Multiple biologic effects of interferon-α (IFN-α), including cell growth inhibition and antiviral protection, are initiated by tyrosine phosphorylation of STAT proteins. Although this signal pathway has been intensively investigated, the relevance of STAT signal persistence has received scant attention. Using paired isogenic lymphoma cells (Daudi), which either are sensitive or resistant to growth inhibition by IFN-α, we found comparable initial tyrosine phosphorylation of multiple STAT proteins; however, the phosphorylation durations and associated DNA-binding activities diverged. Phosphorylation and DNA-binding capacity of STAT1 decreased after 4 to 8 hours in resistant cells, as compared with 24 to 32 hours in sensitive cells, whereas phosphorylation of STAT3 and STAT5b was briefer in both lines. Functional significance of the prolonged STAT1 signal, therefore, was explored by experimental interruption of tyrosine phosphorylation, either by premature withdrawal of the IFN-α or deferred addition of pharmacologically diverse antagonists: staurosporine (protein kinase inhibitor), phorbol 12-myristate 13-acetate (growth promoter), or aurintricarboxylic acid (ligand competitor). Results indicated that an approximately 18-hour period of continued STAT1 phosphorylation was associated with growth arrest, but that antiviral protection developed earlier. These differences provide novel evidence of a temporal dimension to IFN-α signal specificity and show that duration of STAT1 activation may be a critical variable in malignant cell responsiveness to antiproliferative therapy.
LYMPHOPROLIFERATIVE disorders and hematologic malignancies can be complications of immunosuppression or immunodeficiency1-3 and are subject to interferon (IFN) regulation of the abnormal cell growth.3,4 In tissue cultures, the growth of a number of lines of human malignant lymphoma cells can be inhibited by type I IFNs (IFNs-α,β). Several lines of B-lymphoma cells, derived from a Burkitt tumorDaudi5 are highly sensitive to growth arrest by IFN-α. The IFN-α has been shown to drive a pronounced shift in the cell-division cycle of these lymphoma cells, from exponential growth into a G1 arrest,6-8 followed by transition into a prolonged G0-like condition.7,8 Isogenic variants of Daudi cells that resist growth inhibition by IFN-α have been isolated and studied in several laboratories.6,9-11These variants can retain specific and functionally competent receptors for IFNs-α,β (IFNAR).6,9,11 Correlations of molecular and cellular responses to IFN-α in the Daudi lymphoma system thus can be particularly informative with respect to the molecular mechanisms of direct antiproliferative action.6-12
The gene expression leading to major biologic actions of IFNs-α,β and many other cytokines or growth factors is regulated by latent cytoplasmic proteins that serve as dual signal transducers and activators of transcription (STATs).13-17 The primary events are membrane-associated and require no new protein synthesis.18 IFN-α signal transduction is initiated by dimerization of IFNAR subunits,19-21 and rapid autophosphorylation of Tyk2 and Jak120-23 Janus tyrosine kinases is required for tyrosine phosphorylation of STAT proteins in the Jak-STAT pathway.13-16 Although IFN-α can initiate activation of multiple STAT proteins,13,15,16 a decisive role of STAT1 in the biologic responses has been demonstrated by targeted gene disruption in transgenic mice.24,25IFN-α does not induce STAT1 tyrosine phosphorylation in cells that fail to express Jak1 or Tyk2.13,21,22 26
STAT1α (91 kD) and STAT1β (84 kD) are splice variants of a common gene transcript27 with structural elements related to STAT2 (113 kD)28 of separate gene origin. Tyrosine-phosphorylated STAT1 (STAT1-pTyr) is a principal transcriptional regulator induced by IFN-α,13,15,29 but its activation is intimately dependent on a prior tyrosine phosphorylation of STAT2.30,31Tyrosine-phosphorylated STATs form stable dimers that translocate to the nucleus, bind to highly conserved promoter elements of IFN-stimulated genes, and enhance or initiate transcription.13-15,32-35 Specificity of STAT function in transcriptional activation is conferred both by secondary serine phosphorylation15,36,37 and by selective pairings of phosphorylated STATs in homodimers or heterodimers.13-17,38STAT1-pTyr itself can bind to oligonucleotide promoter elements associated with the genes for IRF-1 orFcγR34,38; however, its binding to a highly conserved IFN stimulated response element (ISRE), located upstream to the great majority of known IFN-stimulated genes, depends on association with STAT2-pTyr in a heterocomplex (ISGF3α)39and further protein-protein interaction with a 48-kD polypeptide (ISGF3γ-p48) in the IRF/myb family of transcriptional regulators.40 This multimeric holocomplex constitutes ISGF3.13,14 39 In parallel to tyrosine phosphorylation, the protein interactions of STAT1 thus can specify and enhance its DNA-binding function.
Although phosphorylation events and functional specificities of proteins during stimulation of the Jak-STAT signal pathway have been subjects of intense investigation, the temporal regulation of signal dynamics in relation to major biologic responses has not been extensively explored.33-35 Present interest in antiproliferative signal processes was motivated by preliminary observations that formation of ISGF3 could persist for more than 24 hours when relatively sensitive Daudi cells were continuously exposed to IFNs-α,β, yet disappeared before 8 hours in a derived line of variant cells resistant to the antiproliferative effect of IFNs-α,β. Although earlier results with other cell types had shown that extended times of exposure must precede some antiviral or growth regulatory actions of IFNs-α,β,41-44 those investigations antedated recognition of the Jak-STAT pathway and the evidence for transcriptional downregulation by a nuclear tyrosine phosphatase.33 35 Because the divergence of molecular and biologic effects in parental and variant Daudi cells was dramatic, this lymphoma model offered a unique opportunity to analyze the temporal relationships with the advantage of current methodologies.
The experimental approach was to periodically interrupt IFN-α signal transduction and measure the impact on antiproliferative and antiviral effects. Although this could most simply be accomplished by premature withdrawal of IFN-α, we found that necessary medium changes impeded the resumption of Daudi cell growth and confounded interpretation of results. As alternative means to assess the relevance of STAT activity duration to growth inhibition, we therefore attempted to interrupt the STAT tyrosine phosphorylation with potent pharmacologic antagonists. Three agents with diverse modes of molecular action proved effective, and two of these consistently protected the sensitive Daudi cells from impending growth arrest when added as late as 18 hours during a continuous IFN-α exposure.
MATERIALS AND METHODS
Reagents
Recombinant human IFN-α2a used in all of the biologic experiments was generously donated by Hoffmann-La Roche (Nutley, NJ). Recombinant IFN-α2b, used in tests of IFNAR binding, was a gift from Schering Co (Kenilworth, NJ). These IFN-α subtypes differ in only one amino acid. Both are potent antiproliferative agents, and the original stocks exhibited identical specific antiviral activity (2 × 108 U/mg of protein). Rabbit polyclonal antibodies to peptides representing unique COOH-termini of STATs 1, 2, 3, and 5a/b were generously supplied by A. Larner (Food and Drug Administration, Bethesda, MD) or L. Hennighausen (National Institutes of Health, Bethesda, MD) and described previously.33 45 An antiphosphotyrosine (anti-pTyr) mouse monoclonal antibody (4G10) was obtained from Upstate Biotechnology (Lake Placid, NY). Horseradish peroxidase (HRP)-conjugated goat antibodies to mouse or rabbit IgG came from Transduction Laboratories (Lexington, KY) and were detected by enhanced chemiluminescence substrate (ECL) for Western blotting obtained from Amersham Canada (Oakville, Ontario, Canada). Alkaline phosphatase-conjugated goat antibody to rabbit IgG and a phosphatase substrate detection kit were obtained from Kirkegaard and Perry Laboratory (Gaithersburg, MD). Immobilon-P transfer membranes came from Millipore (Bedford, MA), and Ficoll came from Pharmacia LKB Biotech (Piscataway, NJ).
Staurosporine (STSP; 0.1 mmol/L) and phorbol-12-myristate-13-acetate (PMA; 0.1 mmol/L) were solubilized in pure dimethyl sulfoxide. Aurintricarboxylic acid (ATA; 200 mmol/L) was solubilized in the cell culture medium. These organic reagents were purchased from Sigma Chemical (St Louis, MO).
Cell Cultures
Parental human lymphoma cells of a Daudi line originally sensitive to growth inhibition by IFNs-α,β (designated DWS) and a derived variant line that is resistant to growth regulation by IFNs-α,β10(designated DWR) were a generous gift from A. Kimchi at the Weizmann Institute of Science (Rehovot, Israel); these cells have been passaged in our laboratories for 10 years.46 Consistent with the original IgM-κ Daudi phenotype,5 gene rearrangements of both the J heavy chain (EcoRI) and κ light chain (BamHI) were identical in parental and variant cells (analyzed by J. Sklar, Collaborative Research, Waltham, MA). The suspension growth medium was RPMI 1640-25 mmol/L HEPES with L-glutamine from GIBCO (Grand Island, NY), supplemented with 10% Nuserum from Collaborative Biomedical Products (Bedford, MA). Cell density was regularly readjusted to less than 1 × 106 cells/mL for exponential growth. Stocks of DWR cells were subjected weekly to IFN-α2a (100 U/mL) for a period of 60 hours. Cell viability was monitored by trypan blue (0.2%) exclusion and exceeded 95% at the start of all experiments.
Antiproliferative Assay
Serial twofold dilutions of IFN-α or other reagents in RPMI growth medium were prepared in 96-well tissue culture microtiter plates. Cells were added in a final volume of 100 μL (5 × 104cells). Changes in viable cell mass were measured metabolically by a modified Mossman assay47 with incubation for 3 hours at 37°C in 0.2% of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) in neutral buffered saline (25 μL/well). Cell lysis with complete solubilization of the blue reduction product (formazan) was accomplished by adding 125 μL of 10% sodium dodecyl sulfate (SDS) in 10 mmol/L HCl and incubation for 48 hours at room temperature. Absorbance at 570 nm was quantitated with a Ceres 900 semi-automatic plate reader from Bio-Tek Instruments (Winooski, VT). Assay linearity was controlled both by hemocytometer manual and electronic cell counts with a Coulter ZBI (Coulter Electronics, Hialeah, FL).
Cell Cycle Analysis
Samples of 2 × 106 cells were sedimented at 200g, gently resuspended into 1 mL of an ice-cold hypotonic solution (PBS [1:10], 0.6% NP-40, 10 μg/mL propidium iodide, 100 μg/mL RNAse A), and collected with an Epics Elite Cytometer (Coulter Cytometry, Hialeah, FL). Fluorimetric histograms of stained DNA (15,000 nuclei) represented the intensity of propidium iodide emissions (675 ± 10 nm, excitation 488 nm) and numbers of nuclei per channel (Y-axis). Cell cycle parameters were analyzed by the Multi-Cycle AV program of P.S. Rabinovitch (University of Washington, Seattle, WA).
Antiviral Assay
Edmonston strain measles virus in low passage 7 from human embryonic kidney48 was plaque-purified and amplified in VERO cells. Samples of 5 × 105 Daudi cells/mL were treated with IFN-α2a (100 U/mL) for 18 or 24 hours or left untreated (as parallel controls), then sedimented at 300g, washed twice in RPMI 1640, and infected with a 50% effective tissue culture infectious dose (TCID50) of virus per cell in fresh growth medium. Parallel cell controls were mock-treated without measles virus. After 1 hour at 37°C, cells were pelleted, washed, and resuspended in conditioned growth medium with or without IFN-α (100 IU/mL). In some experiments, ATA (125 μmol/L) was added together with, 4 or 18 hours after, or without IFN-α. Measles virus production was assayed at 48 hours after infection: aliquots of culture fluid supernatant were obtained after cell sedimentation at 300g, and aliquots of lysed cells, obtained after three freeze-thaw cycles, were used to quantitate cell-associated virus. Virus titers were determined in quadruplicate by microtitration on VERO cell monolayers. Virus cytopathic effect was evaluated microscopically and confirmed by staining with 5% glutaraldehyde, 0.1% crystal violet. Reed and Muench end points49 were expressed as TCID50 per milliliter at 6 days. Infectious center assays (ICA) used infected Daudi cells sedimented at 300g and then incubated for 1 hour at 37°C in RPMI 1640 with a 1:30 dilution of human antimeasles serum (neutralization titer of 1:16,000). Neutralized extracellular virus was removed by washing cells in RPMI. Cell aliquots resuspended in fresh growth medium then were inoculated onto indicator monolayers of VERO cells. Infectious centers at 6 days were enumerated after neutral red staining.50
Quantitation of IFNAR
IFN-α2b was radiolabeled with 4 to 5 μCi/μg of [125I]-Bolton-Hunter reagent (Amersham, Arlington Heights, IL).51,52 Aliquots 5 × 106 Daudi cells were centrifuged and resuspended into fresh growth medium (400 μL) and incubated on ice for 90 minutes with incremental concentrations of [125I]-IFN-α2b up to IFNAR saturation with 5,000 U/mL (0.4 nmol/L). In some experiments, the cells were pretreated for 18 hours with IFN-α2a (100 U/mL) or ATA (31 to 150 μmol/L) and then washed in fresh growth medium before the addition of the [125I]-IFN-α2b. In other experiments, serial concentrations of ATA were added at 90 minutes after 5,000 U/mL of [125I]-IFN-α2b had been allowed to equilibrate with the DWS cells. The free and cell-associated [125I]-IFN-α2b were separated by centrifugation through phthalate oil (n-butyl phthalate:bis[2-ethylhexyl]phthalate, 1.1:1.0) at 4°C in an MTX-150 microcentrifuge from TOMY (Tokyo, Japan) at 12,000g for 3 minutes. Supernatants were removed, and the radioactivity associated with cell pellets in the tube bottoms was determined using a 1272 gamma counter from Pharmacia LKB Biotech. Specific [125I]-IFN-α2b binding was defined as the difference between total and nonspecific binding in the presence of greater than 200-fold excess of unlabeled IFN-α2b. Nonspecific binding ranged from 5% to 40% of the total binding at reaction equilibrium on ice. The mean number of receptor binding sites per cell and apparent dissociation constants (kd) were calculated from Scatchard plots of competitive displacement binding data by the LIGAND program of P. Munson (National Institutes of Health, Bethesda, MD).52 For tests of receptor downregulation, cells were treated for 18 hours with IFN-α2a and washed before incubation with the [125I]-IFN-α2b.
Immunoprecipitations and Western Blotting
Samples of 5 × 107 cells were sedimented and resuspended into 1 mL of an ice-cold lysis buffer as previously detailed.45 Microtubes were rotated end over end (1 hour) to ensure complete lysis. After sedimentation at 4°C for 30 minutes (12,000g), supernatant protein was normalized, specific antisera (2 to 3 μL/mL) were added (see Reagents), and samples were rotated as described above (2 hours). Immune complexes captured on protein A sepharose beads from Pharmacia LKB Biotech were eluted with a double-strength SDS-sample buffer45 and heat-denatured before protein resolution in 7.5% SDS-polyacrylamide gels. Proteins were transferred to Immobilon-P, using a Multiphor Novablot semidry unit (Pharmacia LKB Biotech). Membranes were incubated in a blocking buffer45 and exposed to primary antibodies. After reaction with monoclonal (4G10) anti-pTyr (1 μg/mL), or rabbit anti-STAT sera (1:1,000 to 1:10,000), blots were rinsed, reincubated in blocking buffer with matched HRP-conjugated secondary antibody (500 ng/mL), incubated for one minute in HRP-substrate mixture, and exposed to X-Omat film from Eastman Kodak (Rochester, NY). Some blots were developed with an alkaline phosphatase substrate kit.
Electrophoretic Mobility Shift Assay (EMSA)
Assays were performed as previously described33,45 and used double-stranded oligonucleotide probes end-labeled with [γ-32P]-ATP.33,45,53 The DNA probes corresponded to response elements in IFN-α–stimulated genes:IRF-1 pIRE IFN-γ activation sequence (GAS),545′-gatccatttccccgaaatga-3′; an FcγR high-affinity FcRFγ-binding response region (GRR),555′-agcatgtttcaaggatttgagatgtatttcccagaaaag-3′; and the ISGF3-binding ISRE of the ISG15 gene,565′-gatccatgcctcgggaaagggaaaccgaaactgaagcc-3′.33Samples of 5 × 107 cells were sedimented at 200g, transferred to 5 mL of a neutral HEPES buffer with protease inhibitors,45 and disrupted in an ice-cold dounce homogenizer. Centrifugation at 500g for 5 minutes separated a cytoplasmic supernatant. The nuclear pellet was further extracted in 600 μL of HEPES buffer45 with 33 μL of 5 mol/L NaCl and chilled on ice for 10 minutes. Centrifugation at 17,000gproduced a clarified supernatant from which samples of 10 μg protein were incubated with DNA probe (1 to 2 ng) for 10 minutes on ice in 30 μL of an EMSA binding cocktail45 and loaded into nondenaturing polyacrylamide gels (6%) with 0.25× TBE buffer (45 mmol/L Tris-HCl [pH 8.0], 45 mmol/L boric acid, 1 mmol/L EDTA) containing 5% glycerol (gels prerun at 280 V for 1.5 hours at 18°C). After an approximately 3-hour loaded run, gels were dried and exposed to X-Omat film. In reconstitution experiments, samples of nuclear extracts were alkylated with N-ethyl maleimide,33,40,53 quenched for 10 minutes with 20 mmol/L of dithiothreitol (DTT) on ice, and mixed with excess ISGF3γ-p48 that had been translated in vitro with cDNA (clone kindly supplied by D. Levy, New York University, New York, NY)40 in a TNT-reticulocyte lysate system from Promega (Madison, WI). For EMSA supershifts, samples were pretreated with antisera to specific STAT proteins or with 0.5 to 1 μL of preimmune rabbit serum as control.
RESULTS
IFN-α Binding, Cell Growth Inhibition, and Antiviral Activity
The generation time of parental Daudi cells (DWS) that are sensitive to growth inhibition was 26 ± 2.0 hours, and MTT assays showed that IFN-α (100 U/mL) reduced the culture growth by 60% to 80% within 48 hours. After 48 hours, culture growth was completely arrested for at least 4 days. In contrast, the paired variant Daudi cells (DWR) grew continuously in up to 5,000 U/mL of IFN-α, with a generation time of 31 ± 3.4 hours, and remained stably IFN-α-resistant for greater than 10 population doublings without maintenance in IFN-α. Scatchard analyses of specific [125I]-IFN-α2b binding to the variant DWR and parental DWS cells yielded similar numbers of IFNAR and ligand dissociation constants (kd) shown in Table 1. Receptor downregulation after 18 hours in IFN-α occurred with both cell types, but was more pronounced with the DWS cells (Table 1).
Quantitation of Daudi Cell IFNAR and Dissociation Constants (kd)
| Pretreatment . | DWS Cells . | DWR Cells . | ||
|---|---|---|---|---|
| IFNAR/ Cell . | kd (×10−10) . | IFNAR/ Cell . | kd (×10−10) . | |
| None | 8,620 | 5.5 | 8,940 | 5.2 |
| IFN-α2a-150 | 2,660 | 4.9 | 4,730 | 5.6 |
| Pretreatment . | DWS Cells . | DWR Cells . | ||
|---|---|---|---|---|
| IFNAR/ Cell . | kd (×10−10) . | IFNAR/ Cell . | kd (×10−10) . | |
| None | 8,620 | 5.5 | 8,940 | 5.2 |
| IFN-α2a-150 | 2,660 | 4.9 | 4,730 | 5.6 |
Average of two determinations calculated from Scatchard plots of competitive displacement binding with [125I]-IFN-α2b and cold IFN-α2b (see Materials and Methods).
100 U/mL for 18 hours.
Although the DWR cells originally had been selected on the basis of resistance to growth inhibition by IFN-α,10 they also had become resistant to an IFN-α antiviral activity. Table 2 shows that, whereas IFN-α2a was able to suppress measles virus infection in DWS cells, the identically treated DWR cells yielded significant titers both of supernatant-released and cell-associated virus at 48 hours. The DWR susceptibility to measles virus infection in the presence of IFN-α also was shown by an ICA that measured the percentage of cells actually infected by 48 hours and thus normalized for a relative reduction of DWS cell numbers due to IFN-α antiproliferative action (Table 2).
IFN-α2a Effect on Measles Virus Growth Compared in IFN-Sensitive (DWS) and IFN-Resistant (DWR) Daudi Cells
| Virus Assay IFN-α | Virus-Infected Cells | |||||||||||
| DWS | DWR | |||||||||||
| Supernate* | Cell† | ICA‡ | Supernate* | Cell† | ICA‡ | |||||||
| − | + | − | + | − | + | − | + | − | + | − | + | |
| Infectivity | 4.7 | 2.5 | 3.7 | 1.5 | 29 | 6 | 4.2 | 4.5 | 2.7 | 3.0 | 21 | 18 |
| % Inhibition | 99 | 99 | 70 | 0 | 0 | 14 | ||||||
| Virus Assay IFN-α | Virus-Infected Cells | |||||||||||
| DWS | DWR | |||||||||||
| Supernate* | Cell† | ICA‡ | Supernate* | Cell† | ICA‡ | |||||||
| − | + | − | + | − | + | − | + | − | + | − | + | |
| Infectivity | 4.7 | 2.5 | 3.7 | 1.5 | 29 | 6 | 4.2 | 4.5 | 2.7 | 3.0 | 21 | 18 |
| % Inhibition | 99 | 99 | 70 | 0 | 0 | 14 | ||||||
Cells incubated with or without IFNα2a (100 U/mL) for 24 hours before infection with Edmonston strain measles virus (1 TCID50/cell).
Log10 titers (TCID50 per milliliter) of virus released at 48 hours, detected by microtitration of culture fluids on VERO cells for 6 days.
Log10 titers (TCID50 per milliliter) of virus that was cell-associated at 48 hours, detected by microtitration of whole cell lysates on VERO cells for 6 days.
Infectious center assay of Daudi cells infected with measles virus for 48 hours. Data represent the ratio (×100) of virus plaques enumerated per number of the Daudi cells inoculated onto indicator cell (VERO) monolayers.
Activation of STAT1, STAT2, and Associated DNA-Binding Is Abbreviated in IFN-Resistant (DWR) Daudi Cells
Western blots.
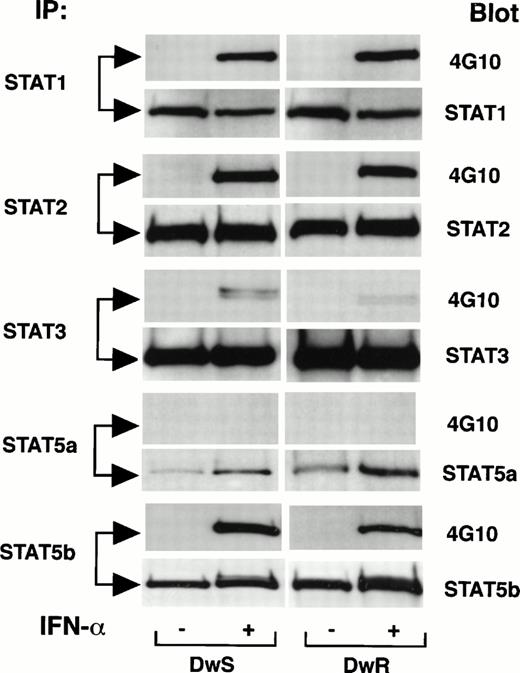
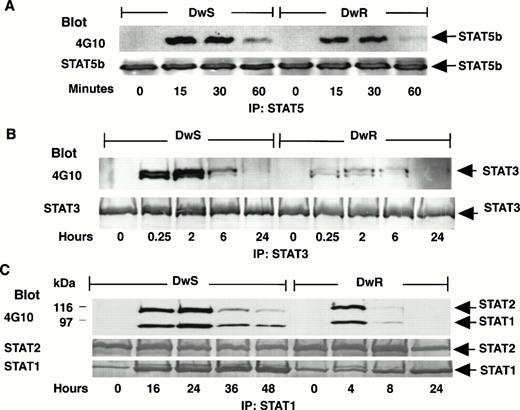
Expression of multiple STATs (1,2, 3, and 5) was abundant both in IFN-sensitive DWS and -resistant DWR cells as analyzed by Western blotting with specific anti-STAT antibodies. Figure 1shows that, with the exception of STAT5a and STAT3, tyrosine phosphorylation induced within 15 minutes after IFN-α treatment was robust in DWS and DWRcells. These results were comparable in a range of 50 to 1,000 U/mL of IFN-α (not shown). STAT3 became weakly phosphorylated with 100 IU/mL of IFN-α, but even at 1,000 U/mL phosphorylation in the DWR cells remained comparatively weak (not shown). As the IFN-α treatment was continued, differences also emerged in the durations of STAT phosphorylation. Figure 2 shows that, although the expression of each STAT protein remained abundant, phosphorylated isoforms (STAT-pTyr) of STAT5b (STAT5b-pTyr) decreased after just 1 hour (Fig 2A). STAT3-pTyr was attenuated by 6 hours (Fig 2B). In the resistant DWR cells, a similar pattern of decline was observed both for STAT1-pTyr and STAT2-pTyr; these were no longer detected after 4 to 8 hours. In the sensitive DWS cells, by contrast, the intensities of STAT2-pTyr and STAT1-pTyr remained stable for up to 24 hours and detectable up to 48 hours (Fig 2C). Gross imbalances in STAT2 or STAT1 availability or in proteolysis through a ubiquitin-proteosome pathway57 were excluded, because total amounts of these STATs remained equivalent to controls in Western blots reprobed with specific anti-STAT antibodies (Fig 2C).
Comparisons of initial tyrosine phosphorylation of STAT proteins induced by IFN-α in sensitive (DWS) and resistant (DWR) Daudi cells. At 15 minutes after identical treatment with IFN-α2a (100 U/mL), cells were extracted for concordant immunoprecipitation (IP) with specific STAT antibodies, gel electrophoresis, and Western blot. Patterns of the STAT-pTyr are shown with 4G10 blots developed by ECL. Total latent and phosphorylated STAT proteins were assessed by reprobing of transfer membranes with specific anti-STAT antibodies and secondary detection by ECL.
Comparisons of initial tyrosine phosphorylation of STAT proteins induced by IFN-α in sensitive (DWS) and resistant (DWR) Daudi cells. At 15 minutes after identical treatment with IFN-α2a (100 U/mL), cells were extracted for concordant immunoprecipitation (IP) with specific STAT antibodies, gel electrophoresis, and Western blot. Patterns of the STAT-pTyr are shown with 4G10 blots developed by ECL. Total latent and phosphorylated STAT proteins were assessed by reprobing of transfer membranes with specific anti-STAT antibodies and secondary detection by ECL.
Comparisons of the persistence of STAT tyrosine phosphorylation induced in sensitive (DWS) or resistant (DWR) Daudi cells continuously exposed to IFN-α (100 U/mL). As in Fig 1, STATs-pTyr were disclosed by IP and 4G10 Western blots developed with ECL substrate. (A) STAT5b-pTyr (92 kD) began to decrease after 1 hour both in sensitive (DWS) and resistant (DWR) cells. Anti-STAT5b was localized by ECL. (B) STAT3-pTyr (89 kD), as indicated in Fig 1, was more strongly phosphorylated in sensitive (DWS) than in resistant (DWR) cells, but, nevertheless, had declined in each cell line by 6 hours. Anti-STAT3 was localized by ECL. (C) STAT2-pTyr (113 kD) and STAT1-pTyr (91 kD) persisted for more than 24 hours in DWS cells, but decreased in DWR cells by 8 hours. Total latent and phosphorylated STATs were assessed by reprobing of transfer membranes with specific anti-STAT antibodies and secondary detection by ECL (STAT5b, STAT3) or phosphatase substrate reaction (STAT2, STAT1).
Comparisons of the persistence of STAT tyrosine phosphorylation induced in sensitive (DWS) or resistant (DWR) Daudi cells continuously exposed to IFN-α (100 U/mL). As in Fig 1, STATs-pTyr were disclosed by IP and 4G10 Western blots developed with ECL substrate. (A) STAT5b-pTyr (92 kD) began to decrease after 1 hour both in sensitive (DWS) and resistant (DWR) cells. Anti-STAT5b was localized by ECL. (B) STAT3-pTyr (89 kD), as indicated in Fig 1, was more strongly phosphorylated in sensitive (DWS) than in resistant (DWR) cells, but, nevertheless, had declined in each cell line by 6 hours. Anti-STAT3 was localized by ECL. (C) STAT2-pTyr (113 kD) and STAT1-pTyr (91 kD) persisted for more than 24 hours in DWS cells, but decreased in DWR cells by 8 hours. Total latent and phosphorylated STATs were assessed by reprobing of transfer membranes with specific anti-STAT antibodies and secondary detection by ECL (STAT5b, STAT3) or phosphatase substrate reaction (STAT2, STAT1).
EMSA.
A close relationship of the STAT1 and STAT2 phosphorylation kinetics to DNA binding activities was verified with nuclear extracts. Two types of oligonucleotide [32P]-labeled probes were used to detect STAT1 binding activity: (1) DNA probes that could be bound by STAT1 homodimers represented the GAS sequence upstream of IRF-1 that is strongly implicated in the antiproliferative and antiviral actions of IFN-α58,59 or the GRR region upstream ofFcγR involved in immune cell functions55; and (2) a DNA probe that was bound most effectively by the ISGF3 heterocomplex (STAT1-STAT2 heterodimer and ISGF3γ-p4832,39) represented the ISRE nucleotide enhancer element upstream of a large number of IFN-stimulated genes.34
Figure 3 shows that, with both types of DNA probes, binding by STAT1 at serial time points generally matched the kinetics of STAT1 tyrosine phosphorylation evident in Fig 2C. In EMSA with nuclear extracts from DWS cells (Fig 3A, top), binding of the IRF-1 GAS probe to STAT-pTyr persisted for up to 48 hours. With parallel extracts from DWRcells, DNA-binding was not detected after 6 hours. Comparable results were obtained with the FcγRγ GRR probe (not shown). With the ISG15 ISRE probe, DNA-binding activity in extracts from DWS cells was maximum at 16 to 32 hours and persisted for 48 hours (Fig 3A, bottom). With parallel extracts from DWR cells, DNA binding activity was negligible after 8 hours. To exclude variable expression of the ISGF3γ-p48 component of ISGF3 as a factor in these differences, parallel samples of nuclear extracts from both types of Daudi cells were alkylated to inactivate the ISGF3γ-p48 and then reconstituted with an excess of recombinant ISGF3γ-p48 protein.40 53EMSA results (not shown) remained consistent with the pattern for the ISRE probe shown in Fig 3A.
DNA-binding by STAT-pTyr proteins in nuclear extracts from resistant (DWR) and sensitive (DWS) Daudi cells. EMSA was performed using indicated [32P]-oligonucleotide probes. (A) After cell treatments with IFN-α (100 U/mL) for the times indicated, mobility shifts were evident for up to 48 hours with DWSextracts, but for a maximum of just 8 hours with DWR extracts. Probes represented either theIRF-1 GAS promoter sequence or an ISG15 ISRE promoter element. (B) The DWS or DWR cells were treated with IFN-α (100 U/mL) for 2 hours. STAT DNA-binding complexes were identified by incubation of nuclear extracts with or without specific rabbit antisera before EMSA with the IRF-1 GAS probe. Supershifts were confined to samples treated with anti-STAT1.
DNA-binding by STAT-pTyr proteins in nuclear extracts from resistant (DWR) and sensitive (DWS) Daudi cells. EMSA was performed using indicated [32P]-oligonucleotide probes. (A) After cell treatments with IFN-α (100 U/mL) for the times indicated, mobility shifts were evident for up to 48 hours with DWSextracts, but for a maximum of just 8 hours with DWR extracts. Probes represented either theIRF-1 GAS promoter sequence or an ISG15 ISRE promoter element. (B) The DWS or DWR cells were treated with IFN-α (100 U/mL) for 2 hours. STAT DNA-binding complexes were identified by incubation of nuclear extracts with or without specific rabbit antisera before EMSA with the IRF-1 GAS probe. Supershifts were confined to samples treated with anti-STAT1.
EMSA with the IRF-1 GAS probe further was conducted in the presence of antibodies specific for STATs 1, 3, or 5. Figure 3B shows that the GAS-binding STAT-pTyr complexes found in extracts from DWS or DWR cells after IFN-α treatment were supershifted only by anti-STAT1. The same result was obtained whether cells were extracted after exposure to IFN-α for 2 hours (Fig 3B) or for 24 hours (DWS cells only; results not shown). These findings were consistent with in vitro and transgenic mouse studies demonstrating a central role of STAT1 in the biologic actions of IFN-α.24,25,29 60
Similar Lability of STAT Tyrosine Phosphorylation Induced by IFN-α in DWS and DWRCells
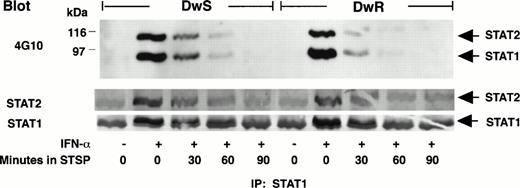
STSP is a multipotent inhibitor of protein tyrosine kinases and serine/threonine kinases61 and inhibits STAT phosphorylation.35,62 Figure 4shows that, in the continuous presence of IFN-α, STAT1-pTyr and STAT2-pTyr disappeared from whole cell extracts within 1 to 2 hours after treatment with STSP (250 nmol/L). Based on previous work, this can be ascribed to the unopposed action of nuclear phosphatase(s) that inactivates translocated STAT-pTyr.33 35 The STAT-pTyr downregulation kinetics were similar for DWSand DWR cells, and retention of total STAT protein during STSP treatment was comparable to controls (Figs 4 and5). These results, along with the spontaneous attenuation of IFN-α–induced STAT5 and STAT3 phosphorylation shown in Fig 2, indicated that the singular persistence of STAT1-pTyr and STAT2-pTyr in DWS cells need not reflect a generalize deficiency in phosphatase-driven downregulation.
Comparison of STAT1-pTyr and STAT2-pTyr phosphorylation downregulation in sensitive (DWS) and resistant (DWR) Daudi cells during tyrosine kinase inhibition. STSP (250 nmol/L) was added at 2 hours after cell treatment with 100 U/mL of IFN-α. Effects on STAT1-pTyr and STAT2-pTyr at 30-minute intervals were disclosed by IP, gel electrophoresis, and 4G10 Western blots developed with ECL. Total latent and phosphorylated STATs were assessed by reprobing of the transfer membranes with specific anti-STAT antibodies and secondary detection by phosphatase substrate reaction.
Comparison of STAT1-pTyr and STAT2-pTyr phosphorylation downregulation in sensitive (DWS) and resistant (DWR) Daudi cells during tyrosine kinase inhibition. STSP (250 nmol/L) was added at 2 hours after cell treatment with 100 U/mL of IFN-α. Effects on STAT1-pTyr and STAT2-pTyr at 30-minute intervals were disclosed by IP, gel electrophoresis, and 4G10 Western blots developed with ECL. Total latent and phosphorylated STATs were assessed by reprobing of the transfer membranes with specific anti-STAT antibodies and secondary detection by phosphatase substrate reaction.
Comparison of effects of pharmacologically diverse antagonists or medium change on the STAT1-pTyr and STAT2-pTyr induced in IFN-sensitive Daudi cells (DWS). After cell exposure to IFN-α2a (100 U/mL) for 18 hours, each antagonist was added to the medium for 1 or 2 hours as indicated. Alternatively, the cells were sedimented twice for a complete medium change (wash). Whole cell extracts were analyzed by IP, gel electrophoresis, and 4G10 Western blots. Total latent and phosphorylated STATs were assessed by reprobing of the transfer membranes with the specific anti-STAT antibodies and secondary detection by ECL.
Comparison of effects of pharmacologically diverse antagonists or medium change on the STAT1-pTyr and STAT2-pTyr induced in IFN-sensitive Daudi cells (DWS). After cell exposure to IFN-α2a (100 U/mL) for 18 hours, each antagonist was added to the medium for 1 or 2 hours as indicated. Alternatively, the cells were sedimented twice for a complete medium change (wash). Whole cell extracts were analyzed by IP, gel electrophoresis, and 4G10 Western blots. Total latent and phosphorylated STATs were assessed by reprobing of the transfer membranes with the specific anti-STAT antibodies and secondary detection by ECL.
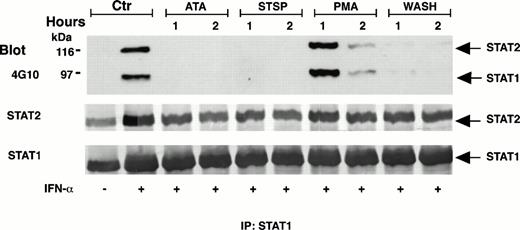
In further efforts to probe the relevance of a prolonged STAT1 and STAT2 DNA-binding capacity to the antiproliferative action of IFN-α, we found that phosphorylation of these STATs also could be interrupted promptly by two antagonists of IFN-α that were not intrinsically cytotoxic: PMA or ATA. PMA is a potent modulator of protein kinase C activity, and in a range of 5 to 20 nmol/L inhibits the antiproliferative action of IFN-α in Daudi cells without disrupting the cell cycle.63,64 ATA is a triphenylmethane derivative that forms polymers in solution65 and competes for the binding of IFN-α to IFNAR.66 Figure 5 shows that both PMA and ATA caused a rapid loss of STAT2-pTyr and STAT1-pTyr when added to DWS cells after 18 hours in the continuous presence of IFN-α. Earlier addition of these agents gave the same results, and ATA proved equally potent to STSP in abrogating the STAT tyrosine phosphorylation within 1 to 2 hours. As shown in Fig 5, PMA acted more slowly.
Antagonism of IFN-α–Induced Growth Inhibition of DWS Cells Is Associated With Deferred Interruptions of STAT1 and STAT2 Phosphorylation
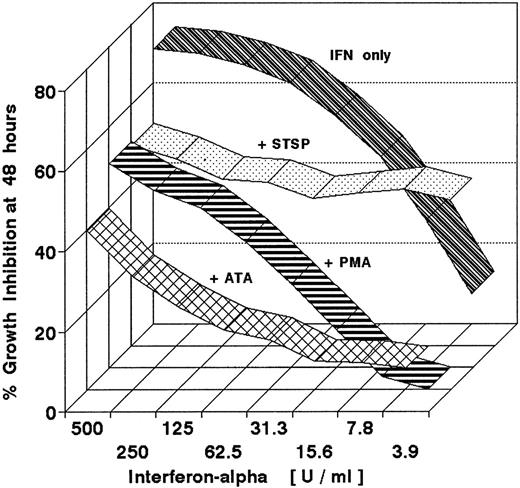
Figure 6 represents a series of experiments designed to test the effects of interrupted STAT1 and STAT2 phosphorylations on the growth inhibition of DWS cells. Each antagonist was added in a fixed concentration to cells treated with serial dilutions of IFN-α in 96-well plates for a predetermined period. IFN-α was not withdrawn, a strategy to ensure consistent measurements of cell growth in a stable culture milieu. Multiple replicate measurements were obtained at different agent ratios using highly reproducible MTT assays with minimal statistical variation (<5% in replicate wells). The end point for all experiments was 48 hours, because repeated experience showed that DWS cell cultures treated with IFN-α for this duration remained growth arrested for at least 4 days after IFN-α withdrawal. Although STSP is a potent inhibitor of multiple protein kinases and by itself was ultimately cytotoxic, its antagonism of STAT phosphorylation, demonstrated in Figs 4 and 5, nevertheless could be shown to interfere with the antiproliferative action of up to 500 U/mL of IFN-α within a useful time frame. Figure6 illustrates the effect when STSP was added as late as 18 hours after the initiation of STAT phosphorylation by IFN-α. It further shows that a deferred addition of essentially noncytotoxic PMA (10 nmol/L) or ATA (125 μmol/L) was similarly antagonistic.
Antagonistic effects of pharmacologically diverse agents on the growth-inhibitory action of IFN-α2a. Sensitive (DWS) Daudi cells were continuously exposed to serial twofold dilutions of IFN-α2a for 48 hours. Growth inhibition was measured by MTT assay. Ribbon graphs show antagonism when STSP (125 mmol/L), PMA (10 nmol/L), or ATA (125 μmol/L) were added to the IFN-α from 18 to 48 hours. Neither the PMA nor ATA themselves inhibited cell growth during this 30-hour period. STSP alone inhibited cell growth by 28%, but was nevertheless effective in decreasing the inhibitory action of IFN-α.
Antagonistic effects of pharmacologically diverse agents on the growth-inhibitory action of IFN-α2a. Sensitive (DWS) Daudi cells were continuously exposed to serial twofold dilutions of IFN-α2a for 48 hours. Growth inhibition was measured by MTT assay. Ribbon graphs show antagonism when STSP (125 mmol/L), PMA (10 nmol/L), or ATA (125 μmol/L) were added to the IFN-α from 18 to 48 hours. Neither the PMA nor ATA themselves inhibited cell growth during this 30-hour period. STSP alone inhibited cell growth by 28%, but was nevertheless effective in decreasing the inhibitory action of IFN-α.
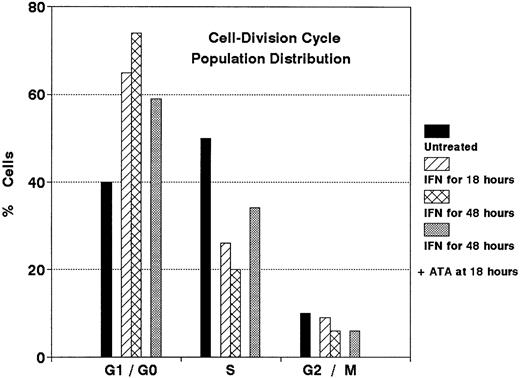
Figure 7 shows that ATA also antagonized the IFN-α arrest of DWS cell cycling. Whereas IFN-α induced a major population shift from exponential growth (with ∼50% of cells in S phase) to G1/G0 phase (with <25% of cells in S phase), an 18-hour deferred addition of ATA (125 μmol/L) initiated a reversal of this imbalance. However, after 24 hours, ATA only partially antagonized the growth inhibition induced by IFN-α (not shown). These effects of ATA may be explained by its ability to competitively displace bound [125I]-IFN-α2b from the IFNAR of DWS cells (Fig 8).
Antagonistic effect of ATA on the cell cycle redistribution induced by IFN-α in sensitive Daudi cells (DWS). Cells were treated with IFN-α (100 U/mL) for 18 hours or 48 hours. ATA (125 μmol/L) was added to samples indicated after 18 hours. Flow cytometric results represented in the DNA histogram show reversal of the G1/G0 shift induced by IFN-α.
Antagonistic effect of ATA on the cell cycle redistribution induced by IFN-α in sensitive Daudi cells (DWS). Cells were treated with IFN-α (100 U/mL) for 18 hours or 48 hours. ATA (125 μmol/L) was added to samples indicated after 18 hours. Flow cytometric results represented in the DNA histogram show reversal of the G1/G0 shift induced by IFN-α.
ATA effect on the binding of [125I]-IFN-α2b to sensitive (DWS) Daudi cells. At the concentrations indicated, ATA was added at 90 minutes after saturation of the IFNAR with [125I]-IFN-α2b (5,000 IU/mL at 4°C for 90 minutes) or at 18 hours before this saturation. Specific binding curves show both competitive binding of the ATA and displacement of bound [125]I-IFN-α2b (mean of duplicated experiments).
ATA effect on the binding of [125I]-IFN-α2b to sensitive (DWS) Daudi cells. At the concentrations indicated, ATA was added at 90 minutes after saturation of the IFNAR with [125I]-IFN-α2b (5,000 IU/mL at 4°C for 90 minutes) or at 18 hours before this saturation. Specific binding curves show both competitive binding of the ATA and displacement of bound [125]I-IFN-α2b (mean of duplicated experiments).
The Antiviral Action of IFN-α Is Less Sensitive Than Growth Inhibition to Deferred Signal Interruption
Pharmacologic actions of STSP or PMA have known direct effects on enzymes or cell functions essential to measles virus replication,67 68 so these antagonists were not considered appropriate for assessing whether interruption of STAT phosphorylation during IFN-α treatment would alter the progress of virus infection. We therefore relied on a delayed withdrawal of IFN-α or a deferred addition of ATA. Figure 5 shows that simple withdrawal of IFN-α and replacement of the growth medium caused a marked reduction of STAT1 and STAT2 phosphorylation. In the infectivity experiments, a delayed resumption of cell proliferation after medium changes was not an obstacle to the measles virus replication, and the ICA, which quantitates the proportion of infected cells, compensates for differences in overall virus production that might be influenced by changes in absolute cell number. Results in Table 3 show that, by the time of IFN-α withdrawal at 18 hours, a significant antiviral state had already developed. Table 4 similarly shows that ATA, which rapidly abrogated STAT phosphorylation (Fig 5) and antagonized the growth inhibition by IFN-α (Fig 6), exerted limited or no impact on the antiviral action of IFN-α at 18 hours.
Effect of Late Withdrawal of IFN-α on Antiviral Activity in IFN-Sensitive (DWS) Daudi Cells Infected With Measles Virus
| Experiment . | Supernatant Virus* . | Cell-Associated Virus* . | |||
|---|---|---|---|---|---|
| IFN-α . | Washed . | Titer . | Inhibition† . | Titer . | Inhibition† . |
| − | − | 5.0 | 4.2 | ||
| +‡ | − | 3.5 | 97% | 3.0 | 94% |
| + | +2-153 | 3.8 | 94% | 3.7 | 68% |
| Experiment . | Supernatant Virus* . | Cell-Associated Virus* . | |||
|---|---|---|---|---|---|
| IFN-α . | Washed . | Titer . | Inhibition† . | Titer . | Inhibition† . |
| − | − | 5.0 | 4.2 | ||
| +‡ | − | 3.5 | 97% | 3.0 | 94% |
| + | +2-153 | 3.8 | 94% | 3.7 | 68% |
Cells infected with measles virus (1 TCID50/cell).
Log10 virus titers (TCID50 per milliliter) of virus released at 48 hours. Culture fluids were microtitrated on VERO monolayers for 6 days.
Inhibition of viral yield, as compared with the controls without IFN-α.
Cells were incubated with IFN-α (100 U/mL) for 18 hours before and throughout virus infection.
After treatment with IFN-α for 18 hours, cells were washed, infected, and maintained in fresh medium without IFN-α.
ATA Reduction of IFN-α Antiviral Activity in IFN-Sensitive (DWS) Daudi Cells
| ATA3-150 @ Time . | Relative Reduction of IFN-α Activity (%) . | |
|---|---|---|
| Virus Assay . | ||
| Supernatant3-151 . | ICA3-152 . | |
| 4 h | 100 | 44 |
| 18 h | 59 | 0 |
| ATA3-150 @ Time . | Relative Reduction of IFN-α Activity (%) . | |
|---|---|---|
| Virus Assay . | ||
| Supernatant3-151 . | ICA3-152 . | |
| 4 h | 100 | 44 |
| 18 h | 59 | 0 |
Cells were incubated in growth medium with IFN-α (100 U/mL) for 24 hours, washed, and infected with the Edmonston strain measles virus (1 TCID50/cell).
ATA (125 μmol/L) was added to the medium with IFN-α at the times indicated.
Log10 virus titers (TCID50) of supernatant released virus at 48 hours after infection of the DWS cells. Yield of infectious virus was determined by microtitration on VERO cells for 6 days. In control DWS cells treated with ATA compared with or without IFN-α at 0 hours of infection, the ATA caused a reduction of 75% to 90% of the IFN-α antiviral activity. For experimental normalization, the 0 hours results define a 100% basis for all data shown in this table. The true effect of ATA added at 0 hours was a 75% reduction in antiviral activity (data not shown).
Data represent the ratio (×100) of viral plaques enumerated on monolayers of indicator cells (VERO) per number of infected Daudi cells inoculated.
DISCUSSION
As direct regulators of hematopoietic cell growth, IFNs-α,β can be advantageous for control of iatrogenic or virus-associated lymphoproliferations,3 and this cytokine family remains an important resource in adjunctive chemotherapy of some hematologic and epithelial malignancies.4,69 Present experiments have established the existence of a prolonged period during which interruption of STAT1 and STAT2 tyrosine phosphorylation by pharmacologically diverse antagonists could avert the growth arrest of malignant lymphoma cells sensitive to IFN-α. Striking differences in the duration of STAT1 activation in isogenic sensitive or resistant cells further suggested that a foreshortened duration of STAT1 tyrosine phosphorylation could be a resistance determinant in some malignancies. Nevertheless, present findings do not exclude an important role of other signal events. Just recently, STAT3 was implicated as a phosphatidylinositol 3-kinase adapter,70 and the relatively low intensity of STAT3 tyrosine phosphorylation induced by IFN-α in IFN-resistant DWR as compared with DWS cells could be an interdependent or independent regulatory factor that deserves further evaluation.
Sensitivity thresholds and amplitudes of the initial STAT1, STAT2, and STAT5b tyrosine phosphorylations, as determined by Western blots, were similar in DWS and DWRcells, and there were no major differences in phosphorylation half lives of these STAT-pTyr as indicated either by spontaneous attenuation or by downregulation after tyrosine kinase activity was inhibited with STSP (Figs 4 and 5). Indeed, the less than 2-hour phosphorylation half life of nuclear-localized STAT1-pTyr and STAT-2pTyr evidenced a continuous activity of nuclear tyrosine phosphatases,33 35 both in the IFN-sensitive and the IFN-resistant cells. It must be inferred that the net increases of STAT-pTyr or IFN-activated DNA binding observed for 16 to 32 hours in DWS cells (Figs 2C and 3A) reflect a perpetuated upstream activity of IFNAR-associated tyrosine kinases.
Relatively early decreases in levels of STAT5b-pTyr and STAT3-pTyr, as compared with STAT1-pTyr and STAT2-pTyr in the DWS cells, suggest some intrinsic and specific asymmetry in the equilibrium of protein tyrosine kinase and phosphatase functions. This could be at the level of receptor-associated molecular phosphorylations18,71; however, assays for SHPTP1 (PTP1C) and SHPTP2 (PTP1D) protein tyrosine phosphatases in the DWS and DWR Daudi cells showed no correlations with the observed differences in STAT1 phosphorylation duration and biologic responses (tested by M. David, personal communication, June 1997). Attempts to modulate phosphorylation responses by phosphatase inhibition with sodium pervanadate were thwarted by excessive cytotoxicity.
Experimental interruption of STAT phosphorylation and IFN-α actions by PMA and ATA proved to be novel and essentially innocuous means for modulating the IFN-α signal in sensitive Daudi cells. A recent investigation of peripheral blood monocytes indicated that PMA can shift the equilibrium of Tyk2 tyrosine phosphorylation by activating a yet obscure receptor-associated phosphatase72; however, identification of such an effect in DWS cells has not been possible. ATA once found major use as an inhibitor of translation in cell free systems,73 but its actions on ligand binding to receptors more recently have been of interest.55,65,74 Because ATA forms polyanionic polymers in solution65 and does not penetrate viable cells,73 we speculate that its strong negative charge may impede the functionally essential dimerization of IFNAR subunits. The ATA reversal of cell cycle changes induced by IFN-α was particularly striking. Conceivably, shifts by ATA in the IFNAR binding equilibrium of IFN-α (Fig 8) may interrupt IFN-α action by displacing ligand from occupied IFNAR or by inhibiting IFN-α ligation to new IFNAR as they regenerate after downregulation. Whereas numbers and IFN-α binding affinity of IFNAR proved similar in DWSand DWR cells (Table 1), IFNAR downregulation occurred to a greater extent in the DWS cells. This was paradoxical, because receptor downregulation can be a mechanism for signal desensitization. Its relationship to ATA effects and observed differences in the membrane fluidity46 or cytoskeletal anchoring75 of IFNAR in cells sensitive or resistant to IFN-α is uncertain.
That DWR cells, isolated for resistance to IFN-α antiproliferative action,10 also proved highly resistant to IFN-α protection during measles virus infection reaffirmed prior evidence for a close linkage between the antiproliferative and antiviral mechanism of IFN-α action,29,60,76 yet even in isogenic Daudi cells the relationship was not simple. Whereas the antiviral action of IFN-α in DWS cells appeared to be less dependent than growth inhibition upon prolonged STAT activation, the STAT1-pTyr in DWR cells also was competent in binding to anIRF-1 promoter element implicated both in antiviral and antiproliferative actions of IFN-α.58,59 The 2′-5′ oligoadenylate system, which is a key mediator of antiviral actions,76 also can be expressed in IFN-resistant cells.6-8,12 Thus, it is possible that a subtle change in the DNA-binding functions of STAT1-pTyr, such as described with the gene 6-16,12 might restrain induction of some biologically critical gene expression in the DWR cells.
Although the precise mechanism of the relatively premature loss of initially robust STAT2 and STAT1 tyrosine phosphorylations in the IFN-resistant DWR cells remains to be resolved, the results with IFN-sensitive DWS cells implicate a temporal dimension of signal transduction as an integral component of cytokine specificity. This needs to be weighed along with other cell-dependent determinants of biologic action, such as receptor density or signal amplitude. Observations that a protracted cell exposure to IFN can be necessary for full expression of biochemical or biological effects41-44 previously were explained by a short half life of some IFN-induced proteins.77 Therefore, it is intriguing that the STAT phosphorylations in DWR terminated before completion of the 18-hour period of continuous STAT1 activation associated with growth arrest of the IFN-sensitive DWS cells. Because bifunctional STAT molecules appear designed to mediate IFN signal transduction and transcriptional activation of immediate early genes particularly rapidly and efficiently,13,15 present evidence of a sustained period of STAT activation related to the antiproliferative action of IFN-α raises the possibility that persistent regulation of some immediate early gene functions by STAT-pTyr may be biologically imperative. Recently, STAT1 activated by IFN-γ78 was reported to be involved in regulation of the G1/S checkpoint inhibitor p21waf1/cip1. One possible inference is that continuous generation of activated STAT1 might be required to sustain the function of checkpoint controls for a period sufficient to stabilize the G0 status demonstrated by Tiefenbrun et al.8 This possibility invites further investigation.
ACKNOWLEDGMENT
The authors thank Dr Andrew Larner, Dr Michael David, and Dr Jeffrey Harmon for helpful discussions during this investigation.
Supported in part by Uniformed Services University of the Health Sciences Grant No. CO-74-EG.
Address reprint requests to Philip M. Grimley, MD, Department of Pathology, USUHS, 4301 Jones Bridge Rd, Bethesda, MD 20814.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
This is a US government work. There are no restrictions on its use.

![Fig. 3. DNA-binding by STAT-pTyr proteins in nuclear extracts from resistant (DWR) and sensitive (DWS) Daudi cells. EMSA was performed using indicated [32P]-oligonucleotide probes. (A) After cell treatments with IFN-α (100 U/mL) for the times indicated, mobility shifts were evident for up to 48 hours with DWSextracts, but for a maximum of just 8 hours with DWR extracts. Probes represented either theIRF-1 GAS promoter sequence or an ISG15 ISRE promoter element. (B) The DWS or DWR cells were treated with IFN-α (100 U/mL) for 2 hours. STAT DNA-binding complexes were identified by incubation of nuclear extracts with or without specific rabbit antisera before EMSA with the IRF-1 GAS probe. Supershifts were confined to samples treated with anti-STAT1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/8/10.1182_blood.v91.8.3017.3017_3017_3027/5/m_blod4083103.jpeg?Expires=1769149572&Signature=e654s8cBFjevTVEl8txzhMI~tU6h2pi2o4iMIITm2t7ps3MpnivLVeaVcGbD4eBOOeu59aF4-8FFlRBXIw9Xn2BoAPuFcl6CH9Byvf7TwYbK4MEcBpVWQVt1GhCpK9EeGKaimMu6MlmvDlWUNlwJ1IMxV3i2JzdBqX5MjKS~3vu78F50lJfIWdN2maHpIv2cjiFu6yx8AFV5Yk4qkrhxM-1CpYy4pTlfOyUSPp4IlY41vCrkxr7YetPO5L~vJbeJgWl53uhorJxpGrCZAdPZ3mhgdyXNK6jbQvJqrhiCD~iul8DjSfU9F48j46a8cVzGTDGlYnbNpe3yKI6cErQiCA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 8. ATA effect on the binding of [125I]-IFN-α2b to sensitive (DWS) Daudi cells. At the concentrations indicated, ATA was added at 90 minutes after saturation of the IFNAR with [125I]-IFN-α2b (5,000 IU/mL at 4°C for 90 minutes) or at 18 hours before this saturation. Specific binding curves show both competitive binding of the ATA and displacement of bound [125]I-IFN-α2b (mean of duplicated experiments).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/8/10.1182_blood.v91.8.3017.3017_3017_3027/5/m_blod4083108.jpeg?Expires=1769149572&Signature=aKhdBDB~3CE~DeON8bxuMYRDW-3AouKAr83YhJPVLGl1r6TqQ4MXB69nK0RSBTJxO6hW~Qbq8jypojvznTvFx7SM-Tlpp1I~-MSnbvknDPQArpLakSnwO2ydzOF3B08m3h9JcxxuFTYWb9SeBpgr9tvpue4Ji897-386J~xyvXTKtty1ncVnx9~mz3PwJALKOnRYcOv2lhdMP~wUjbbbcQvr0W9MyB9005TgsDLZ1Tz7RNPTLPivI0fgJ-ABHxs0ILte4BxqGnj6-i1DOt0syjzpu4PogfRBq~m5tvWhsGt8jvqW-ezVVk4JSTjeJ7~~e4BmHEBgvSCwuiR4zINTvw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal