Abstract
We studied the Hga I polymorphism (46 C/T) in the 5′-untranslated region of the coagulation factor XII (FXII) gene corresponding to four bases upstream from the ATG translation initiation codon. By using allele-specific restriction analysis with restriction endonuclease Hga I, the allele frequency of 46C/T was estimated to be 0.27/0.73 in Orientals (allele number =152), and conversely, 0.8/0.2 in Caucasians (allele number =40). Because it has been reported that plasma levels of FXII were lower in Orientals than in Caucasians, we investigated the relationship between this polymorphism and plasma levels of FXII. As a result, there were significant differences in plasma FXII levels between these three allele types: C/C,170±38% (178±27%); C/T, 141±29% (123±34%); and T/T, 82±19% (61±11%) [FXII activity (FXII antigen levels)]. In heterozygotes of 46 C/T both alleles were equally transcribed in hepatocytes, as determined by reverse transcription polymerase chain reaction (RT-PCR), suggesting little influence of the polymorphism at the level of transcription or on the stability of mRNA. In in vitro transcription/translation analysis, less FXII was produced from cDNA containing 46 T than from that containing 46 C. Therefore, it is highly likely that the 46 T polymorphism in the FXII gene decreased the translation efficiency and led to low plasma levels of FXII activity and antigen, probably due to the creation of another ATG codon and/or impairment of the consensus sequence for the translation initiation scanning model.
PLASMA COAGULATION factor XII (FXII) is a serine protease precursor that is primarily produced and secreted by hepatocytes and is involved in the initiation of the intrinsic coagulation pathway, in fibrinolysis, in the generation of bradykinin, and in the complement system.1 FXII is composed of COOH terminal catalytic domain, two fibronectin-type domains, an epidermal growth factor domain, and a kringle domain that is a component found in most proteins involved in the fibrinolysis system.2 Several patients with inherited FXII deficiency were identified mainly through findings of prolonged activated partial thromboplastin time (APTT) before surgery.3,4 However, these patients did not show the bleeding tendency. Rather, they have been reported to have thromboembolic predisposition.5 6
cDNA7-10 and gene structure11 of the human FXII have been elucidated. The gene spans a total of 12kb and is composed of 14 exons.11 The gene is reported to be located at 5q33-qter in the human chromosome.12 cDNA of FXII is 2044 bp in length and is composed of 49 bases of 5′-untranslated, 1845 bases of amino acid coding, and 150 bases of 3′-noncoding sequences. The transcription was initiated at 49 bases upstream from the translation initiation ATG codon, as determined by the methods of S1 nuclease mapping and primer extension.11
The plasma levels of FXII have been determined to be approximately 30 μg/mL (range: 15 to 47 μg/mL).13 No differences with sex in respect to adult ages have been reported.4,14However, the concentration of plasma FXII has been known to have wide variation between individuals and races.14 Gordon et al15 documented that levels of plasma XII in Orientals were approximately only half of that in Caucasians. However, the reason remains undetermined to date.
Nucleotide sequencing analysis of a Japanese family with congenital FXII deficiency showed 126 Arg to Pro substitution as a responsible mutation for the deficiency.16 On that occasion an unidentified nucleotide polymorphism, the substitution from 46 cytosine to thymidine in exon 1 of the FXII gene, was unexpectedly found. This nucleotide polymorphism is localized at the 5′-untranslated region and is close to the translation initiation ATG codon (4 bases upstream). Hence, it is suspected that this substitution affects the translation efficiency. We report here that a high prevalence of 46 C to T substitution was found in Orientals and led to reduced levels of plasma FXII through low translation efficiency in hepatocytes.
MATERIALS AND METHODS
Subjects.
Blood donors were 76 Orientals (Japanese) and 20 Caucasians. Liver specimens were obtained from patients with hepatic disorders who were undergoing liver biopsy or resection, after informed consent was obtained.
Blood collection and laboratory analysis.
Blood was collected in tubes containing 0.1 volumes of 0.106 mmol/L trisodium citrate. Plasma was prepared by centrifugation for 10 minutes at 2,000g at room temperature and then stored at −70°C in 1.5 mL aliquots.
FXII activity was measured by the APTT method by using a synthetic substrate specific for thrombin (Tos-Gly-Pro-Arg-ANBA-isopropylamide) and FXII-deficient plasma.17 FXII antigen was measured by Laurell's method by using rabbit polyclonal antibody against human FXII.18 Blood for the FXII assays was obtained from healthy adult volunteers (31 Japanese and 20 Caucasians) with age ranges from 15 to 69 years (means 33±12 years). Pooled plasma from 100 healthy Japanese subjects was used as a standard plasma (100% in activity and antigen).
Isolation of DNA and RNA, and polymerase chain reaction (PCR).
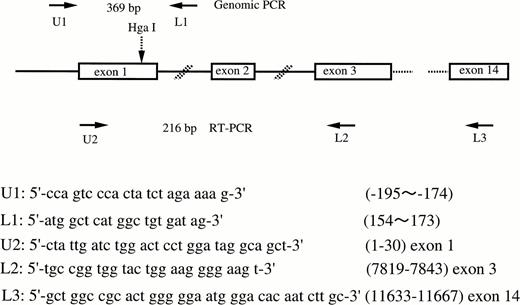
Genomic DNA was isolated from peripheral blood leukocytes by standard procedures.19 Exon 1 of the FXII gene was amplified by PCR by using the primers shown in Fig 1. Total RNA from liver was isolated with an RNA isolation kit (Isogen, Nippon gene, Tokyo, Japan). Ten μg of total RNA was used to synthesize cDNA by using a random hexamer and cDNA synthesis kit (Takara RNA PCR Kit, Takara Shuzo Co, Ltd, Otsu, Japan). Nine μL of this solution was used as a template for PCR. Figure 1 shows the locations and sequences of oligonucleotide primer pairs used in this study. To analyze the polymorphism, PCR products were digested by Hga I (New England BioLabs, Beverly, MA) at 37°C for 3 hours, subjected to 2% agarose gel or 5% polyacrylamide gel electrophoresis, and stained by ethidium bromide (allele-specific restriction analysis).
Schematic illustration of the factor XII gene and the locations and sequences of PCR primers. Arrows show the location of primers. The Hga I site is shown by the broken arrow.
Schematic illustration of the factor XII gene and the locations and sequences of PCR primers. Arrows show the location of primers. The Hga I site is shown by the broken arrow.
Subcloning of PCR products.
The PCR fragments were electrophoresed on agarose gel and purified by using a QIA quick gel extraction kit (Qiagen, Chatsworth, CA), after which they were introduced into the plasmid vector pCRII by using a TA cloning kit (Invitrogen, San Diego, CA). Competent cells (Escherichia coli strain; One Shot competent cell) were transformed with the plasmid, grown in the LB media, and spread on an agar plate containing kanamycin. The plasmids from single colonies were prepared separately by the method of Sambrook, et al.19
Sequencing of subcloned DNA.
The DNA sequencing was performed by using a T7 polymerase DNA sequencing kit (Pharmacia Biotech, Uppsala, Sweden) and universal primers. The reaction mixtures were transferred to an SQ-5500 DNA Sequencer (Hitachi, Tokyo, Japan).
In vitro transcription/translation analysis.
By using reverse transcriptase (RT)-PCR, we constructed two types of FXII cDNA (46C-FXII and 46T-FXII) from the liver biopsy sample of a heterozygote for this polymorphism. The upper primer was designed to be located in the transcription start site (Fig 1). These PCR products were introduced into the plasmid vector pCRII mentioned above. By using nucleotide sequencing, identical sequences in both clones, except in the position 46, were confirmed. mRNA was created from 1μg of plasmid of each construct (46C-FXII and 46T-FXII) by using T7 RNA polymerase, and protein was made by rabbit reticulocyte lysate system and35S-methionine (Promega, Madison, WI). Synthesized proteins were applied on 10% SDS-polyacrylamide gel for electrophoresis under reduced conditions. The gel was dried and exposed on an imaging plate (Fuji Photo Film, Minamiashikaga, Japan) overnight at room temperature and the amounts of FXII synthesized were quantitated by densitometry.
Statistical analysis.
The data were represented as the means±SD. The differences in plasma FXII levels between the three groups were determined by Student's t-test. P values of less than .05 were considered significant.
RESULTS
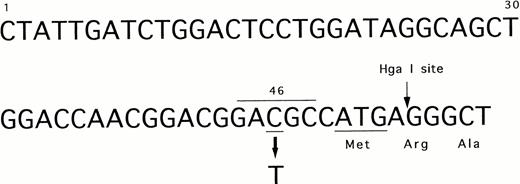
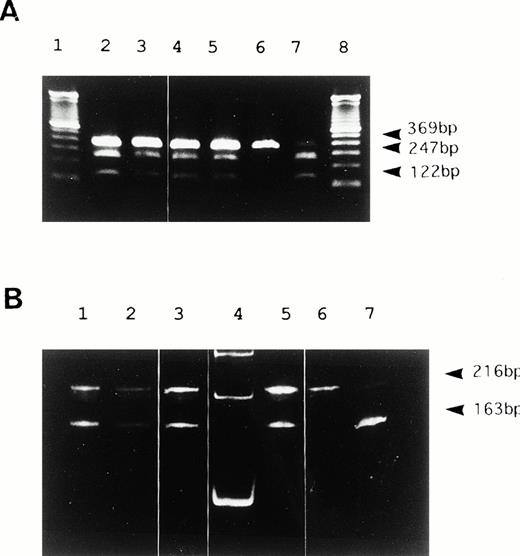
We identified the polymorphism of 46 C/T in exon 1 of the FXII gene as shown in Fig 2. The substitution of 46 C to T was located 4 bases upstream from translation initiation codon ATG (Fig 2). This region corresponded to a Hga I restriction site (GACGC) that was destroyed by this substitution (46T). Therefore, we investigated the allele frequency by PCR amplification (primer, U1, and L1) and Hga I digestion (Fig 3). DNA from peripheral blood leukocytes was obtained from 76 Orientals and 20 Caucasians. The allele frequency of 46 C/T was 0.27/0.73 in Orientals (allele number = 152), and conversely, 0.8/0.2 in Caucasians (allele number = 40).
The nucleotide sequence of 5′ end of exon 1 in the factor XII gene. The polymorphic and Hga I sites are shown by arrows. Underline indicates the translation initiation codon ATG and 46 C (polymorphic site). Overline indicates Hga I recognition sequence.
The nucleotide sequence of 5′ end of exon 1 in the factor XII gene. The polymorphic and Hga I sites are shown by arrows. Underline indicates the translation initiation codon ATG and 46 C (polymorphic site). Overline indicates Hga I recognition sequence.
Hga I restriction analysis of PCR products. (A) 2% agarose gel electrophoresis of PCR products from genomic DNA in 6 representative samples. Lanes 2 to 5 show heterozygosity for 46C/T. Lane 6 shows homozygosity for 46C. Lane 7 shows homozygosity for 46T. Undigested DNA remains as a faint 369 bp in 46C homozygotes even after a long duration of digestion. A DNA size marker (100bp ladder) was applied in lanes 1 and 8. (B) 5% polyacrylamide gel electrophoresis of RT-PCR products from liver mRNA in three genetic types for the polymorphism. Lanes 1 to 3 and 5 show heterozygosity for 46C/T. Lane 6 shows homozygosity for 46T. Lane 7 shows homozygosity for 46C. 53 bp bands digested by Hga I were barely visible in this study (not shown). A DNA size marker (100bp ladder) was applied in lane 4.
Hga I restriction analysis of PCR products. (A) 2% agarose gel electrophoresis of PCR products from genomic DNA in 6 representative samples. Lanes 2 to 5 show heterozygosity for 46C/T. Lane 6 shows homozygosity for 46C. Lane 7 shows homozygosity for 46T. Undigested DNA remains as a faint 369 bp in 46C homozygotes even after a long duration of digestion. A DNA size marker (100bp ladder) was applied in lanes 1 and 8. (B) 5% polyacrylamide gel electrophoresis of RT-PCR products from liver mRNA in three genetic types for the polymorphism. Lanes 1 to 3 and 5 show heterozygosity for 46C/T. Lane 6 shows homozygosity for 46T. Lane 7 shows homozygosity for 46C. 53 bp bands digested by Hga I were barely visible in this study (not shown). A DNA size marker (100bp ladder) was applied in lane 4.
It has been reported that plasma FXII levels in Orientals were much lower than those in Caucasians.15 In addition, because it was suspected that the nucleotide polymorphism close to the translation initiation codon ATG may affect the translation efficiency, plasma levels of FXII activity and antigen were determined in healthy volunteers with three allele types. There were significant differences in plasma levels of FXII between these three types: C/C, 170±38% (178±27%); C/T, 141±29% (123±34%); and T/T, 82±19% (61±11%) [FXII activity in plasma (FXII antigen levels in plasma)] (Fig 4).
Plasma levels of factor XII activity (A) and antigen (B) of individuals with 46C/C, 46C/T, or 46T/T. Thirteen Caucasians were included in C/C, 6 in C/T, and 1 in T/T group. The levels of plasma FXII activity and antigen were 150.0±30.3 (n = 20) and 155.4±36.1 (n = 20) in Caucasians, and 125.4±56.3 (n = 31) and 104.7±57.2 (n=20) in Orientals, respectively.
Plasma levels of factor XII activity (A) and antigen (B) of individuals with 46C/C, 46C/T, or 46T/T. Thirteen Caucasians were included in C/C, 6 in C/T, and 1 in T/T group. The levels of plasma FXII activity and antigen were 150.0±30.3 (n = 20) and 155.4±36.1 (n = 20) in Caucasians, and 125.4±56.3 (n = 31) and 104.7±57.2 (n=20) in Orientals, respectively.
We studied whether the 46 C/T polymorphism modulates either transcription or translation. To investigate the transcription effect of this polymorphism, liver biopsy specimens from seven heterozygotes for 46 C/T were treated for RNA analysis. Subsequently, the amounts of RT-PCR products originating from both alleles were assessed by allele-specific restriction analysis. The 216 and 163 bands were of identical intensity, suggesting nearly equal transcription from both alleles (Fig 3B). Therefore, it is not likely that the polymorphism modulates either transcription in hepatocytes or the stability of mRNA.
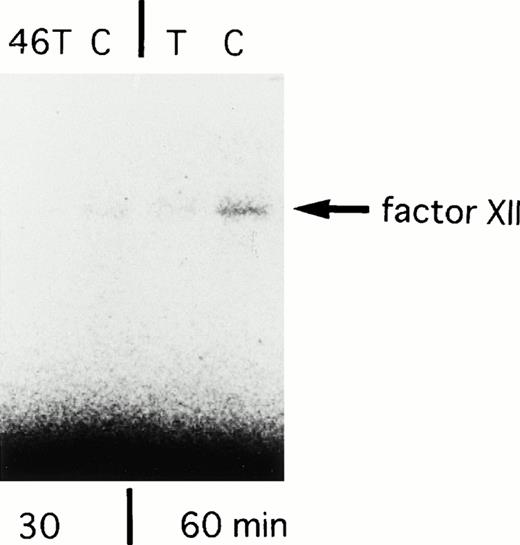
In the next experiment, translational efficiency of the polymorphism was investigated. We constructed two types of FXII cDNA clones (46C-FXII and 46T-FXII) by using RT-PCR (primer, U2 and L3) from the liver biopsy sample of a heterozygote for this polymorphism. In vitro transcription/translation assay showed that the amounts of translated products from 46C-FXII were three times higher than those from 46T-FXII for a translation reaction time of 30 minutes, and 1.6 times higher for the 60-minute reaction (Fig 5). There was no difference between clones in the amounts of translated products in a longer reaction (90 minutes). This result indicated that 46 T reduced the translation efficiency to FXII protein, at least in vitro.
SDS-PAGE analysis of in vitro translated products from 46C- and 46T-FXII cDNA. Incubation times for translation are shown in the lower portion of the panel. The arrow shows the factor XII synthesized in vitro. The experiments were repeated with essentially the same results.
SDS-PAGE analysis of in vitro translated products from 46C- and 46T-FXII cDNA. Incubation times for translation are shown in the lower portion of the panel. The arrow shows the factor XII synthesized in vitro. The experiments were repeated with essentially the same results.
DISCUSSION
We identified the nucleotide polymorphism (46 C/T) in the FXII gene that clearly explained the variety of plasma levels of FXII among individuals. Moreover, 46T was a common polymorphism and was found in 73% of Orientals. Conversely, 80% of Caucasians had the 46C polymorphism. Therefore, the 46 C/T polymorphism could also explain the racial differences in plasma FXII levels.
There are some examples of an association between plasma levels of coagulation factors and polymorphisms in the gene for coagulation factors. There are three polymorphisms in the factor VII gene: decanucleotide insertion in the 5′ region of the gene, repeat variation in intron 7, and 353Arg to Gln.20 In factor V, 1229Hnis to Arg mutation was strongly related to partial deficiency of factor V.21 Also a20210G to A transition in the 3′-untranslated region of the prothrombin gene was associated with elevated levels of plasma prothrombin and an increase in venous thrombosis.22 In these cases, gene polymorphisms and plasma levels have been discussed in relation to the development of thromboembolism. However, the molecular mechanisms of the association of gene polymorphism with plasma levels were not clearly shown.
As found through the genetic analyses for some human protein deficiencies, mutant mRNA levels are decreased in some cases by low transcription efficiency and/or instability of the mutant mRNA.23-25 In our studies, these mechanisms were unlikely because of the presence of approximately equal amounts of mRNA transcribed from both alleles in the livers from heterozygotes for the polymorphism.
The 46T polymorphism creates a codon ATG that is located at 5 bp upstream from original translation initiation codon ATG. Use of the putative ATG codon may make a peptide with only two amino acids (Met-Pro) in a different frame because a TAG stop codon appears 7 bp downstream from this ATG-initiation codon (Fig 2).
An in vitro transcription/translation study showed that FXII was produced in lesser amounts from cDNA containing 46T than from that containing 46C. Even though another ATG codon existed at the 5′-site from the original ATG in 46T-FXII, a second (original) ATG initiation codon was also recognized for translation but the amount of FXII was obviously decreased compared with that obtained with 46C-FXII. Two mechanisms were speculated to explain this phenomenon, according to the theory of Kozak.26 First, because the first ATG codon lies in an unfavorable context for initiation, “leaky scanning” enables some ribosomes to reach and initiate at the second ATG codon. Second, because too little ORF (9bp) is translated in the frame by using the first ATG codon, 40S ribosomal subunits apparently resume scanning and reinitiate translation further downstream. Consequently, the two ATG translation initiation codons in close proximity decreased translation efficiency. Furthermore, 46T destroys the Kozak's consensus sequence (GCCAG CCAUGG) for translation initiation signaling. This may also prevent the proper recognition of the translation initiation site. In the β-globin gene, it has been reported that the G to A transition at 26bp 5′ to the normal ATG codon created a cryptic ATG codon that interfered with correct initiation and caused a β-thalassemia intermedia.27However, to our knowledge, the FXII gene is the first human case in which a polymorphism in the vicinity of the ATG initiating site reduced translation efficiency and regulated the plasma concentration of the translation product.
The prevalence of moderate to severe deficiency of FXII among the normal Caucasian population is quoted as 1.5% to 3%, which is a rather high percentage compared with those of other coagulation and anticoagulation factors.28 We have now proven that the standard values of the FXII were different between Caucasians and Orientals owing to this common polymorphism. Therefore, it can be speculated that the individuals with 46T homozygosity are judged as FXII deficient under the Caucasian standard. Even in heterozygotes for 46C/T, an additional genetic mutation in 46C allele of FXII gene may also induce the FXII deficiency state.
Coagulation factor XII is supposed to act in multiple functional systems involving serine protease activation. In addition, the in vitro growth and mitotic effects of FXII and FXIIa on Hep G2, a hepatoma cell line, have been reported as an additional function.29 30Nevertheless, its physiological roles have remained unknown.
There is controversy over the clinical significance of FXII deficiency because most of subjects with complete deficiency of FXII have no clinical manifestations. Some authors31-33 considered FXII deficiency to be a risk factor for thromboembolism, whereas others34 considered the FXII deficiency to be of minor importance as a thrombotic risk factor. Hence, further large-scale study into the association between the development of thromboembolism and this polymorphism may be needed to elucidate the hemostatic function of FXII. In addition, it may be interesting to investigate the combination effect of such common genetic polymorphisms, including Factor V Leiden and 20210 G to A in prothrombin gene, on the thrombotic predisposition.
Address correspondence to Takashi Okamura, MD, First Department of Internal Medicine,Faculty of Medicine, Kyushu University, 3-1-1 Maidashi Higashi-ku, Fukuoka 812-82, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal