Abstract
T-cell prolymphocytic leukemia (T-PLL) is a rare form of mature leukemia that occurs both in adults as a sporadic disease and in younger patients suffering an hereditary condition, ataxia telangiectasia (AT). The ATM gene, located in the 11q22-23 chromosomal region, is consistently mutated in AT patients. The strong predisposition of AT patients to develop T-PLL and the high frequency of T-cell leukemias/lymphomas observed in atm-deficient mice, together with the known functions of the ATM protein, led us to evaluate the ATM gene as a potential tumor suppressor gene involved in T-PLL. Paired leukemic and nonleukemic cells were obtained from a series of 15 patients suffering sporadic T-PLLs, allowing loss of heterozygosity (LOH) analysis. LOH of the 11q22-23 region was detected in 10 of these 15 cases (67%). The minimal deleted region was defined as an approximately 2.5 Mb interval that contained the ATMgene. No ATM rearrangement or biallelic deletion was detected by Southern blotting in the T-PLL series. However, in five T-PLLs with LOH of the 11q22-23 region, Western blot analysis showed either undetectable (3 cases) or decreased levels (1 case) of ATM protein, whereas ATM was present at high levels in cases without LOH. The protein truncation test (PTT) was then used to search for mutations in the ATM gene. Four mutations (1 nonsense, 2 aberrant splicings, and 1 missense) were detected in patients with LOH and none in patients without LOH of the region. The acquired character of these ATM mutations was demonstrated in three patients. Altogether, allelicATM inactivations by large deletions or mutations were found in approximately two thirds of T-PLL. ATM is thus a tumor suppressor gene whose inactivation is a key event in the development of T-cell prolymphocytic leukemias.
T-CELL PROLYMPHOCYTIC leukemia (T-PLL) is a rare lymphoid neoplasm generally associated with a high count of atypical lymphocytes with a postthymic phenotype, lymphadenopathy, splenomegaly, skin lesions, and an aggressive clinical course.1 Molecular characterization of recurrent chromosomal aberrations associated with T-PLL led to the identification of the MTCP1 and TCL1 genes located on the Xq28 and 14q32.1 regions, respectively.2-4 These genes code for a new class of oncoproteins of, as yet, unknown cellular functions.5,6 On the other hand, the relationship between a rare genetic condition, ataxia telangiectasia (AT) and the occurrence of T-PLL has been progressively recognized.7-9 Four lines of evidence support this link: (1) peripheral blood karyotypes show clonal aberrations in approximately 10% of AT patients.10-14 These aberrations have been shown to occur in lymphoid T-cell populations (named AT clonal proliferations or ATCP), which share many morphologic, immunologic, cytogenetic, and molecular features of T-PLL, except that they are not associated with lymphocytosis or with clinical evidence of malignancy2-4,6,7,15,16 (for reviews, see Stern8 and Taylor et al9). (2) The progression of a few cases of ATCP into a bona fide T-PLL with an aggressive clinical course further argues that ATCP represents the preleukemic stage of T-PLL.9,11,12,17-19 (3) Epidemiological data highlight the strong predisposition of AT patients to develop T-cell leukemias/lymphomas, including T-PLL.9 (4) Knock-out mice with a complete AT-like phenotype consistently develop immature T-cell malignancies in a few months.20-22
Positional cloning has allowed the identification of ATM, a unique gene consistently inactivated by mutation in patients with a classical AT phenotype.23 The ATM gene, located in the 11q22-23 chromosomal region, spans 184 kb of genomic DNA and consists of 66 exons.24,25 The 13-kb ATM transcript encodes a 3056 amino acid (aa) protein belonging to a subgroup of kinases sharing homologies with the catalytic domain of phosphatidylinositol 3-kinase and thought to be involved in DNA metabolism and cell cycle checkpoint control.23 26
The association between ATM deficiency and occurrence of T-PLL prompted us to search for ATM allelic deletions or rearrangements in sporadic T-PLLs with no history of AT. We therefore screened a T-PLL series for allelic loss of the ATM gene region and for abnormalities of the ATM gene.
MATERIALS AND METHODS
Cells.
A series of 15 T-PLL cases with frozen leukemic cells available was assembled by a call to the French Hematologic centers. Patients had major lymphocytosis and no history of ataxia telangiectasia. Diagnosis of T-PLL was established according to the French-American-British (FAB) criteria for chronic (mature) leukemia.27 DNA samples from each of these patients were prepared from buccal cavity epithelial cells, fractionated granular cells, or Epstein-Barr virus (EBV)-transformed cell lines established from blood samples, allowing the analysis of paired normal and leukemic DNAs.
Loss of heterozygosity (LOH) analysis.
Nine polymorphic DNA markers flanking and within the ATM gene on 11q22-q23 were amplified by polymerase chain reaction (PCR), allowing the heterozygosity status analysis. The 11q22-23 microsatellite markers D11S1817, D11S1819, D11S2179, D11S1778, D11S1294, D11S2180, D11S2178, D11S1300, and D11S1391 (from centromere to telomere) were selected, in view of the map described by Savitsky et al23 and by Laake et al28 (see Table 1). Eight additional markers on chromosome 11 were used to explore the extent of the deletions, namely D11S932 (located on chromosomal band 11p15), D11S913 (11q12), D11S901 (11q13.5), D11S917 (11q14.3), D11S938 (11q23), D11S925 (11q23), D11S934 (11q23.3), and D11S910 (11q25). The primer sets were synthesized by Genset (Paris, France). The relative intensities of the two amplified allelic markers were compared after autoradiography. In patients showing a germinal heterozygosity for a given marker, the complete, or near complete (≥ 90% decreased intensity) absence of an allele in the tumor samples consistent with loss of one allele and a low level of nonleukemic cell contamination, was interpreted as LOH.
LOH Analysis in Paired Germline and Leukemic DNAs
| Markers . | Location . | Bat . | Big . | Bul . | Dia . | Imb . | Jun . | Lec . | Mal . | Pot . | Van . | Ber . | Bouq . | Gob . | Mel . | Ver . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D11S932 | p15 | □ | □ | □ | NA | NA | □ | NA | H | NA | □ | NA | H | H | H | NA |
| D11S913 | q12 | ▪ | □ | □ | NA | NA | □ | NA | NA | NA | H | NA | H | H | H | NA |
| D11S901 | q13.5 | ▪ | ▪ | NA | NA | NA | H | NA | NA | NA | ▪ | NA | □ | □ | □ | NA |
| D11S917 | q14.3 | ▪ | ▪ | □ | NA | NA | ▪ | NA | ▪ | NA | H | NA | □ | □ | H | NA |
| D11S1817 | q22-23 | ▪ | ▪ | NA | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ | H | □ | □ | □ |
| D11S1819 | q22-23 | ▪ | ▪ | H | ▪ | ▪ | ▪ | ▪ | H | ▪ | ▪ | □ | □ | □ | □ | □ |
| D11S2179-150 | q23 | ▪ | H | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | H | ▪ | H | H | □ | □ | H |
| D11S1778 | q23 | ▪ | H | ▪ | H | ▪ | ▪ | ▪ | ▪ | H | ▪ | □ | □ | □ | □ | □ |
| D11S1294 | q23 | NA | H | ▪ | ▪ | NA | ▪ | NA | ▪ | H | H | □ | □ | □ | □ | NA |
| D11S2180 | q23 | H | ▪ | ▪ | H | ▪ | ▪ | H | ▪ | NA | H | □ | NA | □ | H | □ |
| D11S2178 | q23 | ▪ | ▪ | NA | □ | ▪ | ▪ | ▪ | ▪ | ▪ | H | □ | □ | □ | □ | H |
| D11S1300 | q23 | ▪ | H | H | □ | ▪ | ▪ | H | ▪ | H | H | □ | NA | □ | □ | H |
| D11S1391 | q23 | ▪ | ▪ | H | □ | H | H | ▪ | ▪ | ▪ | ▪ | □ | □ | H | □ | □ |
| D11S938 | q23 | NA | H | □ | NA | NA | ▪ | NA | NA | NA | ▪ | NA | □ | □ | H | NA |
| D11S925 | q23 | ▪ | NA | □ | NA | NA | ▪ | NA | NA | NA | ▪ | NA | □ | □ | □ | NA |
| D11S934 | q23.3 | H | H | □ | NA | NA | H | NA | ▪ | NA | NA | NA | □ | □ | NA | NA |
| D11S910 | q25 | NA | H | H | NA | NA | □ | NA | NA | NA | NA | NA | NA | NA | □ | NA |
| Markers . | Location . | Bat . | Big . | Bul . | Dia . | Imb . | Jun . | Lec . | Mal . | Pot . | Van . | Ber . | Bouq . | Gob . | Mel . | Ver . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D11S932 | p15 | □ | □ | □ | NA | NA | □ | NA | H | NA | □ | NA | H | H | H | NA |
| D11S913 | q12 | ▪ | □ | □ | NA | NA | □ | NA | NA | NA | H | NA | H | H | H | NA |
| D11S901 | q13.5 | ▪ | ▪ | NA | NA | NA | H | NA | NA | NA | ▪ | NA | □ | □ | □ | NA |
| D11S917 | q14.3 | ▪ | ▪ | □ | NA | NA | ▪ | NA | ▪ | NA | H | NA | □ | □ | H | NA |
| D11S1817 | q22-23 | ▪ | ▪ | NA | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | □ | H | □ | □ | □ |
| D11S1819 | q22-23 | ▪ | ▪ | H | ▪ | ▪ | ▪ | ▪ | H | ▪ | ▪ | □ | □ | □ | □ | □ |
| D11S2179-150 | q23 | ▪ | H | ▪ | ▪ | ▪ | ▪ | ▪ | ▪ | H | ▪ | H | H | □ | □ | H |
| D11S1778 | q23 | ▪ | H | ▪ | H | ▪ | ▪ | ▪ | ▪ | H | ▪ | □ | □ | □ | □ | □ |
| D11S1294 | q23 | NA | H | ▪ | ▪ | NA | ▪ | NA | ▪ | H | H | □ | □ | □ | □ | NA |
| D11S2180 | q23 | H | ▪ | ▪ | H | ▪ | ▪ | H | ▪ | NA | H | □ | NA | □ | H | □ |
| D11S2178 | q23 | ▪ | ▪ | NA | □ | ▪ | ▪ | ▪ | ▪ | ▪ | H | □ | □ | □ | □ | H |
| D11S1300 | q23 | ▪ | H | H | □ | ▪ | ▪ | H | ▪ | H | H | □ | NA | □ | □ | H |
| D11S1391 | q23 | ▪ | ▪ | H | □ | H | H | ▪ | ▪ | ▪ | ▪ | □ | □ | H | □ | □ |
| D11S938 | q23 | NA | H | □ | NA | NA | ▪ | NA | NA | NA | ▪ | NA | □ | □ | H | NA |
| D11S925 | q23 | ▪ | NA | □ | NA | NA | ▪ | NA | NA | NA | ▪ | NA | □ | □ | □ | NA |
| D11S934 | q23.3 | H | H | □ | NA | NA | H | NA | ▪ | NA | NA | NA | □ | □ | NA | NA |
| D11S910 | q25 | NA | H | H | NA | NA | □ | NA | NA | NA | NA | NA | NA | NA | □ | NA |
Symbols: ▪, LOH; □, no LOH.
Abbreviations: H, germinal homozygosity; NA, no data available.
Marker located within the ATM gene.
Southern blot analysis.
High molecular weight DNA was extracted from leukemic samples, digested with BamHI, SacI, or HindIII restriction enzymes and the resulting fragments were separated according to size by electrophoresis through 0.8% agarose gels. After alkaline transfer to Hybond N+ membranes (Amersham, Les Ulis, France) samples were hybridized with 32P radiolabeled probes to partially explore the ATM locus. Four ATMprobes were prepared by PCR amplification of reverse transcribed peripheral blood lymphocyte (PBL) RNA using the following primer pairs: 5′-TTTCCAAGGCTATTCAGTGTGC-3′ and 5′-TGCTCTATTCCCATTTTTACCG-3′ (ATM1, 1.0 kb), 5′-GGGTGTCCTTGGCTGCTACTG-3′ and 5′-TGCGAACTTGGTGATGATTGTC-3′ (ATM2, 1.1 kb), 5′-ATTGTGGTGGAGTTATTGATGACG-3′ and 5′-AGCGAACAATCCCAGCCTAAAA-3′ and 5′-GATGAGGGGATTGCTGTTTCG-3′ (ATM3, 2.1 kb), and 5′-AGCGAACAATCCCAGCCTAAAA-3′ and 5'-ACACCTTCAACACCCGTAATGC-3' (ATM4, 1.8 kb).
Western blotting.
Cellular proteins were extracted with Triple Detergent Lysis Buffer,29 quantified using the BCA kit (Pierce, Rockford, IL), sized fractionated on 5% sodium dodecyl sulfate (SDS)-polyacrylamide gels, and electrotransfered to nitrocellulose membranes. The membranes were blocked overnight in 5% nonfat milk powder in PBST (phosphate-buffered saline, 0.2% Tween 20). The anti-ATM antiserum, pAb 132, was diluted 1:2000 in the blocking buffer and applied to the membranes.30 After a 1-hour incubation, the membranes were washed with PBST and incubated with goat antirabbit IgG-peroxidase conjugate (Boehringer, Meylan, France) for 1 hour. After three washes in PBST, the membranes were overlaid with the chemiluminescent substrate solution and developed according to the instructions of the manufacturer (ECL, Amersham).
Protein truncation test (PTT).
First strand cDNAs were generated from polyA-enriched RNAs using the QuickPrep Micro mRNA Purification and First-Strand cDNA Synthesis kits according to the instructions of the manufacturer (Pharmacia, Orsay, France). The coding region of the ATM gene was divided into seven overlapping regions: region no. 1 (codons 1-533), no. 2 (codons 437-968), no. 3 (900-1388), no. 4 (1321-1818), no. 5 (1760-2177), no. 6 (2107-2618), and no. 7 (2550-3056). Forward primers were designed to include a T7 promoter sequence for the initiation of transcription by T7 RNA polymerase, as well as a translation initiation site. The primer sequences and PCR conditions are available on the Institut Curie (1997) website (http://www.curie.fr/curie/sm/atm) or upon request. PTT analysis was performed from the PCR products using the TNT T7 Coupled Reticulocyte Lysate System as recommended by the manufacturer (Promega, Lyon, France). 35S-labeled protein products were separated on 12% SDS-polyacrylamide gel and detected by autoradiography.
Identification of the mutations.
PCR products showing an abnormal pattern in the PTT analysis were directly sequenced using the dRhodamine Terminator Cycle Sequencing kit on an ABI 377 automatic sequencer (PE Applied Biosystems, Foster City, CA). The relevant genomic regions were sequenced from PCR products using the following primers: for amplification of exon 20 and 29 regions (forward) GTTGTGCCCTTCTCTTAGTGTT (reverse) ACTCATTACATTTAGTCAGCAA, and (forward) CCAGGTCATACAACTTAATGAT (reverse) GAATTTGCTGGCTCATGTAACG, respectively31; for amplification of the exon 45 region, (forward) TAT CTT AGG GTT CTG TTT TTA (reverse) AGT AAC TTT GTC TTT TCA TAA T. Numbering of nucleotides is based on either the ATM transcript sequence (accession number U33841) for the cDNA, or on the full-length ATM gene sequence (accession number U82828) for the genomic DNA.25 32Numbering of codons starts at the initiating ATG.
RESULTS
LOH of the ATM region in T-PLLs.
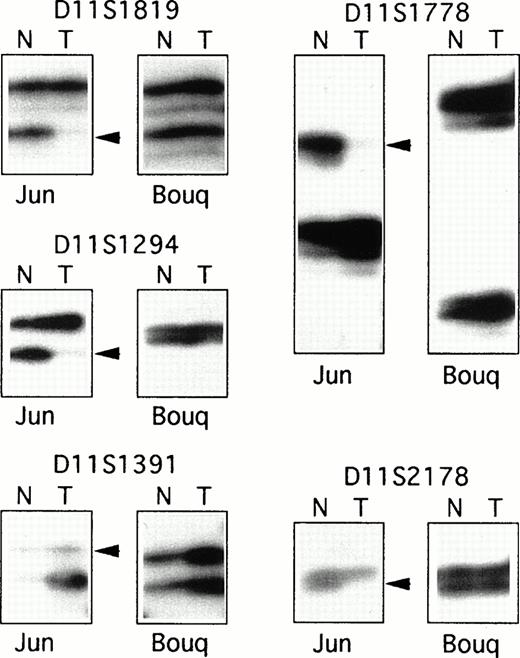
As T-PLL is a rare disease associated with a short survival time, most of the cases included in this series were retrospective, and samples for these cases were obtained from cryopreserved specimens. Therefore, to obtain nonleukemic cells and to assemble a series of paired nonleukemic and leukemic DNAs, EBV-transformed cells lines were derived from patients' frozen blood samples, and buccal cavity epithelial cells or fractionated granular cells were obtained from living patients. Altogether, a series of 15 paired DNAs were assembled from patients with T-PLL. In 10 of the 15 cases (67%), LOH was demonstrated for all the informative markers of the D11S1817-D11S2180 interval (Table 1 and Fig 1), whereas no LOH was shown in five patients for all informative markers. Analysis of eight additional markers covering chromosome 11 showed various sized regions of LOH. The minimal region of LOH was defined by patients “Bul” and “Dia” as the interval of approximately 2.5 Mb between markers D11S917 and D11S2178, which included the ATM locus (Table 1).
PCR analysis of LOH. Examples of the PCR analysis of LOH for two T-PLL cases: “Jun”, which showed a large region of LOH for the 11q22-23 informative markers, and “Bouq,” which showed balanced heterozygosity of the markers. Each autoradiograph includes PCR products from normal (N) and tumor (T) DNAs. Arrowheads indicate allelic loss in tumor DNAs.
PCR analysis of LOH. Examples of the PCR analysis of LOH for two T-PLL cases: “Jun”, which showed a large region of LOH for the 11q22-23 informative markers, and “Bouq,” which showed balanced heterozygosity of the markers. Each autoradiograph includes PCR products from normal (N) and tumor (T) DNAs. Arrowheads indicate allelic loss in tumor DNAs.
Southern blot analysis.
All T-PLLs were analyzed by Southern blotting using at least two restriction digests and four ATM cDNA fragments as probes to partially explore the 184-kb ATM locus. No biallelic deletion or DNA rearrangement was detected in the 15 T-PLL cases (data not shown).
Western blot analysis.
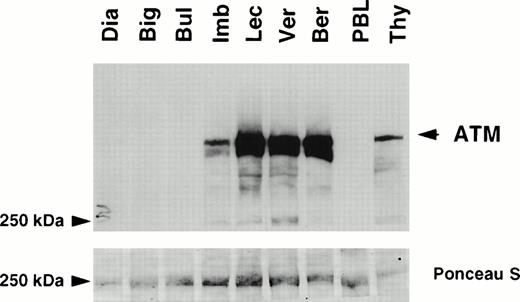
Protein extracts were available from seven T-PLL cases, five with LOH and two without LOH of the 11q22-23 region. The level of expression of the ATM protein was determined by Western blotting using a polyclonal antibody pAB132 directed against amino acid residues 819-844 of ATM (Fig 2).30 ATM was highly expressed in the two patients without LOH, at levels similar to those observed in thymocytes and higher than in normal peripheral blood mononuclear cells. In contrast, ATM was undetectable in three patients with LOH (cases “Dia”, “Big”, and “Bul”), and its level was reduced in patient “Imb”. Expression of ATM was similar in patient “Lec” and in patients without LOH.
Expression of ATM in T-PLLs . Detergent lysates (60 μg/lane) were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) through a 5% gel. Immunoblot analysis was performed using the anti-ATM antiserum pAB 132 (upper panel). Ponceau S staining was used to verify equal loading and protein integrity (lower panel). Samples analyzed were T-PLLs with LOH of the 11q22-23 region: Dia, Big, Bul, Imb, and Lec; T-PLLs without LOH: Ver and Ber; normal peripheral lymphocytes (PBL); normal thymocytes (Thy). Molecular weight standards are given in kD. The ATM specific signal is arrowed.
Expression of ATM in T-PLLs . Detergent lysates (60 μg/lane) were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) through a 5% gel. Immunoblot analysis was performed using the anti-ATM antiserum pAB 132 (upper panel). Ponceau S staining was used to verify equal loading and protein integrity (lower panel). Samples analyzed were T-PLLs with LOH of the 11q22-23 region: Dia, Big, Bul, Imb, and Lec; T-PLLs without LOH: Ver and Ber; normal peripheral lymphocytes (PBL); normal thymocytes (Thy). Molecular weight standards are given in kD. The ATM specific signal is arrowed.
ATM mutations detected by PTT.
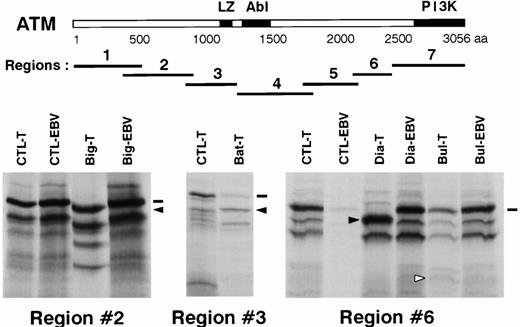
PTT analysis was performed on nine T-PLLs (seven with LOH and two without). The ATM coding sequence was divided into seven overlapping regions, which were amplified by PCR and analyzed after transcription-translation. Four abnormal patterns were shown, occurring in regions no. 2, no. 3, and no. 6 (two cases) (Fig 3). The four relevant PCR products were directly sequenced on both strands.
Mutation screening by PTT. Upper panel, schematic representation of the ATM protein with its presently known domains. LZ: putative leucine zipper domain, Abl: Abl-binding region, PI3K: phospho-inositol 3 kinase family domain. The seven regions analyzed by PTT are shown in relation to the ATM protein. A scale is shown in aa. Lower panel, PTT analysis of the three regions showing abnormal patterns. CTL-T: T-PLL sample showing a normal pattern. CTL-EBV: corresponding EBV-transformed cell line sample. Big-T, Bat-T, Dia-T, Bul-T: T-PLL samples showing abnormal patterns. Big-EBV, Dia-EBV, Bul-EBV: corresponding EBV-transformed cell line samples. Dashes indicate wild-type full-length products and arrows indicate abnormal bands.
Mutation screening by PTT. Upper panel, schematic representation of the ATM protein with its presently known domains. LZ: putative leucine zipper domain, Abl: Abl-binding region, PI3K: phospho-inositol 3 kinase family domain. The seven regions analyzed by PTT are shown in relation to the ATM protein. A scale is shown in aa. Lower panel, PTT analysis of the three regions showing abnormal patterns. CTL-T: T-PLL sample showing a normal pattern. CTL-EBV: corresponding EBV-transformed cell line sample. Big-T, Bat-T, Dia-T, Bul-T: T-PLL samples showing abnormal patterns. Big-EBV, Dia-EBV, Bul-EBV: corresponding EBV-transformed cell line samples. Dashes indicate wild-type full-length products and arrows indicate abnormal bands.
A protein product slightly smaller than normal was observed in region no. 2 for patient “Big” (Fig 3). The sequence of this region showed that a single T nucleotide was inserted at position 2891 [2891insT]. The frameshift created by this insertion led to a stop codon 51 bases downstream. The predicted ATM protein is a truncated protein of 901 aa, plus 17 aa added after the frameshift. The predicted protein product for region no. 2 was compatible with the PTT result. Analysis of nonleukemic cells (an EBV-transformed cell line) from this patient showed a normal pattern in PTT (Fig 3) and the absence of the 2891insT mutation in the amplified cDNA fragment. To exclude an unstable mutated ATM transcript, the corresponding genomic DNA of the EBV cell line was amplified and its sequence was found to be normal.
An approximately 55-kD product was observed in patient “Dia” in the PTT analysis of region no. 6, as compared with the 59.5-kD normal product (Fig 3). The amplified cDNA fragment corresponding to this region was abnormally short and its sequence showed a 105-bp deletion, encompassing exon 46 [6536del105]. This deletion created no frameshift and no abnormal codon, and the predicted protein, 35 aa shorter than the wild-type protein, was compatible with the PTT result. To understand the cause of the direct splicing of exon 45 to exon 47, the fragment containing exon 46 and the adjacent splice donor and acceptor sites was amplified from leukemic genomic DNA and sequenced. A single transition G to A in position +1 of the donor site of exon 46 was detected [IVS46+1G → A], putatively explaining the observed exon skipping (Fig4A). This was an acquired mutation because the analysis of the nonleukemic cells from this patient showed a normal pattern in PTT and a cDNA sequence for region no. 6 identical to the published one. The exon 46 region of genomic DNA was also amplified from “Dia” nonleukemic cells and showed a genomic sequence identical to the published one.
Analysis of ATM mutations. (A) Exon skipping in patient “Dia”. Upper panel, normal partial cDNA sequence demonstrated in the “Dia” EBV cell line sample as compared with the tumor cDNA sequence, which is deleted for the 105-bp of exon 46. Lower panel, genomic sequence of germline (EBV) and tumor (T-PLL) DNAs. Exon 46 sequence is indicated in bold. The G>A mutation is double underlined. (B) Insertion of 29 nt in tumor DNA from patient “Bat”. Tumor cDNA sequences contained, in approximately equal amounts, wild-type and mutated (4182ins29) sequences. The normal corresponding peptide sequence is indicated. The stop codon due to the frameshift as a consequence of the 29 nt insertion is indicated in bold and underlined. Lines indicated the correspondence between the mutated cDNA sequence and the normal published genomic sequence (accession number U82828). The genomic sequence of “Bat” tumor DNA was identical to the published one. The putative splice donor and acceptor sites on each side of the 29 nt insertion are doubly underlined, and the exon 29 sequence is indicated in bold. Numbering of nucleotides is based on either the ATM transcript sequence (accession number U33841) for the cDNA, or on the full-length ATM gene sequence (accession number U82828) for the genomic DNA.25 32Numbering of codons starts at the initiating ATG.
Analysis of ATM mutations. (A) Exon skipping in patient “Dia”. Upper panel, normal partial cDNA sequence demonstrated in the “Dia” EBV cell line sample as compared with the tumor cDNA sequence, which is deleted for the 105-bp of exon 46. Lower panel, genomic sequence of germline (EBV) and tumor (T-PLL) DNAs. Exon 46 sequence is indicated in bold. The G>A mutation is double underlined. (B) Insertion of 29 nt in tumor DNA from patient “Bat”. Tumor cDNA sequences contained, in approximately equal amounts, wild-type and mutated (4182ins29) sequences. The normal corresponding peptide sequence is indicated. The stop codon due to the frameshift as a consequence of the 29 nt insertion is indicated in bold and underlined. Lines indicated the correspondence between the mutated cDNA sequence and the normal published genomic sequence (accession number U82828). The genomic sequence of “Bat” tumor DNA was identical to the published one. The putative splice donor and acceptor sites on each side of the 29 nt insertion are doubly underlined, and the exon 29 sequence is indicated in bold. Numbering of nucleotides is based on either the ATM transcript sequence (accession number U33841) for the cDNA, or on the full-length ATM gene sequence (accession number U82828) for the genomic DNA.25 32Numbering of codons starts at the initiating ATG.
PTT analysis of region no. 3 of case “Bat” showed a weak signal of normal size and a more intense band of smaller size (Fig 3). Direct sequence of PCR products showed a mixture of cDNA species: one corresponding to the normal sequence and the second corresponding to an insertion of 29 nucleotides [4182ins29] (Fig 4B). The predicted protein coded by the transcripts with the 29 nt insertion is a 1332 aa truncated ATM protein, plus 17 aa added after the frameshift. Sequence comparison with published sequences showed that these 29 nucleotides corresponded to nucleotides 75086 to 75114 from the intron 28 of ATM, which are surrounded by potential acceptor and donor splice sites.25 The genomic region containing these 29 bp was amplified from tumoral “Bat” DNA and sequenced. This sequence was identical to the published one, and the cause of the 29 nt insertion remains to be determined.
Although PTT analysis of patient “Bul” showed a normal sized protein product for region no. 6, the pattern of small byproducts was different from that usually observed for this test (Fig 3). The corresponding cDNA fragment was sequenced, showing a transversion C to G at position 7645 in patient “Bul” [7645C → G], whereas the corresponding sequence in nonleukemic cells from this patient was normal. The consequence of this mutation is the replacement of an arginine by a glycine at codon 2486 [R2486G].
DISCUSSION
Here, we tested the hypothesis that ATM alterations could occur at a somatic level and play a role in T-PLL in patients without AT. To show LOH for the ATM gene region in T-PLL, we derived EBV-transformed cell lines from frozen blood samples, or when possible, obtained nonleukemic cells from living patients. Of the 15 cases for whom analysis was possible, LOH was demonstrated in 10 (67%). Deletions were variously sized, and the defined ≈ 2.5 Mb minimal region of deletion contained the ATM gene. No evidence was found in any case for biallelic deletion or rearrangement of theATM locus by Southern blotting. However, in four cases with LOH of the 11q22-23 region, ATM protein was absent or diminished compared with cases without LOH. We thus searched for mutations in the second allele of ATM, using the PTT. This test was chosen considering the previously described germinal mutations of ATM leading to truncated proteins.33,34 Nine cases were analyzed by PTT, seven with LOH, and two without. Abnormal patterns were observed in four cases and further investigated by sequencing both the cDNA and the genomic DNA for each case. Heterogeneous abnormalities were demonstrated: one insertion of a single nucleotide, two splicing aberrations, and one missense mutation. These mutations most probably inactivated the ATM protein. The “Dia” mutation led to transcripts with an in-frame deletion of exon 46, which is associated with undetectable ATM protein by Western blot. Such transcripts are probably not functional because an homozygote mutation has been previously reported in an AT patient (case IARC15/AT4) associated with the same in-frame deletion of exon 46.33The R2486G replacement in patient “Bul” was associated with ATM protein undetectable by Western blotting, which suggests that this protein was unstable. The mutations in cases “Big” and “Bat” led to ATM proteins deleted for the kinase domain, the potential leucine zipper domain, and the binding region for c-ABL kinase product. However, the alternative splicing in patient “Bat” appeared to be leaky, and it is possible that low levels of normal protein exist in this case.
In summary, in the four patients fully investigated, the ATMgene was inactivated by a large deletion of one allele and by a mutation in the other. In three cases (“Dia,” “Big,” and “Bul”), the acquired character of both events was demonstrated, fitting with the model of the inactivation of a recessive gene and the recently refined definition of a tumor suppressor gene for the ATM gene.35-37 Mutations of ATM in T-PLL have been recently demonstrated by Vorechovsky et al38 and by Stilgenbauer et al.39 Our search for LOH of the ATM locus has shown a frequency of ATM deletion of 67% in our patient series, which is similar to the observed frequencies in the other series (44% and 54%, respectively).38,39 Their approaches based on the single strand conformation polymorphism (SSCP) technique, have allowed the demonstration of a higher number ofATM mutations than our approach, PTT, which infrequently detects the missense mutations largely represented in their series (10 of 37 T-PLL cases and four of 24 T-PLL cases, respectively).38 39
A important question raised by the demonstration of inactivation ofATM in T-PLL concerns the germinal or somatic origin of the mutations. Our data showed the ATM mutations were acquired in the three cases fully investigated. The risk of individuals with germline heterozygosity for ATM mutation for the development of T-PLL remains to be established.
The 11q22-23 region has been frequently implicated as the location of a putative tumor supressor gene in various tumors, including breast cancers.40,41 Epidemiological studies have raised the possibility that AT carriers may be at risk of developing breast cancers.42 Despite a high frequency of LOH in the 11q22-23 region, further analyses of the ATM gene did not show biallelic alterations of this gene in breast cancers.28 31 To date, T-PLL is the only cancer for which frequent inactivation of ATMhas been demonstrated.
ATM deficiency causes pleiotropic cellular defects in AT patients, and ATM is likely to interact with many proteins, including p53, c-Abl, and Chk1 (for a review on ATM functions and interactions, see Westphal43). As has been speculated for p53, ATMinactivation effects in tumorigenesis may be either direct, by enhancing cell proliferation and inhibiting apoptosis, or indirect, by enhancing genome instability. The determination of the mechanism(s) by which ATM suppresses tumorigenesis will require more investigations, which could be oriented by the molecular characterization of T-PLL cases without ATM inactivation.
ACKNOWLEDGMENT
The authors are indebted to the following physicians who provided us patient samples: V. Andrieu, M.J. Grange (CHU Bichat, Paris, France); G. Damaj (CHU Henri Mondor, Creteil, France); V. Leblond (CHU La Pitié-Salpétrière, Paris, France); F. Valensi, B. Varet (CHU Necker, Paris, France); B. Cazin (CHRU de Lille, Lille, France); X. Troussard (CHU de Caen, Caen, France); M. Wetterwald (CHG de Dunkerque, Dunkerque, France); M. Tiab, H. Maisonneuve (CHD de La Roche-sur-Yon, La Roche-sur-Yon, France); I. Mahé-Péron (La Verrie, France); S. Daliphard, P. Cornillet (CHRU de Reims, Reims, France). We thank D.A. Tagle for the gift of the anti-ATM antiserum, S. Olschwang for the gift of the polymorphic marker primers, M.T. Daniel for cytological expertise, S.D. Goldstone for critical reading of the manuscript, and A. Baruchel and A. Aurias for helpful discussions.
Supported by INSERM, la Fondation contre la Leucémie, le Comité National, le Comité de Paris et le Comité des Hauts de Seine de la Ligue Nationale Contre le Cancer, and Electricité de France.
Address reprint requests to Marc-Henri Stern, MD, PhD, Unité INSERM U462, Centre Hayem, Hôpital Saint Louis, 75475 Paris Cedex 10, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal