Abstract
The genomic analysis of a 70-year-old man with recurrent deep venous thrombosis having a protein S (PS)-deficient phenotype corresponding to both type III and type II evidenced two different mutations: a +5 g→a mutation in the donor splice site of intron e (ivs e) and a ser 460 to Pro mutation. The propositus' son, who had a type II PS deficiency phenotype, only bore the ivs e +5 g→a mutation. The study of platelet PS mRNA prepared from this subject showed that the ivs e, +5 g→a mutation led to the generation of two abnormal transcripts, one lacking exon 5 and the other lacking exons 5 and 6. The presence of an additional PS band with a decreased molecular mass on immunoblots performed in reducing conditions suggested the presence of truncated PS lacking EGF1 (encoded by exon 5). Two monoclonal antibodies (MoAbs) were used to further characterize the nonfunctional plasma PS. Comparison of PS levels measured with each of these MoAbs and PS levels in conventional assays was consistent with the presence of an abnormal inactive protein in the plasma of both patients bearing the ivs e, +5 g→a mutation, suggesting that variant PS lacking EGF1 is secreted but is devoid of activated protein C cofactor activity.
PROTEIN S (PS), A VITAMIN K-dependent glycoprotein, is a cofactor of the protein C system, an important natural anticoagulant mechanism. In the presence of PS, activated protein C (APC) inactivates factor Va (FVa) and factor VIIIa (FVIIIa) and thereby reduces thrombin generation (for review, see Esmon1). By binding to FVa2 or FXa,3 PS can also directly inhibit prothrombin activation as well as the formation of prothrombinase complexes on phospholipid surfaces.4 In human plasma, approximately 60% of PS is bound to the complement component C4b-binding protein (C4b-BP), and only the free form has APC-cofactor activity. PS deficiency is transmitted as an autosomal dominant trait, and heterozygous subjects belonging to families with the disorder are at risk of recurrent thromboembolic disease in adulthood.5 Severe and early thrombotic complications can occur in patients with very low PS levels. However, only one homozygous PS deficiency associated with severe purpura fulminans in the neonate period has been characterized genotypically.6 According to the International Society on Thrombosis and Hemostasis (ISTH) standardization subcommittee, three types of PS deficiency can be defined on the basis of total PS levels, free PS levels, and APC cofactor activity. Type I deficiency is characterized by low total and free PS antigen levels. Type II deficiency is characterized by normal total and free PS antigen levels and low APC cofactor activity. Type III is defined by selectively reduced levels of free PS antigen and activity. However, two recent reports suggested that type I and type III are two phenotypic expressions of the same genetic defect.5 7
Two homologous genes for PS map to chromosome 3.8 The active gene, PROS1, spans over 80 kb and comprises 15 exons. The second gene, PROS2, has no open reading frame; shows multiple base changes, stop codons, and frameshifts; and is probably a pseudogene.9-11 The 5′ part of the PROS1 gene shows strong homology with other vitamin K-dependent proteins. Exon 1 encodes the signal peptide; exon 2 the propeptide and GLA domain; exon 3 a domain with a high aromatic amino acid content; exon 4 a thrombin-sensitive loop; and exons 5 through 8 four epidermal growth factor-like domains. The 3′ part of the PROS1 gene differs from all other known coagulation proteins, because the last seven exons (9 to 15) encode a sex-hormone-binding globulin-homologous domain.
Ninety mutations, including 2 large deletions, have so far been identified in the PROS1 gene as causal defects.12 Most are point mutations generating nonsense or missense changes in coding or splice site sequences. Microinsertions/deletions have also been reported to result in frameshifts and premature stop codons. Most of these genetic defects cosegregate with type I PS deficiency. So far, few mutations have been shown to give rise to type III PS deficiency.13-16 The single point 460 TCC→CCC mutation in exon 13 of PROS1, leading to the replacement of Ser 460 by Pro and initially described as the Heerlen polymorphism,17 is frequently associated with type III PS deficiency.13
PS type II deficiency is fairly infrequent, and only 5 mutations have been identified in patients with normal PS concentrations and low APC cofactor activity. Interestingly, all 5 mutations giving rise to a type II phenotype are located in the amino-terminal part of PS, which is homologous to that of other vitamin K-dependent proteins and encodes the domains interacting with APC.18 Among the 5 mutations leading to normal expression of nonfunctional PS, 2 are located in the propeptide at positions −2 and −1.19Substitution of Lys 9 by Glu may also modify the conformation and/or carboxylation of the GLA domain.20 The other mutations observed in type II deficiency are substitution of Thr 103 by Asn in the first EGF domain19 and substitution of Lys 155 by Glu in the second EGF domain.21
We report the first case of type II PS deficiency due to a splice site mutation resulting in two alternative splice transcripts lacking either exon 5 or both exons 5 and 6. The variant PS, characterized by plasma immunoblot and monoclonal antibody (MoAb)-based assays, is probably a truncated protein lacking EGF1.
MATERIALS AND METHODS
Blood Sampling
For all coagulation tests, blood was collected in tubes containing 0.11 mol/L trisodium citrate (1/10). Platelet-poor plasma (PPP) was obtained by two centrifugation steps (15 minutes at 4,000 rpm and 4°C) and stored at −80°C. Normal pooled plasma was prepared by mixing equal parts of PPP from 20 healthy subjects (10 males and 10 females) and was stored at −80°C. Blood for DNA or platelet mRNA analysis was collected in 0.05 mol/L EDTA.
Assays for PS
Total (TPS Ag) and free PS antigen (FPS Ag) were measured by using a MoAb-based enzyme-linked immunosorbent assay (ELISA) method (Asserachrom Total Protein S and Asserachrom Free Protein S; Diagnostica Stago, Gennevilliers, France) according to Amiral et al.22 PS activity (PS Act) was assayed with the Staclot Protein S kit (Diagnostica Stago) according to Wolf et al.23
Molecular Biology Techniques
DNA analysis.
Genomic DNA was isolated from peripheral blood leukocytes by using the procedure described by Bell et al.24 The PS genes of patients I1 and II1 were analyzed according to Gandrille et al.19 The 15 exons and intron/exon junctions of PROS1 were selectively amplified in a polymerase chain reaction (PCR) by applying the principle of the amplification-refractory mutation system (ARMS). The sequences of the primers (Genset, Paris, France) used in this study are presented in Table 1. The amplified fragments were submitted to denaturing gradient gel electrophoresis (DGGE). Fragments with abnormal behavior were asymmetrically amplified and sequenced according to the method of Sanger et al,25except for exon 13, which was analyzed by restriction enzyme analysis. PS gene exon 13 was amplified using nucleotides specific for PROS 1. The upstream primer sequence 5′-TGTTAATAATAATTCCTTCTGA-3′ is located in intron l; the downstrean primer 5′-TGATTTTTCAGAGGTGGAGT-3′ is located in the 3′ part of exon 13 and bears a restriction site for HinfI endonuclease. The 221-bp amplified product was digested with 5 U of HinfI and 10 U of Rsa I and then subjected to 6% polyacrylamide gel electrophoresis. The T→C substitution in the codon corresponding to Ser 460 created a new restriction site for the endonuclease Rsa I and thus gave rise to fragments of 21, 80, and 120 bp, whereas only fragments of 21 and 200 bp were obtained in the absence of the mutation. Mutations are designated as recommended in the PS database of mutations.12
Nucleotide Sequences of the Primers Used for cDNA Amplification and Sequencing
| Primer . | Primer Sequence . |
|---|---|
| PS 2a (s) | CTGGTTAGGAAGCGTCGTGCA |
| PS 4a (s) | ACTGGGTTATTCACTGCTGC |
| PS 5a (s) | GCCATTCCAGACCAGTGTAGT |
| PS 5b (a) | ACTACACTGGTCTGGAATGGC |
| PS 6b2 (a) | CCATTTTTACAGGAACAGTGG |
| PS 7b (a) | CCTTCGGGGCATTCACATTC |
| PS 7b2 (a) | TTCACATTCAAAAATCTCCTG |
| PS 10b (a) | GCTGAGTGATCGATAGATTC |
| Junction 4/5 (s)-150 | TGTGTCAATGCCATTCCAGAC |
| Junction 4/6 (s)-150 | CTGTGTCAATGACATAAATGAA |
| Junction 4/7 (s)-150 | TGTGTCAATGATGTGGATGA |
| Primer . | Primer Sequence . |
|---|---|
| PS 2a (s) | CTGGTTAGGAAGCGTCGTGCA |
| PS 4a (s) | ACTGGGTTATTCACTGCTGC |
| PS 5a (s) | GCCATTCCAGACCAGTGTAGT |
| PS 5b (a) | ACTACACTGGTCTGGAATGGC |
| PS 6b2 (a) | CCATTTTTACAGGAACAGTGG |
| PS 7b (a) | CCTTCGGGGCATTCACATTC |
| PS 7b2 (a) | TTCACATTCAAAAATCTCCTG |
| PS 10b (a) | GCTGAGTGATCGATAGATTC |
| Junction 4/5 (s)-150 | TGTGTCAATGCCATTCCAGAC |
| Junction 4/6 (s)-150 | CTGTGTCAATGACATAAATGAA |
| Junction 4/7 (s)-150 | TGTGTCAATGATGTGGATGA |
Abbreviations: (a), antisense; (s), sense.
Bold letters correspond to exon 4.
RNA extraction.
Platelet-rich plasma (PRP) was prepared from 60 mL of whole blood collected in EDTA by centrifugation at 125g for 10 minutes. Platelets were collected by centrifugation at 1,800g for 10 minutes, and after elimination of the supernatant, RNA was extracted by using a monophasic solution containing phenol and guanidinium isothiocyanate (Trizol; GIBCO BRL, Cergy-Pontoise, France) according to the manufacturer's instructions and kept at −80°C in the presence of diethylpyrocarbonate (DEPC). Liver specimens consisted of normal tissue excised from an individual undergoing liver surgery who had no personal or familial history of thrombosis. They were immediately frozen in dry ice and then homogenized in Trizol solution. Extraction was performed as for platelets.
Reverse transcription (RT) and cDNA amplification.
Platelet or liver mRNA (500 to 1,000 ng) was added to 750 ng of hexamers (dp N6; Pharmacia Fine Chemicals, Uppsala, Sweden) and distilled water to a final volume of 20 μL. The mixture was heated to 75°C for 3 minutes and then kept at +4°C. Four microliters of 5× RT buffer (250 mmol/L Tris HCl, pH 8.3, 375 mmol/L KCl, 15 mmol/L MgCl2), 20 U of ribonuclease inhibitor (Promega, Madison, WI), and 1 μL of dATP, dCTP, dGTP, and dTTP (10 mmol/L) from Pharmacia were added and incubated at 42°C for 10 minutes before the addition of 5 U of reverse transcriptase (Moloney murine leukemia virus [M-MLV] Reverse Superscript; GIBCO BRL). After 50 minutes at 42°C, the enzyme was denatured at 94°C for 5 minutes. Then, 20 μL of the RNA-DNA hybrid solution was added to 80 μL of the PCR mixture containing 50 pmol of upstream and downstream primers, 1× PCR buffer (10 mmol/L Tris HCl, pH 8.3, 150 mmol/L KCl, 0.01% [wt/vol] gelatin, and 1.5 mmol/L MgCl2), and 2.5 U of Taq polymerase (Perkin Elmer Cetus Instruments, Norwalk, CT), and the mixture was subjected to PCR. PCR products were analyzed on 6% polyacrylamide gel stained with ethidium bromide solution. Amplified fragments with abnormal molecular weights were excised from the gel and heated at 85°C for 10 minutes in PCR 1× buffer. The genetic material was sequenced as described by Sanger et al.25
Plasma Studies
Polyacrylamide gel electrophoresis and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in 4% to 12% gradient gels according to Laemmli.26 After electrophoresis in 0.02 mol/L Tris, 0.2 mol/L glycine, 0.5% SDS, proteins were transferred to 0.45-μm nitrocellulose membranes (Biorad, Hercules, CA) using the transblot electrophoretic transfer cell (Biorad) at 150 V for 2 hours. The membranes were then incubated overnight with 5% (wt/vol) nonfat dry milk and 0.5% (vol/vol) Tween 20 in 100 mmol/L Tris, 150 mmol/L NaCl, pH 7.5 (TBS). PS was detected by using polyclonal rabbit anti-PS serum (Assera PS; Stago) and shown with biotinylated antirabbit IgG (Vector Laboratories, Burlingame, CA) diluted according to the manufacturer's instructions. The Vectastain ABC Elit kit (Vector Laboratories) was used to increase the sensitivity of the method.
Assay of PS using the MoAb S-12.
We used an ELISA method described by Duchemin et al,13with slight modifications. Control and patient plasma was tested at two dilutions (1:2,000 and 1:4,000). The standard curve was established by using serial dilutions of 1:1,000 to 1:32,000 of normal pooled plasma.
Assay of PS using the MoAb HPS 54.
Plates were coated overnight at room temperature with 100 μL of 1:40 rabbit polyclonal anti-PS antibody (Assera PS; Stago) in 50 mmol/L Na2CO3, pH 9. After this step and all the following steps, extensive washing in 20 mmol/L Tris, 150 mmol/L NaCl, pH 7.5, 1% (vol/vol) Tween 20 (washing buffer) was performed. TBS-bovine serum albumin (BSA; 5%, 200 μL) was added to each well for 2 hours at room temperature, and then 100 μL of samples diluted in 0.1% TBS-BSA (1:400 and 1:800) was allowed to react for 18 hours at room temperature. A pool of normal plasma diluted 1:200 to 1:6,400 was used for the calibration curve. Bound PS was allowed to react for 4 hours with 100 μL of HPS 54 MoAb (50 μg/ml) and then 100 μL of goat antimouse IgG antibody conjugated to peroxidase (Biorad; 1:5,000) was added for 1 hour, followed by 100 μL of 3,3′,5,5′ tetramethylbenzidine solution. The reaction was stopped after 15 minutes with 50 μL of 3 mol/L H2SO4 and absorbance was read at 450 nm in a Multiskan Multisoft apparatus (Labsystems, Helsinki, Finland).
RESULTS
Case Report
The proband (I1), a 70-year-old man with a history of deep vein thrombosis, pulmonary embolism, and myocardial infarction since the age of 23, had been on oral anticoagulants for the previous 10 years. He agreed to switch to low molecular weight (LMW) heparin for 1 month to allow us to measure plasma PS concentrations in the absence of vitamin K antagonist. In the two different blood samples obtained during this time, the total PS concentration was borderline (70%), the free PS concentration was moderately decreased (50%), and PS activity was unambiguously below normal (27%). One of his sons (II1), who had had an episode of deep vein thrombosis at the age of 35, was available for laboratory explorations. He repeatedly had a typical type II phenotype with a total PS concentration of 100% and a free PS concentration of 97%, whereas the functional activity was 30% of normal. Neither subject bore the factor V Arg 506 to Gln mutation that might interfere with PS activity measurement.27
Identification of Two Different Mutations in Genomic DNA
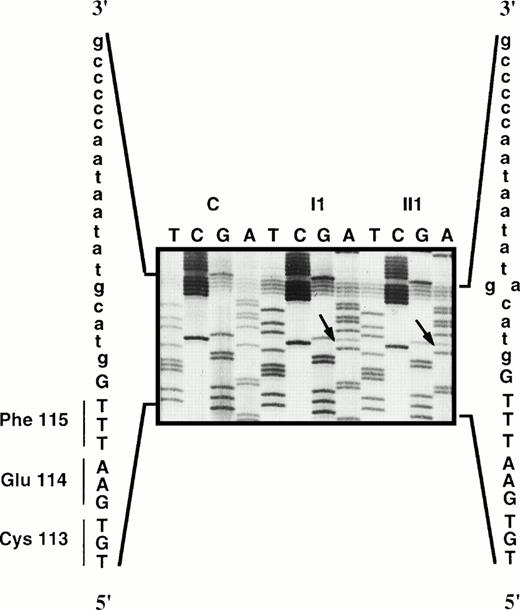
DNA coding regions and exon/intron junction sequences from the propositus (I1) and his son (II1) were screened by DGGE. In patient I1, abnormal migration was observed with exon 5 and exon 13 of the PROS1 gene. Direct sequencing of exon 5 and its flanking regions showed a g→a transition in the fifth basepair of the donor splice site of intron e (ivs e, +5 g→a; Fig1). The DGGE profile of exon 13 was that observed in subjects bearing the Ser 460→Pro mutation in previous studies (data not shown). Cleavage of the amplified fragment by the restriction endonucleaseRsa I13 confirmed the T→C transition at the first nucleotide of codon 460, leading to the replacement of Ser by Pro in the protein sequence (data not shown).
Direct sequencing of the ivs e, +5 g→a mutation in the members of family J compared with a control (C).
Direct sequencing of the ivs e, +5 g→a mutation in the members of family J compared with a control (C).
In the propositus' son (II1), the only DNA abnormality was located in the flanking sequence of exon 5. Sequencing of this part of the gene confirmed that, like his father, he bore the ivs e +5 g→a substitution. He did not carry the Ser 460→Pro mutation, indicating that the two mutations identified in the father were transmitted by different chromosomes.
Identification of Abnormal cDNA Transcripts
To determine whether the ivs e, +5 g→a substitution was responsible for the phenotypic expression of a plasma variant, we searched for mutated transcripts in platelet PS mRNAs from patient II1, who bore only this mutation and had a type II PS phenotype.
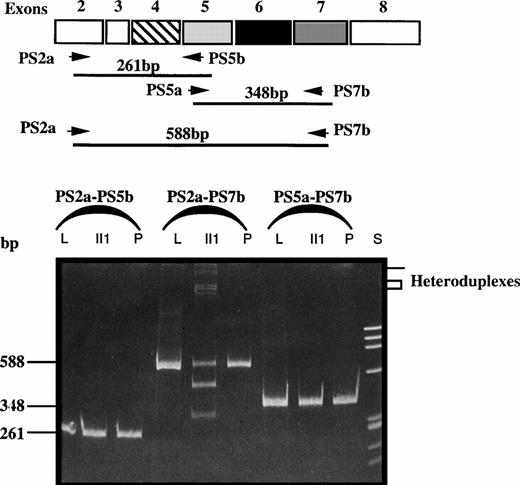
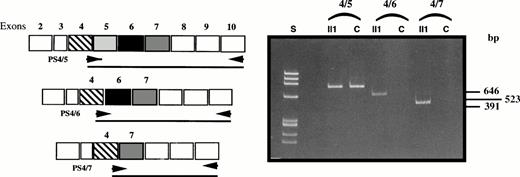
Platelet PS mRNAs were prepared from patient II1, from a normal control (P), and from homogenized liver (L) and then reverse transcribed and PCR-amplified using sets of primers matching the 5′ and 3′ regions of exon 5 (Fig 2).
Comparison of RT-PCR products of fragments PS2a-PS5b, PS2a-PS7b, and PS5a-PS7b of liver (L) and platelet (P) mRNA from two controls and platelet mRNA from patient II1. S, size standard; bp, basepair.
Comparison of RT-PCR products of fragments PS2a-PS5b, PS2a-PS7b, and PS5a-PS7b of liver (L) and platelet (P) mRNA from two controls and platelet mRNA from patient II1. S, size standard; bp, basepair.
As depicted in Fig 2, in normal platelet mRNA and normal liver mRNA, each set of primers resulted in the amplification of one fragment: amplification of the sequence spanning exon 2 to 5 yielded a 261-bp fragment, that of exons 5 to 7 a 348-bp fragment, and that of exons 2 to 7 a 588-bp fragment. In patient II1, amplification of exons 2 to 5 and exons 5 to 7 yielded two fragments of 261 and 348 bp, respectively, as expected. However, after amplification of the transcript spanning exons 2 to 7, two bands of approximately 465 bp and 330 bp were produced in addition to the normal fragment of 588 bp. These additional fragments pointed to the presence of a genetic defect within exon 5 flanking sequences. Interestingly, the electrophoretic pattern showed fragments of high molecular weight compatible with the presence of heteroduplexes generated by reannealing of the normal strand with the mutated strand.
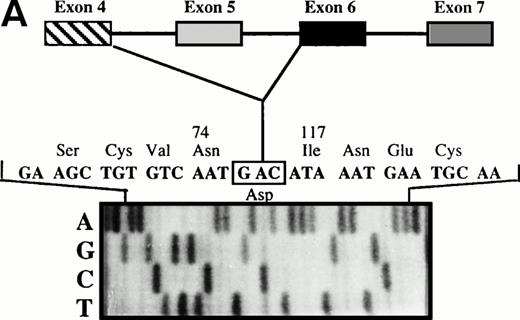
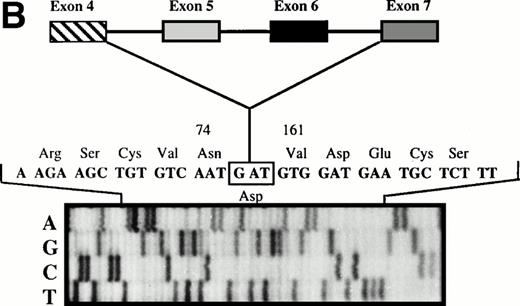
The 465-bp fragment was excised and extracted from the polyacrylamide gel for nested PCR with a set of oligonucleotides located in exon 4 (PS 4a) and exon 6 (PS6b2), surrounding the cDNA region between exons 4 and 6. Direct sequencing of the subsequent fragment showed an exon 4/6 junction, thus indicating exon 5 skipping (Fig 3A). Similarly, the 330-bp fragment was excised and extracted from the gel and subjected to nested PCR with a set of oligonucleotides located in exon 4 (PS 4a) and exon 7 (PS7b2). Direct sequencing of the subsequent fragment showed an exon 4/7 junction, indicating that exons 5 and 6 were skipped (Fig 3B).
(A) Direct sequencing of amplified fragment PS4a-PS6b2 showing the loss of exon 5. (B) Direct sequencing of amplified fragment PS4a-PS7b2 showing the loss of exons 5 and 6.
(A) Direct sequencing of amplified fragment PS4a-PS6b2 showing the loss of exon 5. (B) Direct sequencing of amplified fragment PS4a-PS7b2 showing the loss of exons 5 and 6.
To confirm whether the exon skipping affected exon 5 alone or both exons 5 and 6, we used an antisense primer located in exon 10 (PS10b) and 3 oligomers specifically recognizing the exon 4/5, exon 4/6, and exon 4/7 junctions for RT-PCR with platelet mRNA from patient II1 and six controls (Fig 4). Amplification with the junction 4/5 primer gave rise to a fragment of 646 bp in the controls, whereas no amplification products were observed with the primers matching exon 4/6 and 4/7 junctions. In the patient, cDNA transcripts were observed with the normal 4/5 junction primer and with the 4/6 and 4/7 junction primers, yielding two additional fragments of 523 bp (loss of 123 bp) and 391 bp (loss of 255 bp), respectively.
Confirmation of the double exon skipping in patient II1 by analysis of fragments PS4/5-PS10b, PS4/6-PS10b, and PS4/7-PS10b compared with a control (C).
Confirmation of the double exon skipping in patient II1 by analysis of fragments PS4/5-PS10b, PS4/6-PS10b, and PS4/7-PS10b compared with a control (C).
Identification of Abnormal PS in Plasma
The propositus (I1) was a compound heterozygote, one allele carrying the ivs e, +5 g→a substitution and the other allele the Ser 460→Pro mutation; the latter is frequently associated with a type III phenotype.13 The subnormal total PS (70%) and low free PS (55%) concentrations observed in this patient are consistent with such a phenotype. However, PS activity (27%) was lower than expected from the free PS concentration, suggesting that half the circulating PS molecules were nonfunctional. The propositus' son, who only carried the ivs e, +5 g→a substitution, had a typical type II phenotype, which suggests the expression of a nonfunctional protein by the mutated allele.
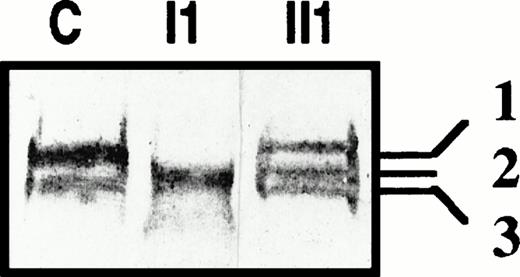
To determine whether the abnormal transcripts due to the exon skipping associated with the ivs e, +5 g→a substitution encoded truncated protein(s), we searched for PS of lower molecular weight in the patients' plasma. Figure 5 shows immunoblots of PS performed in the presence of a reducing agent obtained from a normal control (C), propositus (I1), and propositus' son (II1). In normal plasma, the thrombin-sensitive region of about half the PS molecules is cleaved, which results in the presence of a second band with a lower molecular mass than the native form (bands 1 and 3, Fig 5). Interestingly, in the plasma of the propositus' son, who bore only the splice site mutation, an additional band was detected, possibly corresponding to the reduced form of cleaved PS lacking EGF1. In the propositus' plasma (I1), only one band was observed, suggesting that he did not express normal PS. Because the propositus is a compound heterozygote, he might express both Heerlen PS and a PS lacking EGF1, and the single band observed on immunoblotting with a reducing agent suggests that, after cleavage, Heerlen PS and the putative truncated PS migrated with the same molecular mass.
Immunoblots of plasma PS from a normal control (C), patients II1 and I1 of family J, in reducing conditions.
Immunoblots of plasma PS from a normal control (C), patients II1 and I1 of family J, in reducing conditions.
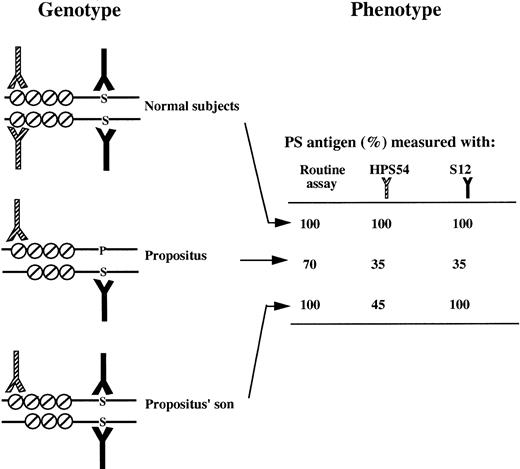
To confirm the presence in plasma of PS lacking EGF1, we performed ELISAs with S12, a conformational MoAb that does not recognize Heerlen PS,17 and HPS 54, the epitope of which encompasses EGF1.28 The concentration observed with each MoAb was compared with that observed in a routine assay based on 2 other MoAbs recognizing all PS forms (Fig 6).
Diagrammatic representation of binding of MoAbs binding to the two PS species present in the plasma of a normal subject, the propositus, and the propositus' son (deduced from the subject genotype). The figure shows PS domains significant for MoAb binding. The EGF domains are symbolized by a circle with a diagonal slash, and the amino acid at position 460 (P, Pro; S, Ser) is indicated by an uppercase letter. The MoAb recognizing EGF1 (HPS 54) is represented by a half-shaded symbol, and the MoAb containing Ser at position 460 (S12) of its epitope is in black. PS concentrations (percentage of the value for a normal pool) observed with the routine assay and two MoAb-based assays.§
Diagrammatic representation of binding of MoAbs binding to the two PS species present in the plasma of a normal subject, the propositus, and the propositus' son (deduced from the subject genotype). The figure shows PS domains significant for MoAb binding. The EGF domains are symbolized by a circle with a diagonal slash, and the amino acid at position 460 (P, Pro; S, Ser) is indicated by an uppercase letter. The MoAb recognizing EGF1 (HPS 54) is represented by a half-shaded symbol, and the MoAb containing Ser at position 460 (S12) of its epitope is in black. PS concentrations (percentage of the value for a normal pool) observed with the routine assay and two MoAb-based assays.§
The proband had a PS concentration at 35% with S12 and at 35% with HPS 54; this suggested the expression of two different PS molecules, half having the Ser 460→Pro mutation (detected by HPS 54) and the remaining half (detected by MoAb S12) corresponding either to a normal transcript (wild-type PS) or to the transcript lacking exon 5. The absence of 73-kD wild-type PS on the immunoblots favors the second possibility.
The propositus' son (II1) was heterozygous for the mutation that potentially results in the expression of a truncated PS. The observation that this patient had normal PS concentrations in the routine assay and low APC cofactor activity strongly suggests the presence of nonfunctional truncated molecules in the circulation; this was supported by the abnormal band with a lower molecular mass observed on immunoblots in reducing conditions. The PS concentration measured with MoAb HPS 54 was 45%, inferring that about half the PS molecules were not recognized. The lack of recognition of PS by MoAb HPS 54 indirectly indicates the presence of truncated PS lacking EGF1 in patient II1.
DISCUSSION
DNA analysis of the 15 exons and intron/exon junctions of the PROS1 gene of a PS-deficient patient showed two different genetic abnormalities located on different alleles. The first mutation was a g to a substitution on the fifth nucleotide of the donor splice site of intron e (ivs e, +5, g→a). The second mutation was a T to C transition resulting in a Ser 460 to Pro mutation. The propositus' son, who had a qualitative (type II) PS deficiency phenotype, only bore the ivs e, +5, g→a substitution.
Usually, splice site mutations are associated with quantitative deficiencies, resulting either in reduced amounts of mature mRNA or in abnormal maturation of the mutant mRNA.29 Two mechanisms (exon skipping and cryptic splice site activation) have been recognized as being responsible for this abnormal maturation, the latter being responsible for inclusion of intronic material that mainly disrupts the open reading frame. The effect of the splice site mutation depends on its position in the consensus splice site.29-32Replacements of G residues at position +5 of donor splice sites, such as those present in the family we studied, are predicted to reduce significantly the stability of base-pairing of the splice site with the complementary region of U1 snRNA.33
Three exon skips have been described in the PROS1 gene of patients with a quantitative (type I) PS deficiency phenotype. The first patient bore a mutation located on the first invariant nucleotide of the donor splice site of intron d,14 generating two abnormal mRNA species: the predominant abnormal transcript resulted from activation of a cryptic splice site located 48 bp downstream of the normal donor splice site, whereas the other transcripts lacked exon 4. The second patient bore a nonsense mutation in codon 62 leading to exon 4 skipping.34 The third patient bore a mutation located in the fourth nucleotide of the donor splice site of intron a,35 generating multiple abnormal RNA species by exon skipping and cryptic splice site activation. No open reading frame was conserved in the abnormal transcripts of either patient, and this probably explains the type I PS deficiency phenotype observed.
Because our patient had an unusual association of a qualitative deficiency phenotype and a splice site mutation, we studied platelet mRNA species in the propositus' son. In addition to the normal transcript generated by the normal PROS1 allele, at least two abnormal transcripts were easily detected, one corresponding to mRNA lacking exon 5 and the other to mRNA lacking both exons 5 and 6. The presence of several splice products corresponding to different exon-skipped products, all with skipping of the exon located upstream of the donor splice site mutation (here exon 5) is more the rule than the exception, and the present observation is thus consistent with previous studies.36 37
These abnormal transcripts resulted in exon 4/exon 6 and exon 4/exon 7 direct junctions, as confirmed by using oligomers that specifically recognized these junctions. The absence of the normal donor splice site of intron e probably leads to the use of the donor splice site of intron d. The acceptor splice sites may be those of intron e and intron f. The same mechanism of alternative exon skipping has been reported for a β hexosaminidase (HEXA) gene mutation causing Tay-Sachs disease,38 for the Bruton tyrosine kinase (Btk) gene mutation causing X-linked agammaglobulinemia,37and for type V collagen gene (COL5A1) mutation causing Ehlers-Danlos syndrome.39
With the PROS1 ivs e +5 g→a mutation described here, an open reading frame was conserved in both abnormal transcripts, theoretically allowing the synthesis of truncated PS species lacking EGF1 or both EGF1 and EGF2. Because the propositus' son's PS deficiency phenotype was consistent with the presence of an abnormal PS species, we searched for such an abnormal plasma protein. The presence of circulating protein S lacking EGF1 would be consistent with the plasma phenotype, as it has previously been shown with different approaches that EGF1 is involved in the PS-APC interaction. Indeed, using MoAbs or chimeric recombinant PS,28,40,41 it was demonstrated that EGF1 is involved in the PS-APC interaction. Using the specificity of APC cofactor activity in various species of PS, combined with sequence variation comparison, the critical role of this EGF in the PS-APC interaction was confirmed.42
The propositus being a compound heterozygote, his plasma should contain two PS species, one generated by the Heerlen allele and the other by the ivs e, +5 g→a mutated allele. It has been shown that the Heerlen mutation leads to a partially deglycosylated PS migrating with a lower MW than the wild-type PS. Immunoblotting of the propositus' plasma performed in the presence of a reducing agent showed that no PS with the normal MW was present, suggesting that the ivs e +5 g→a mutation precludes detectable expression of normal MW PS. Because the MW of Pro 460 PS is similar to that of the putative EGF1-lacking mutant PS, the two molecular PS species could not be distinguished in the propositus' plasma. That the allele bearing the ivs e +5 g→a mutation only produces mRNAs with exon skipping could have been checked in this patient whose other allele bears the Ser 460 →Pro mutation. However, the patient, a 70-year-old man who had already accepted a switch from oral anticoagulants to LMW heparin for 1 month for laboratory investigations, was reluctant to give more blood for platelet mRNA extraction and we could not obtain this crucial information.
Immunoblotting of plasma from the propositus' son (who only bore the ivs e +5 g→a mutation) performed in reducing conditions evidenced an additional band at a molecular mass corresponding to truncated PS lacking EGF1. Because the presence of this molecule with a lower molecular mass did not prove the presence of such a truncated variant, we used ELISAs with MoAbs recognizing wild-type but not Heerlen (S12) PS and the EGF1 domain of PS (HPS 54). Using MoAb S12, the propositus' PS concentration was 35%, compared with 70% with the assay recognizing both wild-type and Heerlen PS. Using MoAb HPS 54, the propositus' plasma PS concentration was 35%. These results are consistent with the expression of two different PS molecules, half bearing the Heerlen mutation (detected by MoAb HPS 54) and half being normal or lacking EGF1 (detected by S12 only). The absence of normal MW wild-type PS on the immunoblots favors the second possibility.
The propositus' son bore only the ivs e +5 g→a mutation. In standard assays he had normal PS concentrations (100% total and 97% free antigen) but low activity (30%), which strongly suggests the presence of a nonfunctional PS. Using MoAb S12, his PS plasma concentration was normal (100%), whereas using MoAb HPS 54, the concentration was only 45%, which strongly suggests that 55% of his PS molecules lacked EGF1. The lack of PS recognition by MoAb HPS 54 further suggests the presence of truncated PS lacking EGF1 in the propositus' son.
All mutations so far identified in patients with the type II PS deficiency phenotype have been missense mutations. We report the first PROS1 splice site mutation associated with hereditary type II PS deficiency. Contrary to previous splice site mutations identified in the PROS1 gene, which were associated with a quantitative deficiency, this mutation was associated with a qualitative deficiency. Synthesis of a truncated protein is strongly suspected, because the exon skipping observed in the mRNA transcript led to the conservation of an open reading frame. The skipped exon encoded EGF1, and the plasma phenotypes established using MoAbs with different target epitopes were consistent with the presence of a truncated circulating PS lacking EGF1. This would indicate that the EGF1 domain is not an absolute requirement for the correct folding, stability, and secretion of PS.
ACKNOWLEDGMENT
The authors are grateful to Prof B. Dahlbäck for providing the HPS 54 MoAb and to Prof R. Bertina for providing the S12 MoAb.
Address reprint requests to S. Gandrille, PhD, INSERM U. 428, UFR des Sciences Pharmaceutiques et Biologiques, 4, Avenue de l'Observatoire, 75006 Paris, France; e-mail:gandrill@pharmacie.univ_paris5.fr.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal