Clonal chromosomal changes in multiple myeloma (MM) and related disorders are not well defined, mainly due to the low in vivo and in vitro mitotic index of plasma cells. This difficulty can be overcome by using comparative genomic hybridization (CGH), a DNA-based technique that gives information about chromosomal copy number changes in tumors. We have performed CGH on 25 cases of MM, 4 cases of monoclonal gammopathy of uncertain significance, and 1 case of Waldenstrom's macroglobulinemia. G-banding analysis of the same group of patients demonstrated clonal chromosomal changes in only 13 (43%), whereas by CGH, the number of cases with clonal chromosomal gains and losses increased to 21 (70%). The most common recurrent changes detected by CGH were gain of chromosome 19 or 19p and complete or partial deletions of chromosome 13. +19, an anomaly that has so far not been detected as primary or recurrent change by G-banding analysis of these tumors, was noted in 2 cases as a unique change. Other recurrent changes included gains of 9q, 11q, 12q, 15q, 17q, and 22q and losses of 6q and 16q. We have been able to narrow the commonly deleted regions on 6q and 13q to bands 6q21 and 13q14-21. Gain of 11q and deletion of 13q, which have previously been associated with poor outcome, can thus be detected by CGH, allowing the use of this technique for prognostic evaluation of patients, without relying on the success of conventional cytogenetic analysis.
MULTIPLE MYELOMA (MM) is a malignancy of clonal plasma cells with a wide variability in clinical features, responses to treatment, and survival times among patients. Although it accounts for 10% of hematologic malignancies, it represents less than 1% of chromosomally abnormal hematologic disorders reported.1-6 This lack of correlation between incidence and information on chromosomal changes in MM is due to the low mitotic index of plasma cells that reduces the availability of analyzable metaphases. Clonal chromosomal changes have been detected in approximately 40% of MM cases; and they show clustering of rearrangement breakpoints at bands 14q32, 16q11, and 22q11.6 In addition, duplication of the long arm of chromosome 1 and deletions affecting the long arms of chromosomes 6 and 13 have been frequently noted.2-6
The usefulness of karyotypic analysis in the prognostic evaluation of MM patients has been recently studied.5-7 Univariate and multivariate survival analyses have shown that hypodiploidy, partial or complete deletion of chromosome 13, and abnormalities of 11q and 22q have been significantly associated with an adverse outcome.7,8 However, the potential value of cytogenetic analysis is limited to the subset with karyotypic data. The molecular cytogenetic technique comparative genomic hybridization (CGH) enables identification of chromosomal copy number changes in tumors without the need to perform conventional cytogenetic analysis.9-11 This approach can thus be applied to all MM cases to obtain the prognostically relevant chromosome gain/loss information. We used CGH to analyze chromosome copy number changes in a panel of 30 patients with either MM, monoclonal gammopathy of uncertain significance (MGUS), or Waldenstrom's macroglobulinemia (WM). We found that 70% of the cases showed clonal changes that included aberrations previously noted to be of prognostic value as well as those so far not identified as recurring changes in MM.
MATERIALS AND METHODS
Tumor ascertainment and cytogenetics.
Bone marrow samples were collected from 30 patients, 25 with MM, 4 with MGUS, and 1 with WM. Of these, 17 were ascertained at the University Clinic of Navarra (UCN; Pamplona, Spain), and the remaining 13 were ascertained at the Memorial Sloan-Kettering Cancer Center (MSKCC; New York, NY). The samples were derived from 22 at diagnosis and 8 previously treated patients (Table 1). The ages of the patients ranged from 33 to 77 years, with a median of 59 years. Among the 30 patients, 16 were women and 14 were men. Cytogenetic analysis at UCN included unstimulated short-term and B-cell–stimulated 48-hour cultures,12 whereas cytogenetic analysis at MSKCC was performed on unstimulated short-term cultures of biopsy samples. G-banding by standard procedures was used in both laboratories. Karyotypes of cases no. 16, 17, and 19 were previously reported.6
Treatment Status, Percentage of Plasma Cells in Bone Marrow, and G-Banded and CGH Karyotypes of Tumor Samples Studied
| Tumor No. . | % Plasma Cells in Bone Marrow . | G-Banded Karyotype . | Loss and Gain Karyotype . |
|---|---|---|---|
| MM untreated | |||
| 02 | 81 | 50-52,+5,del(6)(q15q23),+9,+11×2,+15, −16,+17,+19,−20[cp10]/46,XY[5] | +5,−6q21-qter,+9,+11,+15q23-qter,−16q,+19,−20q |
| 03 | 30 | 76-79,XXX,+1,+3,+6,del(7)(p15),+9, del(9)(p13), del(9)(q22), +10,+11, del(11)(q23),−13,−14, −15,−17,+18,+19,−20,del(20)(q11.2), del(22)(q11.2)[cp5]/46,XX[13] | +5p15,−11q21-qter,−13q21-qter,+19p |
| 04 | 35 | 46,XY | +19 |
| 05 | 31 | 46,XY | +1p21-31,+4,+5q21,+6q,+8q21-24,+11q21-23, +13q21-qter,+18q |
| 06 | 54 | 46,XX,del(12)(p13)[5]/46,XX [20] | −1p31-pter,+3,−4q,−13q11-14,−17p |
| 11 | 63 | 46,XY | None |
| 12 | 30 | 46,XY | None |
| 16 | 98 | 46,XX,del(16)(q22)[27]/46,XX [13] | +9,−13 |
| 18 | 60 | 46,XX | −6q21,+9q,+11q13,−13q21-22,+15,+19 |
| 21 | 2 | 46,XX | None |
| 22 | 47 | 44,XY,−16,−17[5]/46,XY[15] | +1p36,+12q24,−16q11-22,+17q,+19p,−Y |
| 25 | 3 | 46,XX | None |
| 26 | 74 | 46,XY,der(14)t(11;14)(q13;q32)[25] | +11q13-qter,−16q |
| 27 | 80 | 45,X,−Y[3]/46,XY[21] | +12q24,−13q14-32,+17,−18,+19,+22,−Y |
| 28 | 20 | 46,XY | +12q24,+22q11-qter,−Y |
| 29 | 15 | 46,XX | None |
| 30 | 16 | 46,XY,del(11)(q23),der(14)t(11;14)(q13;q32)[7]/46,XY[13] | +11q13-21 |
| MM treated | |||
| 01 | 85 | 81-83,X,−X×3,der(14)t(11;14)(q13;q32),inc[cp10] | +4p,+6p,+11q23,−13,+15q24-qter,−16q,−17p, +17q21 |
| 07 | 90 | 45,X,−X[5]/46,XX[23] | +19 |
| 08 | 62 | 46,XX | +1p31-pter,+1q21,−6q12-21,+16p,+17,+19,+22 |
| 10 | 80 | 46,XX | None |
| 14 | 98 | 46,XX | −3,−13q22 |
| 17 | 90 | 46,XX,add(4)(p15),del(10)(q22),der(14)t(14;7?) (q32;q?)[25] | −4p,+7q31-qter,−8p |
| 19 | 90 | 46-47,XY,del(1)(q21),add(14)(q32),inc[32] | +1q24-25,+8q24,+11q13-14,−13q21-qter,−16q22-23 |
| 24 | 51 | 46,XY | None |
| MGUS | |||
| 09 | 47,XX,+mar[7]/46,XX[22] | −1,−2,−4p,−5p15,−9p23-pter,−13q14-qter,+19 | |
| 13 | 46,XX | None | |
| 15 | 46,XX | −4q23-28,−6q23 | |
| 20 | 46,XX | None | |
| WM | |||
| 23 | 46,XY | −4q33-qter |
| Tumor No. . | % Plasma Cells in Bone Marrow . | G-Banded Karyotype . | Loss and Gain Karyotype . |
|---|---|---|---|
| MM untreated | |||
| 02 | 81 | 50-52,+5,del(6)(q15q23),+9,+11×2,+15, −16,+17,+19,−20[cp10]/46,XY[5] | +5,−6q21-qter,+9,+11,+15q23-qter,−16q,+19,−20q |
| 03 | 30 | 76-79,XXX,+1,+3,+6,del(7)(p15),+9, del(9)(p13), del(9)(q22), +10,+11, del(11)(q23),−13,−14, −15,−17,+18,+19,−20,del(20)(q11.2), del(22)(q11.2)[cp5]/46,XX[13] | +5p15,−11q21-qter,−13q21-qter,+19p |
| 04 | 35 | 46,XY | +19 |
| 05 | 31 | 46,XY | +1p21-31,+4,+5q21,+6q,+8q21-24,+11q21-23, +13q21-qter,+18q |
| 06 | 54 | 46,XX,del(12)(p13)[5]/46,XX [20] | −1p31-pter,+3,−4q,−13q11-14,−17p |
| 11 | 63 | 46,XY | None |
| 12 | 30 | 46,XY | None |
| 16 | 98 | 46,XX,del(16)(q22)[27]/46,XX [13] | +9,−13 |
| 18 | 60 | 46,XX | −6q21,+9q,+11q13,−13q21-22,+15,+19 |
| 21 | 2 | 46,XX | None |
| 22 | 47 | 44,XY,−16,−17[5]/46,XY[15] | +1p36,+12q24,−16q11-22,+17q,+19p,−Y |
| 25 | 3 | 46,XX | None |
| 26 | 74 | 46,XY,der(14)t(11;14)(q13;q32)[25] | +11q13-qter,−16q |
| 27 | 80 | 45,X,−Y[3]/46,XY[21] | +12q24,−13q14-32,+17,−18,+19,+22,−Y |
| 28 | 20 | 46,XY | +12q24,+22q11-qter,−Y |
| 29 | 15 | 46,XX | None |
| 30 | 16 | 46,XY,del(11)(q23),der(14)t(11;14)(q13;q32)[7]/46,XY[13] | +11q13-21 |
| MM treated | |||
| 01 | 85 | 81-83,X,−X×3,der(14)t(11;14)(q13;q32),inc[cp10] | +4p,+6p,+11q23,−13,+15q24-qter,−16q,−17p, +17q21 |
| 07 | 90 | 45,X,−X[5]/46,XX[23] | +19 |
| 08 | 62 | 46,XX | +1p31-pter,+1q21,−6q12-21,+16p,+17,+19,+22 |
| 10 | 80 | 46,XX | None |
| 14 | 98 | 46,XX | −3,−13q22 |
| 17 | 90 | 46,XX,add(4)(p15),del(10)(q22),der(14)t(14;7?) (q32;q?)[25] | −4p,+7q31-qter,−8p |
| 19 | 90 | 46-47,XY,del(1)(q21),add(14)(q32),inc[32] | +1q24-25,+8q24,+11q13-14,−13q21-qter,−16q22-23 |
| 24 | 51 | 46,XY | None |
| MGUS | |||
| 09 | 47,XX,+mar[7]/46,XX[22] | −1,−2,−4p,−5p15,−9p23-pter,−13q14-qter,+19 | |
| 13 | 46,XX | None | |
| 15 | 46,XX | −4q23-28,−6q23 | |
| 20 | 46,XX | None | |
| WM | |||
| 23 | 46,XY | −4q33-qter |
CGH.
Tumor DNA was extracted from bone marrow samples and subjected to CGH analysis essentially as described.11 Briefly, the tumor (test) and normal (reference) DNAs were labeled by nick-translation with fluorescein-12-dUTP and Texas Red-5-dUTP (NEN-DuPont, Boston, MA), respectively. Equal amounts (200 ng) of tumor and normal DNAs were coprecipitated with 10 mg of human Cot-1 DNA (GIBCO/BRL, Gaithersburg, MD) and resuspended in the hybridization mix before in situ hybridization to human metaphase chromosome spreads prepared from phytohemagglutinin-stimulated lymphocytes from normal individuals. After hybridization, the slides were washed and the chromosomes were counterstained with 4,6-diamino-2-phenylindole (DAPI) to enable identification of the chromosomes. Fluorescent hybridization signals and DAPI-staining patterns were captured with a cooled charge-coupled device (CCD) camera (Photometrics, Tuscon, AZ) attached to a Nikon Microphot-SA microscope and processed using an image analysis system (Quips, Vysis, IL). The software performed a calculation of the green (tumor DNA) to red (normal DNA) fluorescent ratios along the length of each chromosome. The average of readings from eight chromosomes were graphed for each chromosome and compared with the profile for the same chromosome in a reference DNA/reference DNA hybridization to set the boundaries of gain and loss. Ratios greater than 1.20 and less than 0.80 were considered to represent chromosomal gain and loss, respectively. These threshold levels were tested as reported previously by us.12 CGH detects DNA sequence copy number relative to the average copy number in the tumor but not the ploidy level of the tumor.10 Therefore, the ploidy level of the tumor in this study was determined by conventional cytogenetics (Table 1). Chromosomal regions near the centromeres of 1, 9, 13-16, 21, and 22 were not scored for CGH analysis because of the highly repeated sequences in these regions. In the present study, control DNA was always obtained from a male donor, and the ratios for gains and losses of sex chromosomes were appropriately adjusted when test DNA was not matched. Recurrence of a change was defined by its presence in 2 or more tumors.
RESULTS
G-banded cytogenetic analysis.
Thirteen of the 30 (43%) patients showed clonal chromosomal abnormalities by G-banding (Table 1). Karyotypes were hyperdiploid (30%), pseudodiploid (47%), or hypodiploid (23%). A 14q+ marker chromosome was detected in 5 cases (38%); in 3 of them, it was derived from the t(11;14)(q13;q32) translocation. Among the remaining chromosomal abnormalities seen, recurrent changes comprised del(11) and del(16) in 3 cases each and −X and +1q in 2 cases each.
CGH analysis.
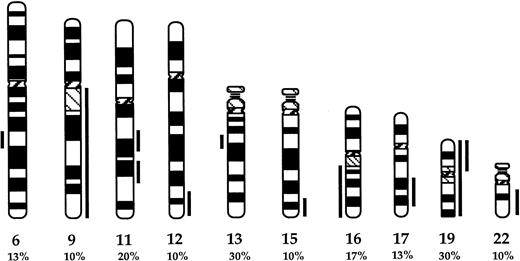
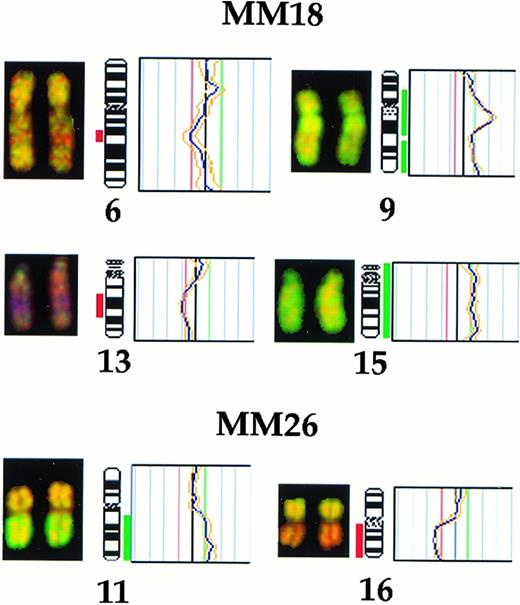
The proportion of patients with chromosomal aberrations increased to 21 by CGH (70%; Table 1). The clonally abnormal cases included 18 MM, 2 MGUS, and the single WM cases. The proportion of bone marrow plasma cells in cases that were clonally abnormal by CGH ranged between 16% to 98%. Overall, 10 chromosomal changes were recurrent (Fig 1). The most frequent changes were complete or partial deletions of chromosome 13 and gain of chromosome 19 or 19p (30%). Other frequent losses were complete or partial deletions of 6q (13%) and 16 (17%), with the commonly deleted regions spanning bands 6q21 and 13q14-21 (Fig 1). The partial karyotypes displayed in Fig 2 show representative losses of 13q, 6q, and 16q. None of the cases studied showed any of the recurrent losses detected to be unique aberrations (Table 1). Recurrent gains included partial gains of 11q (20%) and, less frequently (∼10%), gains of 9q, 12q24, 15q23-qter, 17q22-24, and 22q. Among gains, 2 were unique changes: +19 in cases no. 4 and 7 and +11q13-21 in case no. 30. Representative partial karyotypes of cases no. 18 and 26, showing +9, +11q, and +15, are shown in Fig 2.
Partial ideogram showing recurrent DNA copy number changes detected by CGH in the MM cases studied. Vertical lines on the right and left of each chromosomal ideogram identify gains and losses, respectively. The proportions of cases showing each of the changes are noted. For chromosomes 6, 9, 11, 13, 15,17, and 22, the lines delineate only the commonly gained or deleted regions. For chromosome 19, both complete and partial gains represented were recurrent.
Partial ideogram showing recurrent DNA copy number changes detected by CGH in the MM cases studied. Vertical lines on the right and left of each chromosomal ideogram identify gains and losses, respectively. The proportions of cases showing each of the changes are noted. For chromosomes 6, 9, 11, 13, 15,17, and 22, the lines delineate only the commonly gained or deleted regions. For chromosome 19, both complete and partial gains represented were recurrent.
Partial CGH karyotypes (left) and corresponding ratio profiles (right) observed in cases no. MM18 and MM26. Case no. MM18 showed deletions of chromosomes 6q21 and 13q14-22 and gains of 9q and chromosome 15. Case no. MM26 showed gain of 11q and loss of 16q. Hybridized tumor DNA was visualized via fluorescein isothiocyanate (green) and control DNA was visualized via Texas Red (red). The averaged green to red fluorescent signal ratio along the length of the chromosome is shown. The blue line in the ratio profile represents the mean of 8 to 10 chromosomes and the yellow line represents the standard deviation. The vertical red and green bars on the right of the ideogram indicate threshold values of 0.8 and 1.20 for loss and gain, respectively.
Partial CGH karyotypes (left) and corresponding ratio profiles (right) observed in cases no. MM18 and MM26. Case no. MM18 showed deletions of chromosomes 6q21 and 13q14-22 and gains of 9q and chromosome 15. Case no. MM26 showed gain of 11q and loss of 16q. Hybridized tumor DNA was visualized via fluorescein isothiocyanate (green) and control DNA was visualized via Texas Red (red). The averaged green to red fluorescent signal ratio along the length of the chromosome is shown. The blue line in the ratio profile represents the mean of 8 to 10 chromosomes and the yellow line represents the standard deviation. The vertical red and green bars on the right of the ideogram indicate threshold values of 0.8 and 1.20 for loss and gain, respectively.
Comparison of abnormalities detected by G-banding versus CGH in the cases in which both types of data were available (Table 1) showed a variation in the degree of concordance between the two methods of analysis. A subset of 6 cases (1, 2, 3, 17, 26, and 30) showed a high level of concordance between the abnormalities detected by the two techniques; in contrast, the remaining 7 cases (6, 7, 9, 16, 19, 22, and 27) showed discrepancy in almost every single abnormality. In addition, the types of abnormalities detected in different proportions of cases also seemed to depend on the technique used. Thus, rearrangements of 11q were observed by G-banding in 5 of the 6 cases that showed this anomaly by CGH; in contrast, only 1 case with abnormal karyotype by G-banding showed monosomy 13 compared with 9 cases in which this anomaly was detected by CGH.
DISCUSSION
Cytogenetic analysis of MM and related disorders has so far been hampered by the low proliferative activity of plasma cells, with abnormal karyotypes having been noted in 30% to 50% of cases.2-6,13 Interphase fluorescence in situ hybridization studies using centromeric probes have shown numerical chromosomal aberrations in 80% to 90% of cases, suggesting that clonal chromosomal abnormalities are frequent in these disorders.14,15 Therefore, we reasoned that application of CGH would provide new information on gains and losses of chromosomal regions in MM and related disorders, because this technique is performed using small amounts of DNA rather than cultured cells. In the present study, 70% of tumors showed clonal chromosomal changes. Thus, CGH was more sensitive in detecting chromosomal copy number changes than conventional G-banding. The 6 cases (12, 13, 20, 21, 25, and 29) that did not show clonal chromosomal abnormalities by CGH shared some features: they were obtained at diagnosis, the plasma cell infiltration in them was either less than 30% or they were diagnosed as MGUS, and they showed normal karyotype by G-banding analysis. We have previously described a similar association between absence of cytogenetic aberrations and lack of prior treatment or less than 30% plasma cell infiltration.6 12 A possible explanation for this association, and thus the trend seen in the CGH results, is that at the early stages of the disease, genetic changes may not involve recognizable chromosomal alterations. However, cases no. 10, 11, and 24, which presented a high proportion of plasma cells in the bone marrow, are not compatible with this explanation. These tumors may carry a chromosomal translocation(s) and/or small deletion (<2 kb) that cannot be detected by CGH.
Overall, we observed a variable correlation between the results of G-banding and CGH analyses. Thus, we noted 10 recurrent changes (Fig1), all of which have been previously described by us and others in MM.1-6 13 We have identified 9 cases (30%) that showed total or partial gains of chromosome 19 by CGH. This change has been one of the most frequent in our series and it has been noted as a unique anomaly in 2 cases. However, by G-banding, this change was detected only in 2 cases. The significance of this trisomy in MM development remains to be elucidated. Partial loss of chromosome 13 (also seen in 30% of the cases) was missed by conventional cytogenetic analysis in 8 of the 9 cases in which it was detected by CGH. We found some other discrepancies also in the results of the 13 cases in which both techniques were successful (Table 1). These discrepancies can be explained by the fact that G-banding analysis is based on the study of chromosomes of the clone proliferating in vitro, which may or may not be the predominant tumor clone, and hence may or may not be representative of the CGH result.
The CGH analysis allowed us to narrow the chromosome 13 deletion to a common region that spans the bands 13q14-21. The same region has previously been reported to be frequently deleted in chronic lymphocytic leukemia (CLL) and in 1 case of MM.16-18 Therefore, the candidate tumor-suppressor gene proposed in CLL and other lymphoid malignancies may be involved in the genesis and/or progression of MM as well. In the same way, deletions affecting 6q have been reported in MM and other lymphoid malignancies.19 We have identified the common region of deletion at 6q21, a region that has also been identified in a subset of non-Hodgkin's lymphoma by G-banding and LOH studies.20Finally, 16q loss has been observed in a subset of patients in our series; del(16)(q22) has previously been reported as unique change in MM.6 Therefore, candidate tumor-suppressor genes at 6q21 and 16q22 may be of importance in the pathogenesis of MM.
Using CGH, we have shown that 70% of MM biopsies present recurrent chromosomal gains and losses. These included deletions of 13q and gains of 11q that have previously been associated with a poor outcome.7 8 These abnormalities can be easily detected by CGH, without relevance to G-banding analysis, thus providing a valuable approach to identifying prognostically significant lesions using small amounts of DNA from nondividing cells.
Supported by Grants No. CA-34775 and CA-66999 from the National Institutes of Health/National Cancer Institute (to R.S.K.C.).
Address reprint requests to R.S.K. Chaganti, PhD, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10021.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal