini, F. Incitti, S. Fazzitta, M. Chiodi, R. Marinelli, A. Marini, P. Di Zeo, and S. Alessandroni for handling the Cynomolgus colony.
Supported by a grant from the “Animal Model Development Project” of the Italian Ministry of Health, Istituto Superiore di Sanità, Rome, Italy (to P.V.) and the IX AIDS Research Project, ISS No. 9403-13 (to C.D.B.).
Address reprint requests to Fausto Titti, PhD, Laboratory of Virology Istituto Superiore di Sanità, Viale Regina Elena, 299, 00161 Rome, Italy.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal