Abstract
Ferrochelatase (E.C. 4.99.1.1), the enzyme that catalyzes the terminal step in the heme biosynthetic pathway, is the site of defect in the human inherited disease erythropoietic protoporphyria (EPP). Previously it has been demonstrated that patients with EPP may have missense mutations leading to amino acid substitutions, early chain termination, or exon deletions. While it has been clearly demonstrated that two missense mutations result in lowered enzyme activity, it has never been shown what effect specific exon deletions may have. In the current work, recombinant human ferrochelatase has been engineered to have individual exon deletions corresponding to exons 3 through 11. When expressed in Escherichia coli, none of these possesses significant enzyme activity and all lack the [2Fe-2S] cluster. One of the human missense mutations, F417S, and a series of amino acid replacements at this site (ie, F417W, F417Y, and F417L) were examined. With the exception of F417L, all lacked enzyme activity and did not contain the [2Fe-2S] cluster in vivo or as isolated in vitro.
THE HUMAN GENETIC disease erythropoietic protoporphyria (EPP) has been shown to occur when the enzyme ferrochelatase, the terminal step in the heme biosynthetic pathway, has decreased activity.1 The major clinical symptom of the disorder is cutaneous photosensitivity of a relatively benign nature.2 However, in approximately 5% to 10% of the reported cases, one finds significant hepatobiliary accumulation of inclusion bodies composed of protoporphyrin crystals. In these cases, the inclusion may lead to biliary obstruction, liver cirrhosis, and eventual liver failure.3 While the disease exhibits an autosomal dominant mode of transmission in most cases, evidence exists suggesting that additional factors may affect the severity of the disorder and possibly explain the commonly reported variable penetrance of the disease symptoms.
Since the cloning of the cDNA for human ferrochelatase4 and the determination of the chromosomal location and genomic structure,5,6 a number of investigators have identified a variety of mutations in the gene encoding ferrochelatase. Two general categories of mutations have been found. In one, an intron/exon splicing error occurs resulting in an internal exon deletion. In the case of an exon 2, 4, or 6 deletion, this will result in an early termination of translation. In the second class of mutation, a missense mutation is present that results in an amino acid replacement or in early polypeptide chain termination. In three instances the recombinant missense proteins have been expressed with a demonstrated decrease of in vitro activity.7 8 To date, however, there have been no biophysical explanations for the decreased enzyme activity. Interestingly, the majority of EPP patients studied contain an exon deletion rather than a missense mutation.
One of the interesting findings among EPP patients is that the residual amount of ferrochelatase activity has usually been reported to be about 25% or 50%. Because EPP is a dominantly inherited disorder, one would expect a residual activity of about 50%, as is found for other dominantly inherited porphyrias. This variability in residual activity cannot be attributed to exon deletion mutations versus missense mutations because individual cases of both high and low residual activity exist within each general category. Possible explanations for the 25% versus 50% reduction are that ferrochelatase is a dimer,9 that cellular conditions (for example, derangement in lipid metabolism)10-12 might secondarily effect residual measured activity, or, that there is unequal expression of alleles.5,10,11 13
Because of the unexplained variability in cellular levels of activity and its potential relevance to clinical management of EPP, we have undertaken to characterize the biochemical and biophysical nature of some of the known human EPP mutations. In the present study, we present data on one previously identified missense mutation, F417S, and a series of possible exon deletions. The data below show that the mutation F417S results in the synthesis of an enzyme in which the [2Fe-2S] cluster, which is present at the carboxyl terminal end of animal ferrochelatase,14 15 is either not assembled or is highly unstable. Furthermore, we show that any exon deletion will result in an inactive enzyme, which lacks the [2Fe-2S] cluster.
MATERIALS AND METHODS
Strains and cell culture.
Recombinant mutant and wild-type human ferrochelatases were expressed in Escherichia coli (E coli) (strain JM109 and ferrochelatase-deficient E coliΔhem H as described elsewhere.7 For whole-cell electron paramagnetic resonance (EPR) investigations, all plasmids were expressed in E coli strain DW35 ΔfrdABCD andsdhC::kan as described previously.16 These cells were grown on a minimal glucose/fumarate medium.17
Plasmids.
To construct several individual mutations at Phe 417 (eg, F417L, F417S, F417W, and F417Y), cassette mutagenesis was used, taking advantage of aPst I endonuclease restriction site just upstream from F417 and a downstream HindIII site in the nearby polylinker region of the plasmid.7 For each mutation, synthetic sense and antisense oligos, both containing the desired base pair changes, were annealed to form a cassette. The resulting double-stranded cassette also contained the appropriate ends corresponding to the Pst I and HindIII site overhangs. The cassettes were first ligated into the mouse ferrochelatase expression vector, pHDTF2, as described earlier.7
To produce these same mutations in human ferrochelatase, the mutant mouse ferrochelatase plasmids were digested to remove the entire mouse ferrochelatase coding sequence leaving only the mutated cassette still intact in the linearized vector. An EcoRI site upstream from the start site and the Pst I were used to excise the main portion of mouse ferrochelatase, leaving only the short carboxy-terminal tail containing the desired mutation. The human ferrochelatase plasmid, pHDTF20, containing an additional ribosomal binding site for increased protein production,7 was correspondingly digested with EcoRI and Pst I. The digested human ferrochelatase insert and mutant-containing mouse ferrochelatase vectors were isolated by agarose gel electrophoresis, purified by Geneclean (BIO 101, Inc, Vista, CA) and ligated.
The resulting recombinant proteins are actually chimeric, containing 350 amino acids from the wild-type mature length human ferrochelatase and 12 carboxy-terminal amino acids, including the desired mutation, from the mouse mutant ferrochelatases. Because the final 12 amino acid residues are completely identical in wild-type mouse and human ferrochelatases, no further modifications were necessary. Mutations were confirmed using the fmol DNA Sequencing System (Promega Corp, Madison, WI).
To create the exon 3 and exon 11 deletions, polymerase chain reaction (PCR) was employed using the plasmid described above, pHDTF20, as a template. Specifically, for the exon 3 deletion the sense primer was 5′ ATATACC ATGGCTAAG CTG GCA CCA TTC ATC 3′. The double underline denotes the unique Nco I site used for cloning purposes, and the underline corresponds to the amino acid sequence KLAPFI, the first six amino acids of exon 4. The antisense primer used for the exon three deletion was 5′ CA GAC CGC TTC TGC GTT CTG 3′, corresponding to a sequence in the TF20 plasmid outside the coding region. This primer pair was used to amplify the coding region of ferrochelatase from the indicated beginning of exon 4 through to the termination codon and including the Nco I/HindIII cloning sites. The PCR conditions used were 3 minutes at 95°C (1 minute at 95°C, 1 minute at 52°C, and 2 minutes at 72°C) × 30, 7 minutes at 72°C with 2.5 U TAQ polymerase (Promega), 1 μmol/L final primers, and 100 ng template. The PCR product was Magic PCR prepped (Promega), and the PCR product and TF20 were digested with Nco I and HindIII, the digestion products were run out on a 1% tris acetic acid EDTA (TAE) gel, and the exon 3 deleted piece was exchanged for the wild-type ferrochelatase piece.
For the exon 11 deletion, the following antisense primer was used: 5′ TAACCGGC AAGCTT CACTT AGA GAA CAA TGG ATT 3′. The HindIII site is indicated by the double underline and the underline, corresponding to the amino acid sequence NPLFSK, is the carboxyl terminal end of exon 10. This primer was used in conjunction with a sense primer contained in the vector of the TF20 plasmid upstream of the coding region, 5′ GTG TGG AAT TGT GAG CGG ATA AC 3′. The PCR product amplification was identical to that described for the exon 3 deletion. The resulting PCR product was Magic PCR prepped (Promega) and digested with EcoRI/HindIII, and the corresponding wild-type piece from TF20 exchanged for the exon 11 deletion.
The remainder of the exon deletions, exons 4 through 10, were created using the method of Deng and Nickoloff.18 In each case, the selection primer used was 5′ GCT CAT CAT TGG ATA TCG TTC TTC GGG 3′, which mutated a unique EcoRI site to an EcoRV site. The mutagenic primers used were:
In each case the * indicates the boundary between the exon preceding the one deleted and the exon after the one deleted. The template contained the EcoRI/HindIII fragment from pHDTF20 in the corresponding sites of pGEM-3Z. The mutated ferrochelatases in each case were digested with EcoRI/HindIII and exchanged with the wild-type ferrochelatase in the plasmid TF20. All exon deletions were confirmed by fmol sequencing (Promega).
Purification.
Recombinant wild-type and F417 mutant ferrochelatases were purified as described previously.7 The procedure involves solubilization of the protein, ammonium sulfate precipitation, and an affinity column. It was not possible to purify the exon deletion mutants according to this method, so cell lysates of E coliΔhem H expressing these proteins were used for characterization of these mutants.
Characterization.
For wild-type and F417 mutant ferrochelatases, protein purity was determined by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis, and protein concentration was quantitated by UV-visible absorption at 278 nm with ε = 46,900 M−1cm−1. Ferrochelatase activity was assayed using iron and either protoporphyrin or mesoporphyrin (Porphyrin Products, Logan, UT) as substrates. Product was quantitated as the pyridine hemochromogen.19
For whole-cell EPR, cells were harvested by centrifugation at low speed, washed, and transferred to an anerobic chamber. Cell pellets were resuspended in a buffer containing 100 mmol/L Tris morpholino-propane sulfonic acid (MOPS), pH 8.1, and excess sodium dithionite. Samples were placed in EPR tubes, and a small amount of chloroform was mixed into the tube before freezing.16
Western blot analysis.
For Western blot analysis, mutant and wild-type ferrochelatases were expressed in E coliΔhem H and in E colistrain DW35 ΔfrdABCD and sdhC::kan. Rabbit antihuman ferrochelatase antibody was used after standard procedures with the ProtoBlot Western Blot AP System (Promega).
Instrumentation.
Oligonucleotides were synthesized using an Applied Biosystems Model 391 DNA Synthesizer (Applied Biosystems, Foster City, CA). UV-visible absorption spectra were recorded on a Varian 219 Spectrophotometer (Varian Australia Pty Ltd, Victoria, Australia). EPR spectra were recorded using a Bruker ESP-300E spectrometer (Bruker Instruments, Billerica, MA) equipped with an Oxford Instruments ESR-9 liquid helium flow cryostat (Oxford Instruments, Oxfordshire, UK).
RESULTS
Exon deletion mutations.
A series of ferrochelatase mutations were made that correspond to deletion of exons 3 through 11. In case of exons 4 and 6 where a natural exon deletion would result in a downstream frameshift with early termination, we instead engineered a simple deletion of the sequence coded by exon 4 or exon 6. This was done to produce a protein that would allow us to determine if activity remained and if the [2Fe-2S] cluster was still present. Figure 1 shows the sequence of human ferrochelatase with the positions of all intron-exon boundaries noted.
Full-length sequence of human ferrochelatase. The full-length amino acid sequence of human ferrochelatase is shown with intron/exon boundaries specified by **. Phenylalanine at position 417 is denoted by the double underline.
Full-length sequence of human ferrochelatase. The full-length amino acid sequence of human ferrochelatase is shown with intron/exon boundaries specified by **. Phenylalanine at position 417 is denoted by the double underline.
All engineered deletions were expressed using a previously described expression vector that contains a Tac promotor with T7 enhancer and an optimally spaced ribosomal binding site.7 When these constructs were expressed in E coli strain JM109 and in E coli strain DW35 ΔfrdABCD and sdhC::kan, all produced proteins of the appropriate size as determined by Western blot analysis (Fig 2), although exon 3 and 4 deletions did not yield bands in the blots. Because all other exon deletions, truncations, and site directed mutants in this and other studies (Kools and Dailey, in preparation) yielded positive Western blots and because cDNA sequencing of all vectors confirmed the proper open reading frame on Δ exon 3 and 4, we believe that exons 3/4 contain the antigenic epitopes of ferrochelatase recognized by this antibody preparation.
Western blot analysis of exon deletion ferrochelatase mutants. Cell extracts of the exon deletion and wild-type human ferrochelatases containing about 5 μg were separated by SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and treated with human ferrochelatase antiserum. Procedures are as stated in Materials and Methods. From right to left, lanes 1 and 2, wild-type human ferrochelatase used as control; lanes 3 through 11, ▵exon 3 through ▵exon 11 in sequential order. Exons 3 and 4 are believed to contain the epitope for antibody recognition. The size differences of these proteins are evident as the mature-length wild-type human ferrochelatase (from pHDTF20) has a molecular weight of 42 kD and contains 363 amino acid residues, ▵exon 5 contains 320 residues, ▵exon 6 contains 331 residues, ▵exon 7 contains 333 residues, ▵exon 8 contains 330, ▵exon 9 contains 310, ▵exon 10 contains 347, and ▵exon 11 contains 318 residues.
Western blot analysis of exon deletion ferrochelatase mutants. Cell extracts of the exon deletion and wild-type human ferrochelatases containing about 5 μg were separated by SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and treated with human ferrochelatase antiserum. Procedures are as stated in Materials and Methods. From right to left, lanes 1 and 2, wild-type human ferrochelatase used as control; lanes 3 through 11, ▵exon 3 through ▵exon 11 in sequential order. Exons 3 and 4 are believed to contain the epitope for antibody recognition. The size differences of these proteins are evident as the mature-length wild-type human ferrochelatase (from pHDTF20) has a molecular weight of 42 kD and contains 363 amino acid residues, ▵exon 5 contains 320 residues, ▵exon 6 contains 331 residues, ▵exon 7 contains 333 residues, ▵exon 8 contains 330, ▵exon 9 contains 310, ▵exon 10 contains 347, and ▵exon 11 contains 318 residues.
Because it was not possible to purify the exon deletion ferrochelatases using the usual purification procedure, the proteins were expressed in Δhem H cells, which lack endogenous E coli ferrochelatase. As expressed in these cells, none of the deletion mutants have any enzyme activity. These proteins were also expressed in DW35 ΔfrdABCD and sdhC::kan, which is a strain of E coli that lacks the subunit of succinate dehydrogenase containing the [2Fe-2S] cluster.17 Thus, it is possible in this strain to see by in situ EPR if an expressed protein contains a [2Fe-2S] cluster. Expression of normal recombinant human ferrochelatase yielded a strong EPR signature for the cluster, but none of the expressed, exon deleted ferrochelatases had such a signal, thus strongly suggesting that an intact [2Fe-2S] cluster does not exist in vivo in these mutants.
F417 mutants.
Previously, F417S, F417W, F417Y, and F417L mutants were reported.7 At the time of the earlier publication, the presence of the [2Fe-2S] cluster had not been unequivocally demonstrated and the expression system for mammalian ferrochelatase produced significantly lower quantities of protein than the current expression system. Because of this we reexamined these mutant forms of ferrochelatase. It was found that F417S, the human EPP mutation identified by Brenner et al,5 lacks the spectral characteristics of an intact [2Fe-2S] cluster (Fig 3) and has extremely low enzyme activity. Likewise, F417W and F417Y have no cluster and no activity (data not shown). However, F417L, which previously had been suggested to have low activity,7 has near normal levels of enzyme activity and a spectrum characteristic of the [2Fe-2S] cluster. This mutant does appear to have a cluster that is less stable than the normal, which explains why lower activity was observed in the older expression system where significantly lower (ca. 10-fold) level of protein expression was achieved. Growth of F417S, F417W, or F417Y in DW35 ΔfrdABCD and sdhC::kan does not result in the generation of a [2Fe-2S] cluster EPR signature, whereas F417L does (Fig 4).
UV/visible spectrum of F417S recombinant human ferrochelatase. The spectrum of purified recombinant F417S ferrochelatase in elution buffer7 was obtained with a Varian 219 spectrophotometer. Not present are characteristic features resulting from the [2Fe-2S] cluster, which are normally present at 460 nm, 420 nm, and 325 nm in spectra from the wild-type protein.14 A minor absorbance peak at 410 nm is attributed to small amounts of residually bound porphyrin.
UV/visible spectrum of F417S recombinant human ferrochelatase. The spectrum of purified recombinant F417S ferrochelatase in elution buffer7 was obtained with a Varian 219 spectrophotometer. Not present are characteristic features resulting from the [2Fe-2S] cluster, which are normally present at 460 nm, 420 nm, and 325 nm in spectra from the wild-type protein.14 A minor absorbance peak at 410 nm is attributed to small amounts of residually bound porphyrin.
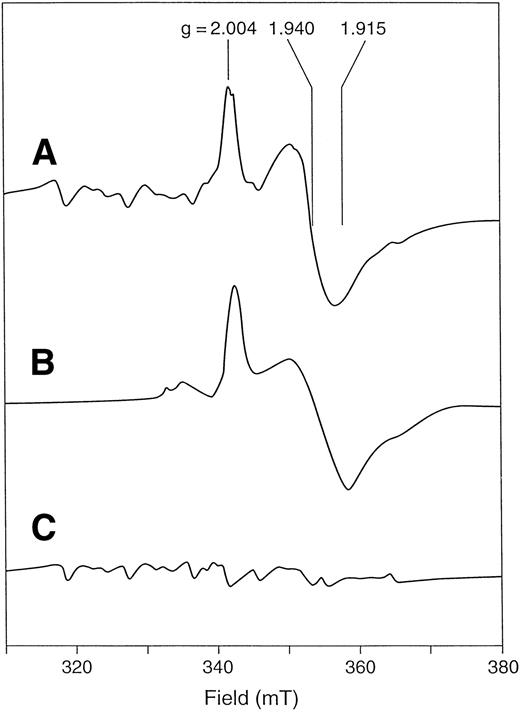
X-band EPR spectrum of F417L recombinant human ferrochelatase. Spectra are from whole cells of E coli DW35 (A) with the plasmid encoding wild-type human ferrochelatase, (B) with the plasmid encoding F417L human ferrochelatase, and (C) alone, containing no plasmid. Cell pellets were washed and suspended anerobically in 0.1 mol/L Tris MOPS, pH 8.1, containing an excess of sodium dithionite. Immediately before freezing, approximately 10% (vol/vol) chloroform was added. EPR conditions: temperature, 35°K; microwave power, 10 mW; microwave frequency, 9.60 Ghz; modulation amplitude, 0.64 mT.
X-band EPR spectrum of F417L recombinant human ferrochelatase. Spectra are from whole cells of E coli DW35 (A) with the plasmid encoding wild-type human ferrochelatase, (B) with the plasmid encoding F417L human ferrochelatase, and (C) alone, containing no plasmid. Cell pellets were washed and suspended anerobically in 0.1 mol/L Tris MOPS, pH 8.1, containing an excess of sodium dithionite. Immediately before freezing, approximately 10% (vol/vol) chloroform was added. EPR conditions: temperature, 35°K; microwave power, 10 mW; microwave frequency, 9.60 Ghz; modulation amplitude, 0.64 mT.
DISCUSSION
Erythropoietic protoporphyria is a puzzling genetic disease because of the wide and currently unexplained variability in its clinical and biochemical expression. Clinically, symptomatic expression of photosensitivity is highly variable between individuals and may even vary within affected siblings. Biochemically one finds that levels of residual ferrochelatase activity reported in the literature for heterozygous individuals may vary from 11% to 50%.20 21Interestingly, the clinical expression of the disease may not correlate well with the level of enzyme activity except in individuals with extremely low levels of residual activity. At the molecular level, the disease is heterogeneous in nature and may be attributable to nucleic acid mutations resulting in either exon deletions or missense mutations. These differences in types of mutations, however, do not explain the variability in residual enzyme activity.
Previously, we examined two different ferrochelatase missense mutations by studying enzymatic properties of the purified recombinant proteins.7 Because EPP patients are heterozygous for the dominantly inherited disorder, the ability to express recombinant, homogenous mutant ferrochelatase protein in E coli allowed us to study properties of just the mutant form of the enzyme. In these earlier studies, it was found that one, M267I, resulted in a thermolabile protein, whereas the other, F417S, was reported to possess minimal enzyme activity.7 After that report, we determined that mammalian ferrochelatase possesses a [2Fe-2S] cluster at the carboxyl terminal end of the protein and that in the absence of the cluster, the apoprotein has greatly diminished activity.14With this background, along with a higher yielding expression system, we chose to reexamine the F417S mutation.
The data presented above clearly shows that the F417S mutation results in a protein whose [2Fe-2S] cluster is unstable. The fact that this construct is able to modestly complement a ferrochelatase deficient strain of E coli7 suggests that in vivo there may exist a small amount of enzyme with an intact cluster or that the small residual ferrochelatase activity remaining is sufficient for cell growth. Purification of the mutant protein yielded enzyme without a detectable cluster. Thus, the F417S human protoporphyric mutation results from the synthesis of a mutant protein that possesses diminished enzyme activity because it lacks the [2Fe-2S] cluster.
Three additional mutations were made at F417; F417W, F417Y, and F417L. Interestingly, both the tryptophan and tyrosine replacements, which would seem to be much less drastic than the serine replacement, had no activity and lacked the characteristic spectra of the [2Fe-2S] cluster. The leucine replacement, however, retained the cluster and enzyme activity. This suggests that the region normally occupied by F417 is hydrophobic in nature, but is spatially too small to accommodate the bulk of a tryptophan side chain.
Taken as a whole, these data would indicate that F417 plays no essential role in substrate binding or catalysis, but it apparently has some role in stabilization of the [2Fe-2S] cluster. Because it has been shown that this cluster is involved in the response to nitric oxide (NO),22 any mutation that would destabilize this feature may result in an enzyme that, even with residual activity, would be more sensitive to NO and any other potential effectors. Recent data imply that only three of the four amino acids, which coordinate the [2Fe-2S] cluster, are located in the carboxyl terminal region in exon 11.16 If the fourth ligand to the cluster is located in an earlier exon, this could explain the lack of the cluster in other exon deletion mutants. This also supports the idea that the metal center may function to regulate the activity of ferrochelatase through maintaining the structure of the protein.22
To date ferrochelatase exon deletions of exons 2,233,24 7,25 9,26 and 1021,27 have been reported. With the exception of exon 2 skipping, none of these exon mutations introduces a frame shift, so it would be expected that in these cases a protein should be produced that is missing the sequence corresponding to the exon that is skipped. Such would not be the case for skipping of exons 2, 4, and 6 where frame shift mutations occur, which result in truncated proteins with an altered carboxyl termini. It is clear that the deletion of exon 1 would result in a protein that would, at best, be truncated with a new amino terminus at met 73, about a dozen residues downstream from the putative proteolytic processing site, and it would not be targeted to its proper cellular location.28 While this alone may not cause significant problems, the absence of two conserved amino acid residues (Ile and Leu) just upstream from the new start site may result in an inactive protein, or equally likely the elimination of 5′UTR in exon 1 would result in an unstable or nonfunctional mRNA.
Before the current report, there had not been any characterizations of ferrochelatases resulting from an exon deletion. The data presented above clearly show that any exon deletion will result in an inactive enzyme and that none of these proteins appear to possess the [2Fe-2S] cluster. This suggests that any of these deletions affects the protein structure in a significant fashion. One point of interest from the current patient data base is that the large majority of EPP patients suffer from an exon deletion rather than a missense mutation. This is much different from what has been found to date in other porphyrias such as acute intermittent porphyria (AIP) where a large number of different missense mutations have been identified. Also of interest is that none of these missense mutations occur at a highly conserved residue.
ACKNOWLEDGMENT
The authors thank Elizabeth A. Strum for production of the antibody and for performing the Western blot.
Supported by Grants No. DK 32303 and DK 35898 from the National Institutes of Health (to H.A.D.) and by the National Science Foundation Training Group Award to the Center for Metalloenzyme Studies (DIR9014281).
Address reprint requests to H.A. Dailey, PhD, Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA 30602-7229.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.



![Fig. 3. UV/visible spectrum of F417S recombinant human ferrochelatase. The spectrum of purified recombinant F417S ferrochelatase in elution buffer7 was obtained with a Varian 219 spectrophotometer. Not present are characteristic features resulting from the [2Fe-2S] cluster, which are normally present at 460 nm, 420 nm, and 325 nm in spectra from the wild-type protein.14 A minor absorbance peak at 410 nm is attributed to small amounts of residually bound porphyrin.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/10/10.1182_blood.v91.10.3980/3/m_blod41005003x.jpeg?Expires=1763755572&Signature=p8Ic1UF47eI2MVmOtRLs1UP8WqoIWsMB3wpjvMsBXhWDUBfWGB573kxnu3iGL5kMpdj4r2x~97WOWAm7sz~e5SYoQ229KWa-KjkbJpweYTBHpNWyL7j3RNmEwAk~DcE3X6FX9tirtHQxv~8Oo1iRURhqe2P-20b1BAvcgn3c3Ixl9U7ITe8aP2BseCJGZeZiVjvm3h406w87psaeAX7~-uTo~oV0iEexjlIAh56PlASQZtSuXv5Zjv9FACIQESXlkwmbJebQg5OufajxOgz9dUXw2HhwJ4Is7bW5WHciH5cncnVzNO7phDofsNkUWyrDSp3oNYaB64tAm1OHSfVzLg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal