Abstract
The ets family transcription factor PU.1 is expressed in monocytes/macrophages, neutrophils, mast cells, B cells, and early erythroblasts, but not in T cells. We have recently shown that PU.1 gene disruption results in mice with no detectable monocytes/macrophages and B cells but T-cell development is retained. Although neutrophil development occurred in these mice, it was delayed and markedly reduced. We now proceed to demonstrate that PU.1 null hematopoietic cells fail to proliferate or form colonies in response to macrophage colony-stimulating factor (M-CSF), granulocyte CSF (G-CSF), and granulocyte/macrophage CSF (GM-CSF). In contrast, PU.1 null cells did proliferate and form colonies in response to interleukin-3 (IL-3), although the response was reduced as compared with control littermates. Compared with control cells, PU.1 null cells had minimal expression of G- and GM-CSF receptors and no detectable M-CSF receptors. The size of individual myeloid colonies produced from PU.1 null primitive and committed myeloid progenitors in the presence of IL-3, IL-6, and stem cell factor (SCF) were reduced compared with controls. Under these conditions, PU.1 null progenitors produced neutrophils but not monocytes/macrophages. These observations suggest that PU.1 gene disruption induces additional cell-autonomous effects that are independent of the alterations in myeloid growth factor receptor expression. Our results demonstrate that PU.1 gene disruption affects a number of developmentally regulated hematopoietic processes that can, at least in part, explain the changes in myeloid development and reduction in myeloid and neutrophil expansion observed in PU.1 null mice.
MATURE BLOOD CELLS are continually replenished throughout life. This requires establishment of a pool of pluripotent stem cells early in embryogenesis from which various hematopoietic lineages are derived by a tightly regulated developmental process. One level of regulation of this process occurs via transcription factors that direct the expression of genes controlling commitment and/or differentiation. This pattern of lineage-specific gene expression is complex, most likely requiring interactions between lineage-specific and broadly expressed transcription factors. These critical transcriptional events result in the expression of various cytokine receptors, adhesion molecules, and other key cellular proteins in pluripotent cells, thus providing the means for individual cells to survive, proliferate, and differentiate.1-3
PU.1 is a member of the ets family of transcription factors4,5 recognizing a purine-rich DNA sequence containing the core sequence 5′-GGAA/T-3′.6 Expression of PU.1 is limited to hematopoietic cells, including primitive CD34+ cells, macrophages, B lymphocytes, neutrophils, mast cells, and early erythroblasts.7-11 In vitro studies suggest that PU.1 regulates the activity of a number of myeloid- and lymphoid-specific promoters and enhancers.12 Recent evidence also suggests that promoters for the genes encoding receptors for macrophage colony-stimulating factor (M-CSF),13granulocyte/macrophage CSF (GM-CSF),14 and granulocyte CSF (G-CSF)15 are regulated by PU.1. CSF receptors have been proposed to be critical for survival, proliferation, and terminal maturation of myeloid cells.16 17
We18 and others19 have shown that the loss of myeloid lineages in PU.1 gene–disrupted mice implicates PU.1 in the regulation of myeloid development. Fetal18,19 or newborn18 PU.1 null mice have no detectable monocytes/macrophages or neutrophils. However, within 2 to 3 days after birth, neutrophils, as defined by multisegmented nuclei, expression of Gr-1, and chloroacetate esterase (CAE) staining, can be detected within the liver, bone marrow, and spleen.18 Although these cells express CD18, they fail to express CD11b. In contrast to neutrophils, monocytes/macrophages18 and osteoclasts20 have not been detected in older PU.1 null mice. This loss of normal myelopoiesis in PU.1 gene–disrupted mice could be due to a cell-autonomous defect. That is, the loss of PU.1 in myeloid progenitors may directly affect development, producing the observed multiple myeloid lineage defects. Alternatively, PU.1 gene disruption may alter the hematopoietic microenvironment, for example, by loss/dysfunction of stroma, resulting in the absence or reduction of requisite signals for developmental progression of myeloid progenitors. This is not without precedent, given the absence of osteoclasts in the PU.1 null mouse and the subsequent failure of bone marrow cavity formation.20 These changes caused by PU.1 gene disruption are not mutually exclusive, and each could contribute to the myeloid defects observed.
To investigate cell-autonomous defects produced by the loss of PU.1, we assessed hematopoietic cell responses to M-CSF, G-CSF, GM-CSF, and interleukin-3 (IL-3) or stem cell factor (SCF), IL-3, and IL-6 in vitro. We show that PU.1 null hematopoietic cells do not respond to M-, G-, and GM-CSF. Concomitant with the loss of response to these cytokines, M-CSF receptors were not detected and minimal levels of G- and GM-CSF receptors were detected on fresh or cultured PU.1 null cells. When PU.1 null progenitor cells were assessed in clonogenic assays using only SCF, IL-3, and IL-6, neutrophils were produced, whereas monocytes/macrophages were absent. These results convincingly demonstrate that PU.1 gene disruption affects a number of developmentally regulated hematopoietic events that can, at least in part, explain the alterations in myeloid development observed in PU.1 null mice. Thus, PU.1 appears pivotal in regulating myelopoiesis. PU.1 null mice should be useful for studying mechanisms controlling lineage determination during myelopoiesis.
MATERIALS AND METHODS
Mice.
C57BL/6x129 PU.1 gene–disrupted mice were produced as previously reported.18 PU.1 gene–disrupted hemizygous mice, F6 generation, were bred to produce PU.1 null homozygous neonates. Homozygous neonates were identified as PU.1 null mice by the absence of neutrophils/monocytes in blood. Tissue obtained from the tail was used to confirm the genotype as previously reported.18 PU.1 null neonates all developed septicemia within 24 hours and died by 48 hours if not placed on enrofloxacin treatment (2.5 mg/kg/d). PU.1 null mice treated in this manner have survived up to 20 days.
Isolation of hematopoietic cells.
Livers or spleens of the mice were aseptically removed, and a single-cell suspension was generated. For bone marrow, femurs were removed, stripped of soft tissue, and then crushed to release cells within the marrow cavity. When required, red blood cells were lysed with a 0.15-mol/L solution of ammonium chloride. For some studies, low-density mononuclear cells were isolated by purification over a density gradient (Histopaque 1083; Sigma, St Louis, MO) per the manufacturer's instructions.
Cell proliferation.
Proliferation was assessed using an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide)-based colorimetric assay (Cell Proliferation Kit I; Boehringer, Mannheim, Germany) as directed. Mononuclear cells (104 to 105 cells per well) from control and PU.1 null neonate liver and spleen were grown in a 96-well plate in triplicate under the following conditions: Iscove's media with 20% fetal calf serum (medium) with no added factors (as a negative control); IL-3 only at 0.01%, 0.1%, and 1.0% conditioned media (from X63 cells; a kind gift from F. Melchers); and GM-CSF or G-CSF (R&D Systems, Minneapolis, MN) only at 0.1, 1.0, and 10.0 ng/mL for 4 days at 37°C and 5% CO2. M-CSF was used at 10% to 30% L cell–conditioned media or 5,000 U/mL rmM-CSF (a kind gift from D. Hume). Cell viability was assessed on the basis of trypan blue exclusion in a duplicate plate.
Enzyme histochemistry and immunohistochemistry.
Flow cytometric analysis.
Isolated hematopoietic cells were either directly analyzed or cultured for 2 to 3 weeks in medium plus 1% IL-3–conditioned media, 10 ng/mL G-CSF, and 10 ng/mL GM-CSF before flow cytometric analysis. Protocols for staining the various cell populations for flow cytometric analyses were as previously described.18 To determine cell surface G- and GM-CSF receptor expression, we used commercially available phycoerythrin (PE)-labeled cytokines (Fluorokines; R&D Systems) with PE-streptavidin as a control, as directed by R&D Systems. Flow cytometric analysis used a Becton Dickinson FACScan with Cell Quest acquisition and analysis software (Becton Dickinson, Franklin Lakes, NJ).
Colony-forming assays.
Assays for hematopoietic progenitor cells were performed as described previously24 with some modification. For generation of committed progenitor colonies and high–proliferative potential colony-forming cells (HPP-CFC), low-density mononuclear cells were seeded at 5 × 103/mL in commercially available methylcellulose media containing SCF, IL-3, IL-6, and erythropoietin (MethoCult GF M3434; Stem Cell Technologies, Vancouver, BC, Canada). This combination has been shown to be sufficient for generation of both HPP-CFC and committed progenitor colonies.25 Assays were performed in triplicate at 1 mL/35-mm2 petri dish. Single-factor colony assays used either 1% IL-3–conditioned media (from X63 cells; gift from F. Melchers) or 300 U/mL rmIL-3 (gift from D. Hume), 10 ng/mL rmGM-CSF (R&D Systems), 10 ng/mL rhG-CSF (R&D Systems), or 30% L929 cell–conditioned media (as a source of M-CSF). Cultures were maintained at 37°C in a humidified atmosphere of 5% CO2 in air. Colonies were scored as clusters containing more than 50 cells. HPP-CFC were identified as tight colonies with a diameter greater than 0.5 mm2 containing greater than 50,000 cells. Committed progenitor and single-factor colonies were scored at 7 to 9 days and HPP-CFC at 21 days by in situ observation with an inverted microscope. Colonies were routinely evaluated by cytologic examination after aspiration of individual colonies with a Gilson pipetman followed by cytospin preparation and Wright-Giemsa (Sigma, St Louis, MO) staining to determine phenotype.
RESULTS
Impaired response of hematopoietic cells from PU.1 null mice to M-CSF, G-CSF, or GM-CSF.
We have previously shown that PU.1 gene targeting results in the absence of detectable monocyte/macrophage development, reduced and delayed neutrophil generation, and generalized reduction of hematopoietic cell numbers in PU.1 null mice.18 Given these findings, the demonstration of PU.1 binding sites in the promoter region of M-, G-, and GM-CSF receptor genes,12 and the possible role of PU.1 in regulating these receptors genes, we examined whether alterations in myeloid development in PU.1 null mice were a consequence of the inability to respond to each of these cytokines.
The ability of neonatal PU.1 null (deficient) hematopoietic cells to use M-, G-, GM-CSF or IL-3 was first assessed in clonogenic assays. Cells removed from the bone marrow, spleen, or liver of PU.1 null neonatal mice failed to produce colonies when cultured with M-, G-, or GM-CSF, but generated colonies when cultured with IL-3. Although the level of response to IL-3 varied in the studies (Table1, and data not shown), at no time did we observe M-, G-, or GM-CSF supporting colony generation from PU.1-deficient hematopoietic cells. In contrast, cells from similar hematopoietic compartments of control littermates generated colonies in response to M-, G-, GM-CSF or IL-3.
Hematopoietic Cells From PU.1 Null Mice Are Unable to Generate Colonies in Response to GM-CSF, G-CSF, or M-CSF
| Mouse . | No. of CFC (×101)/Input Hematopoietic Cells From Various Hematopoietic Compartments* . | |||
|---|---|---|---|---|
| Cytokine . | Phenotype . | Bone Marrow . | Spleen . | Liver . |
| IL-3 | Control-151 | ND | 22.3 ± 9.0 | 7.7 ± 4.5 |
| PU.1 null | ND | 19.7 ± 3.8 | 5.0 ± 3.6 | |
| GM-CSF | Control | 17.3 ± 7.5 | 20.7 ± 2.3 | 7.1 ± 1.1 |
| PU.1 null | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| G-CSF | Control | 4.0 ± 2.0 | 10.0 ± 2.6 | 1.3 ± 1.1 |
| PU.1 null | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| M-CSF | Control | 40.7 ± 4.9 | 21.7 ± 3.0 | 18.0 ± 5.6 |
| PU.1 null | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| Mouse . | No. of CFC (×101)/Input Hematopoietic Cells From Various Hematopoietic Compartments* . | |||
|---|---|---|---|---|
| Cytokine . | Phenotype . | Bone Marrow . | Spleen . | Liver . |
| IL-3 | Control-151 | ND | 22.3 ± 9.0 | 7.7 ± 4.5 |
| PU.1 null | ND | 19.7 ± 3.8 | 5.0 ± 3.6 | |
| GM-CSF | Control | 17.3 ± 7.5 | 20.7 ± 2.3 | 7.1 ± 1.1 |
| PU.1 null | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| G-CSF | Control | 4.0 ± 2.0 | 10.0 ± 2.6 | 1.3 ± 1.1 |
| PU.1 null | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
| M-CSF | Control | 40.7 ± 4.9 | 21.7 ± 3.0 | 18.0 ± 5.6 |
| PU.1 null | 0 ± 0 | 0 ± 0 | 0 ± 0 | |
*CFC includes all hematopoietic colonies of more than 50 cells; values are reported as the mean ± SD. Input cell numbers for clonogenic assay from neonates: bone marrow, 20,000 cells; spleen, 100,000 cells; liver, 50,000 cells. Two studies are shown; these experiments are representative of other evaluations of PU.1 null mice.
PU.1 null connotes mice with both alleles of the PU.1 gene disrupted, whereas control connotes mice with 1 or no alleles of PU.1 disrupted. Mice within an experiment were littermates. All groups, either control or PU.1 null, were composed of 1 to 3 mice.
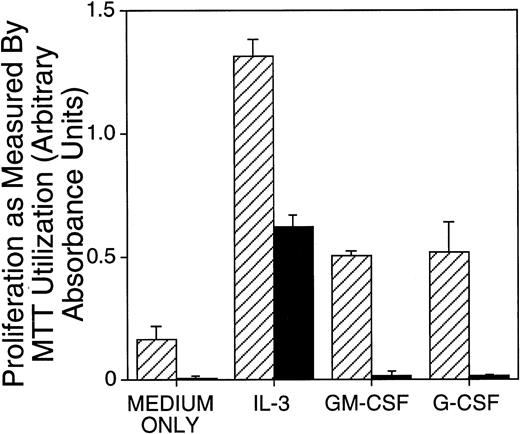
Next, the ability of neonatal PU.1-deficient liver cells to proliferate in response to IL-3, G-CSF, or GM-CSF stimulation was assessed in short-term assays. Proliferation in these cytokines was assessed at the end of a 4-day culture. There was a readily detectable difference between cells from control and PU.1 null mice: cells from PU.1 null mice cultured with G- or GM-CSF did not proliferate above the baseline values observed for cultures without addition of cytokines (Fig1). M-CSF was also found not to support proliferation of PU.1 null cells (data not shown). Although hematopoietic cells from PU.1 null mice exhibited baseline values when cultured with G- or GM-CSF, they proliferated above baseline values when cultured with IL-3. These results were common to all experiments. However, as shown in Fig 1 and in other experiments (data not shown), hematopoietic cells from PU.1 null mice compared with control littermates proliferated suboptimally in IL-3. To assess whether M-, G-, or GM-CSF might function as PU.1 null cell survival factors rather than proliferation factors, PU.1 null cells established in parallel to proliferation assays were tested for viability at the end of these short-term cultures by trypan blue exclusion. We could not detect an increase in survival of PU.1 null cells in M-, G-, or GM-CSF as compared with controls without cytokines (data not shown).
Proliferation of PU.1 deficient cells was reduced in IL-3 and absent in G-CSF and GM-CSF as compared with control cells. Mononuclear cells isolated from neonates were incubated in IL-3 (1% SN), G-CSF (10 ng/mL), or GM-CSF (10 ng/mL) for 4 days. Proliferation was measured by colorimetric assessment of MTT reduction and by counting viable cells at the end of the culture period. Results for cellular proliferation are presented as the mean ± SD of absorbance. Similar results were obtained for spleen and bone marrow cells (not shown). Note an approximately threefold reduced proliferation of PU.1 deficient cells (▪) in IL-3 compared with control (▨) and no proliferation in G-CSF or GM-CSF detectable above the baseline (medium only conditions).
Proliferation of PU.1 deficient cells was reduced in IL-3 and absent in G-CSF and GM-CSF as compared with control cells. Mononuclear cells isolated from neonates were incubated in IL-3 (1% SN), G-CSF (10 ng/mL), or GM-CSF (10 ng/mL) for 4 days. Proliferation was measured by colorimetric assessment of MTT reduction and by counting viable cells at the end of the culture period. Results for cellular proliferation are presented as the mean ± SD of absorbance. Similar results were obtained for spleen and bone marrow cells (not shown). Note an approximately threefold reduced proliferation of PU.1 deficient cells (▪) in IL-3 compared with control (▨) and no proliferation in G-CSF or GM-CSF detectable above the baseline (medium only conditions).
To determine if G-CSF– or GM-CSF–responsive cells were present at low frequency and could be expanded in the absence of IL-3, freshly isolated PU.1 null neonatal liver cells were plated in triplicate at 500,000, 50,000, and 5,000 cells/well in 1% IL-3–conditioned medium (as positive controls) or in 20 ng/mL G- or GM-CSF. After 1 week, few viable cells (< 1%) other than adherent cells remained in either the G-CSF– or GM-CSF–containing wells, and at 3 weeks, only the cells in IL-3–containing medium remained viable and continued to expand (data not shown).
Our results demonstrate that PU.1 disruption imparts a cell-autonomous defect in myeloid cells that manifests as an inability to use M-, G-, or GM-CSF for proliferation and colony formation. Since survival and proliferation were evaluated after 4 days, our studies cannot fully eliminate the possibility that progenitors that use M-, G-, or GM-CSF are present at very low frequency and survive for very brief periods in the presence of these cytokines. Lastly, it should be noted that at the population level, hematopoietic cells from PU.1-deficient liver, spleen, and bone marrow did not respond as well to IL-3 as did populations from control mice.
Hematopoietic cells from PU.1 null mice bound minimal amounts of PE-labeled G-CSF and GM-CSF.
The lack of clonogenic growth and in vitro proliferation in response to M-, G-, or GM-CSF by hematopoietic cells obtained from PU.1 null mice could be due to the absence of cytokine receptors or the inability of the receptors to respond to their respective growth factors. Given our results, we next assessed whether these receptors were present. Both G- and GM-CSF receptors have been reported to be present on early progenitors and highly expressed during various stages of myeloid development.16,17 Since low numbers of myeloid cells were observed in liver, spleen, and bone marrow of PU.1 null neonates18 (and data not shown), we reasoned that in vitro expansion of hematopoietic cells from the liver for 1 or 2 weeks in cultures containing IL-3, G-CSF, and GM-CSF should allow for sufficient numbers of cells for receptor analyses. This combination of cytokines normally allows for both expansion of myeloid cells and G- and GM-CSF receptor expression. Furthermore, as we have shown, hematopoietic cells from PU.1 null mice do not expand in either G- or GM-CSF alone. Assessment of M-CSF receptors on PU.1 null cells is presented elsewhere in this report.
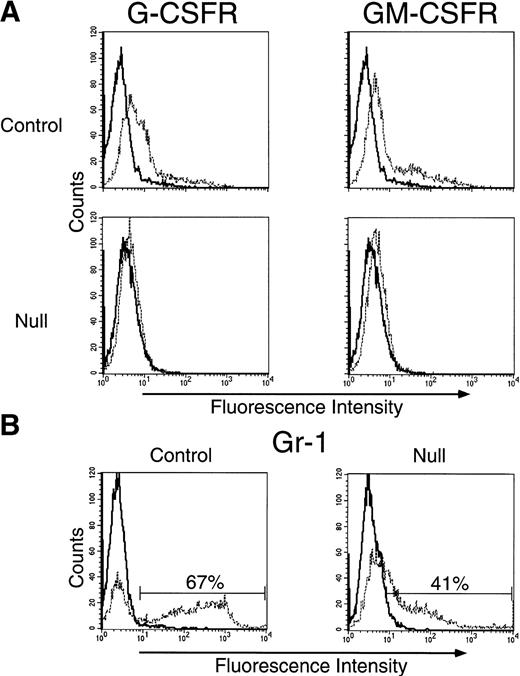
Representative results from flow cytometric analyses for expression of G- and GM-CSF receptors on neonatal PU.1 null and wild-type myeloid cells after short-term culture are presented in Fig2A. Cytokine-expanded PU.1 null cells were found to bind reduced amounts of PE-labeled G-CSF (PE control mean fluorescence, 4.2; mean fluorescence for PE G-CSF binding, 4.9) as compared with binding of PE-labeled G-CSF to cytokine-expanded cells from wild-type (PE control mean fluorescence, 3.2; mean fluorescence for PE G-CSF binding, 17.1) littermates. As seen for G-CSF binding to PU.1 null cells, PE-labeled GM-CSF binding to PU.1 null cells was also reduced (PE control mean fluorescence, 4.2; mean fluorescence for PE GM-CSF binding, 5.5) as compared with wild-type cells (PE control mean fluorescence, 3.2; mean fluorescence for PE GM-CSF binding, 24.1). These results demonstrate the absence of detectable PU.1 null cells capable of binding high levels of PE-labeled G- and GM-CSF after culture (note the right shoulder of both the G- and GM-CSF binding curves merge with controls). Furthermore, the vast majority of cultured PU.1 null cells were incapable of binding detectable amounts of G- and GM-CSF, as compared with cells from wild-type littermates.
Binding of the PE-labeled myeloid growth factors G- and GM-CSF to cultured PU.1 null cells was minimal. Cells were prepared directly from liver of control and PU.1 null neonatal mice and cultured in the presence of IL-3, GM-CSF, and G-CSF for 9 days to expand populations of Gr-1+ cells. Wild-type and PU.1 deficient cells were harvested, live cells enriched over a density gradient and then analyzed for the presence of receptors for G-CSF (G-CSFR), and for GM-CSF (GM-CSFR) by the binding of PE-conjugated G-CSF or GM-CSF (A) or for the neutrophil specific marker Gr-1 (B). (A) PE-control staining (—); PE-conjugated G-CSF or GM-CSF binding (- - -). (B) Irrelevant control antibody staining (—); specific anti-Gr-1 antibody staining (- - -).
Binding of the PE-labeled myeloid growth factors G- and GM-CSF to cultured PU.1 null cells was minimal. Cells were prepared directly from liver of control and PU.1 null neonatal mice and cultured in the presence of IL-3, GM-CSF, and G-CSF for 9 days to expand populations of Gr-1+ cells. Wild-type and PU.1 deficient cells were harvested, live cells enriched over a density gradient and then analyzed for the presence of receptors for G-CSF (G-CSFR), and for GM-CSF (GM-CSFR) by the binding of PE-conjugated G-CSF or GM-CSF (A) or for the neutrophil specific marker Gr-1 (B). (A) PE-control staining (—); PE-conjugated G-CSF or GM-CSF binding (- - -). (B) Irrelevant control antibody staining (—); specific anti-Gr-1 antibody staining (- - -).
We next analyzed the PU.1 null and wild-type cultured cells, used for G- and GM-CSF receptor assessment, for Gr-1 expression. Gr-1 has been used as a marker of mouse granulocyte differentiation.21Gr-1 expression is highest on terminally differentiated neutrophils, lower on myeloblasts, transient on differentiating monocytes and not detectable on primitive progenitors.21,26 IL-3, G-CSF, and GM-CSF all induce Gr-1, progenitor colony formation, and neutrophil production, whereas proliferation to these cytokines is inversely related to Gr-1 expression.21 Cultured PU.1 null cells were found to be 41% Gr-1+ (as well as CAE+, data not shown), as compared with 67% Gr-1+ for wild-type cells (Fig 2B). Cells expressing intermediate and high levels of Gr-1 are known to bind G- and GM-CSF.21 We find that 43% of the Gr-1+PU.1 null cells are intermediate to high as compared with 86% of the Gr-1+ wild-type cells (channel values from 50 to 10,000). These results suggest that sufficient PU.1 null myeloid cells are present after culture that have the potential to express G- and GM-CSF receptors.
Since culture conditions might alter G- and GM-CSF receptor expression on neonatal PU.1 null cells we directly assessed fresh pooled bone marrow and liver cells from 9-day-old PU.1 null (n = 2) and control (n = 1) mice for binding of PE-labeled G- and GM-CSF. We have previously demonstrated neutrophil development in older PU.1 null mice18; therefore, differentiated myeloid cells should be present for G- and GM-CSF receptor analysis. Binding of PE-labeled G-CSF to fresh PU.1 null cells was extremely reduced (PE control mean fluorescence, 5.1; mean fluorescence for PE G-CSF binding, 6.0), as compared with the binding of PE-labeled G-CSF to cells from a wild-type mouse (PE control mean fluorescence, 5.5; mean fluorescence for PE G-CSF binding, 21.9). Receptors for GM-CSF binding were also reduced on fresh PU.1 null cells (PE control mean fluorescence, 5.1; mean fluorescence for PE GM-CSF binding, 7.8), as compared to cells from a wild type mouse (PE control mean fluorescence, 5.5; mean fluorescence for PE GM-CSF binding, 31.5).
In summary, PE-labeled G- and GM-CSF binding to cells taken directly from PU.1 null mice or after short-term culture was minimal, although myeloid cells were present that should bind higher amounts of G- and GM-CSF. Although our results do not allow us to quantify the number of receptors remaining on cells after PU.1 gene disruption, the lack of G- and GM-CSF-induced proliferation and colony formation is consistent with too few receptors for normal function or the loss of normal receptor function for the remaining receptors.
Hematopoietic cells from PU.1 null mice generate myeloid colonies that are reduced in cell number and are lacking monocytes/macrophages.
We have demonstrated the delayed appearance of neutrophils, the absence of monocytes/macrophages in vivo,18 and the inability of myeloid cells from PU.1 null mice to use M-, G-, and GM-CSF in vitro (Table 1 and Fig 1). We next investigated the effects of PU.1 gene disruption on progenitor expansion and development in assays that do not rely on M-, G-, or GM-CSF. For these studies, we used a methylcellulose media containing SCF, IL-3, and IL-6 since this has been shown to be sufficient for the assessment of colony-forming progenitors.25 The number of total colonies produced from the PU.1 deficient bone marrow was substantially lower, 53- to 86-fold, compared with the number of colonies produced from the control bone marrow (Table 2). Progenitor cells from PU.1 deficient liver were reduced threefold to 22-fold compared with control liver.
Progenitor Cells Are Reduced in the Bone Marrow and Liver of PU.1 Null Neonates
| Experiment . | Cells From . | No. of Progenitors per Liver or Bone Marrow ×102 . | |
|---|---|---|---|
| CFC* . | HPP-CFC* . | ||
| 1 | PU.1 null bone marrow† | 0.26 ± 0.16 | 0.23 ± 0.13 |
| Control bone marrow | 22.48 ± 0.58 | 6.40 ± 0.25 | |
| PU.1 null liver | 4.51 ± 0.16 | 1.66 ± 0.03 | |
| Control liver | 40.22 ± 0.29 | 14.6 ± 0.80 | |
| 2 | PU.1 null bone marrow | 0.28‡ | 0.06‡ |
| Control bone marrow | 15.08 ± 0.33 | 5.56 ± 0.13 | |
| PU.1 null liver | 21.38 ± 0.95 | 11.87 ± 0.73 | |
| Control liver | 62.90 ± 19.14 | 39.24 ± 0.99 | |
| 3 | PU.1 null liver | 2.41 ± 0.35 | ND |
| Control liver | 52.00 ± 1.61 | ND | |
| Experiment . | Cells From . | No. of Progenitors per Liver or Bone Marrow ×102 . | |
|---|---|---|---|
| CFC* . | HPP-CFC* . | ||
| 1 | PU.1 null bone marrow† | 0.26 ± 0.16 | 0.23 ± 0.13 |
| Control bone marrow | 22.48 ± 0.58 | 6.40 ± 0.25 | |
| PU.1 null liver | 4.51 ± 0.16 | 1.66 ± 0.03 | |
| Control liver | 40.22 ± 0.29 | 14.6 ± 0.80 | |
| 2 | PU.1 null bone marrow | 0.28‡ | 0.06‡ |
| Control bone marrow | 15.08 ± 0.33 | 5.56 ± 0.13 | |
| PU.1 null liver | 21.38 ± 0.95 | 11.87 ± 0.73 | |
| Control liver | 62.90 ± 19.14 | 39.24 ± 0.99 | |
| 3 | PU.1 null liver | 2.41 ± 0.35 | ND |
| Control liver | 52.00 ± 1.61 | ND | |
*CFC includes all hematopoietic colonies of more than 50 cells, whereas HPP-CFC (colonies >0.5 mm) are as defined in text. Clonogenic assays were established in triplicate, except where noted. Values are reported as the mean ± SD. These experiments are representative of other evaluations of PU.1 null mice.
PU.1 null connotes mice with both alleles of the PU.1 gene disrupted, whereas control connotes mice with 1 or no alleles of PU.1 disrupted. Mice within an experiment were littermates. All groups, either control or PU.1 null, were composed of 2 to 5 mice.
Too few bone marrow cells were obtained from individual neonates; therefore, 3 bone marrow samples were pooled for analysis, yielding 1 clonogenic plate for progenitor analysis.
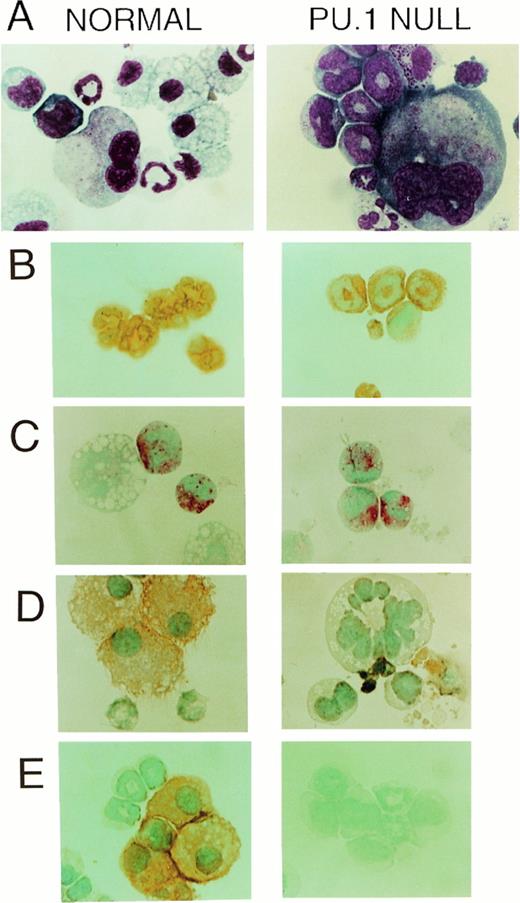
Differences other than the total number of colonies between PU.1 null and control mice were readily apparent. First, it was noted that colonies produced from PU.1 deficient cells were reduced in cell number. To confirm this observation colonies were aspirated from selected clonogenic dishes and counted demonstrating that the average number of myeloid cells present in colonies from PU.1 deficient liver was approximately sixfold lower (Fig 3A).Secondly, it was clear that differences existed in the cell types present in colonies produced from PU.1 null mice (Table3). Based on morphology, it was apparent that PU.1 deficient progenitor cells did not differentiate into monocytes/macrophages. These observations were confirmed by the absence of the macrophage-associated marker F4/80 and the M-CSF receptor after immunohistochemical staining of cells generated from PU.1 null mice in clonogenic assays (Fig 4A, D, and E).
Hematopoietic colony size was reduced in 7-day CFC and 21-day HPP-CFC colonies established from livers of neonatal PU.1 null mice. Individual colonies were aspirated and counted from plates established in triplicate at 5,000 input cells per plate (see Table 2). Results are presented as the mean ± SD of the number of cells per colony. Note that the average PU.1 deficient CFC colony size (▪) at 7 to 10 days was approximately sixfold reduced compared to control (▨) littermates (A). PU.1 deficient HPP-CFC (▪) were reduced in size 3.5-fold compared with control (▨) littermates (B). Colony size = number of cells per colony. These colony sizes were obtained from CFC and HPP-CFC shown in experiment 2, Table 2, and are representative of colony sizes from other experiments.
Hematopoietic colony size was reduced in 7-day CFC and 21-day HPP-CFC colonies established from livers of neonatal PU.1 null mice. Individual colonies were aspirated and counted from plates established in triplicate at 5,000 input cells per plate (see Table 2). Results are presented as the mean ± SD of the number of cells per colony. Note that the average PU.1 deficient CFC colony size (▪) at 7 to 10 days was approximately sixfold reduced compared to control (▨) littermates (A). PU.1 deficient HPP-CFC (▪) were reduced in size 3.5-fold compared with control (▨) littermates (B). Colony size = number of cells per colony. These colony sizes were obtained from CFC and HPP-CFC shown in experiment 2, Table 2, and are representative of colony sizes from other experiments.
Neutrophils But Not Monocytes/Macrophages Are Present in Clonogenic Colonies From the Liver of Neonatal PU.1 Null Mice
| Experiment No. . | Cells From . | Percentage of Colonies Containing Indicated Myeloid Cell Lineages* . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAC . | NEU . | MAST . | MAC NEU . | NEU MAST . | MAC MAST . | NEU MAC MAST . | NEU MAST MK . | NEU MAC MAST MK . | ||
| 1 | PU.1 null mice | 0 | 41 | 9 | 0 | 27 | 0 | 0 | 23 | 0 |
| Control mice | 4 | 0 | 29 | 8 | 8 | 8 | 13 | 0 | 30 | |
| 3 | PU.1 null mice | 0 | 42 | 0 | 0 | 29 | 0 | 0 | 29 | 0 |
| Control mice | 0 | 4 | 0 | 62 | 8 | 0 | 0 | 0 | 26 | |
| Experiment No. . | Cells From . | Percentage of Colonies Containing Indicated Myeloid Cell Lineages* . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAC . | NEU . | MAST . | MAC NEU . | NEU MAST . | MAC MAST . | NEU MAC MAST . | NEU MAST MK . | NEU MAC MAST MK . | ||
| 1 | PU.1 null mice | 0 | 41 | 9 | 0 | 27 | 0 | 0 | 23 | 0 |
| Control mice | 4 | 0 | 29 | 8 | 8 | 8 | 13 | 0 | 30 | |
| 3 | PU.1 null mice | 0 | 42 | 0 | 0 | 29 | 0 | 0 | 29 | 0 |
| Control mice | 0 | 4 | 0 | 62 | 8 | 0 | 0 | 0 | 26 | |
Abbreviations: NEU, neutrophil; MAC, macrophage; MAST, mast cell; MK, megakaryocyte.
All cells from individual colonies were removed from representative clonogenic dishes (see Table 2), counted, and fixed to slides for staining and morphologic identification. Percentages of lineages calculated as number of colonies containing an observed myeloid cell type per total number of colonies.
CAE+ and Gr-1+ cells (neutrophils) but not F4/80+ or M-CSF receptor (c-fms)+ cells (monocyte/macrophages) developed in vitro from PU.1 deficient progenitors when cultured in SCF, IL-3, and IL-6. (A) Wright-Giemsa-stained cytospins of representative multilineage colonies from control and PU.1 null individuals revealed a conspicuous absence of macrophages in PU.1 deficient colonies. However, neutrophils and megakaryocytes were evident in both panels (1,150×). (B) The neutrophil enzyme CAE, demonstrated by a pink reaction product, was evident in PU.1 deficient and control polymorphonuclear cells. Note the larger, nonstaining macrophage in the control individual (1,150×). (C) Both PU.1 deficient and control polymorphonuclear cells expressed the cell surface marker Gr-1 which was demonstrated by immunoperoxidase staining. Detection was with diaminobenzidine (DAB) which yields an orange-brown reaction product (1,150×). (D) M-CSF receptor immunostaining revealed no M-CSF receptor-positive cells in PU.1 deficient colonies. Cell debris was present that stained nonspecifically with DAB in PU.1 null cultures (1,150×). (E) Immunocytochemical staining for the macrophage marker F4/80 revealed no positive cells in PU.1 deficient colonies, whereas many orange-staining F4/80+ cells were found in control colonies (1,150×).
CAE+ and Gr-1+ cells (neutrophils) but not F4/80+ or M-CSF receptor (c-fms)+ cells (monocyte/macrophages) developed in vitro from PU.1 deficient progenitors when cultured in SCF, IL-3, and IL-6. (A) Wright-Giemsa-stained cytospins of representative multilineage colonies from control and PU.1 null individuals revealed a conspicuous absence of macrophages in PU.1 deficient colonies. However, neutrophils and megakaryocytes were evident in both panels (1,150×). (B) The neutrophil enzyme CAE, demonstrated by a pink reaction product, was evident in PU.1 deficient and control polymorphonuclear cells. Note the larger, nonstaining macrophage in the control individual (1,150×). (C) Both PU.1 deficient and control polymorphonuclear cells expressed the cell surface marker Gr-1 which was demonstrated by immunoperoxidase staining. Detection was with diaminobenzidine (DAB) which yields an orange-brown reaction product (1,150×). (D) M-CSF receptor immunostaining revealed no M-CSF receptor-positive cells in PU.1 deficient colonies. Cell debris was present that stained nonspecifically with DAB in PU.1 null cultures (1,150×). (E) Immunocytochemical staining for the macrophage marker F4/80 revealed no positive cells in PU.1 deficient colonies, whereas many orange-staining F4/80+ cells were found in control colonies (1,150×).
In contrast to colonies derived from cells of PU.1 null mice, macrophages were identified in 63% and 87% of all control colonies assessed in two separate experiments (Table 3 and Fig 4; Control; A and D). Neutrophils as identified by morphologic criteria (Fig 4, PU.1 null; A), CAE staining (Fig 4, PU.1 null; B), and Gr-1 immunostaining (Fig 4, PU.1 null; C) were present in 91% and 100% of all PU.1 null colonies compared with 58% and 93% of all control colonies tested in two separate experiments. Mast cells were present in 59% and 28% of all PU.1 null colonies as compared with 87% and 33% of all control colonies, and megakaryocytes were found in 23% and 14% of PU.1 null and 30% and 25% of all control colonies in these experiments (Table3).
These in vitro results revealed that disruption of the PU.1 gene results in an intrinsic defect in committed myeloid progenitor cells that precludes monocyte/macrophage development from progenitors. The inability to detect M-CSF receptors on cells produced in clonogenic assays provides an explanation for the loss of M-CSF response demonstrated in Table 1. Given that SCF, IL-3, and IL-6 were used in the clonogenic assays it is most likely that absence of monocyte/macrophage development is independent of the loss of hematopoietic cell response to M-, G-, or GM-CSF. Finally, these results also confirm that the effect of PU.1 gene disruption is selective: PU.1 appears not to be absolutely necessary for mast cell or neutrophil development from myeloid progenitors, but is essential for monocyte/macrophage generation (Table 3).
Neonatal bone marrow and liver of PU.1 null mice have fewer HPP-CFC.
To determine whether PU.1 gene disruption only targeted more committed progenitors (Table 2) and selected myeloid lineages (Table 3), such as monocytes/macrophages, we next assessed whether myeloid HPP-CFC cells were affected by PU.1 gene disruption. HPP-CFC have been proposed as candidates for a primitive myeloid progenitor cell with some stem cell properties.28 These cells are capable of giving rise to CFU-S, marrow repopulating cells, erythroid and megakaryocyte reconstituting cells in lethally irradiated mice.29 In in vitro assays, these cells form large colonies, greater than 0.5 mm2 and greater than 50,000 cells.30 As for the CFC assessment we relied on methylcellulose media containing the growth factors SCF, IL-3, and IL-6. It must be noted that SCF, IL-3, and IL-6 have been shown to give rise to HPP-CFC, although the frequencies of colonies produced per number of input cells were not as numerous as when M-CSF and/or GM-CSF were included.28 Scoring of colony production from bone marrow at 21 days of culture revealed 28- to 93-fold fewer HPP-CFC from PU.1 null mice than found in control cultures (Table 2). The number of HPP-CFC colonies from PU.1 deficient liver was found to be threefold to ninefold less than control cultures (Table 2). As was the case for committed progenitors, counts of cells per HPP-CFC colony from the liver revealed lower numbers of cells from PU.1 null mice, as compared with control littermates (Fig 3B).
In summary, a reduced number of HPP-CFC-derived colonies were obtained from PU.1 deficient hematopoietic cells as compared with control hematopoietic cells. The reduced number of cells in HPP-CFC colonies is consistent with a cell-autonomous defect limiting colony expansion of primitive myeloid progenitor cells as the result of PU.1 gene disruption. Taken together the CFC and HPP-CFC assays demonstrate that disruption of PU.1 results in an intrinsic defect in committed and primitive myeloid cells in mice that reduces the expansion of primitive and committed myeloid progenitors and disrupts development to monocytes/macrophages.
DISCUSSION
In this study, we investigated the mechanisms responsible for myeloid disruption in PU.1 null mice. Our previous studies demonstrated that PU.1 gene disruption results in the delay of neutrophil development and the absence of monocytes/macrophages18 and osteoclasts.20 Lymphopoiesis was also affected with the loss of B cells, but not T cells. Other lineages, such as megakaryocytes and erythrocytes, were minimally affected.18Our initial studies suggested that PU.1 gene targeting imposed lineage-specific alterations in hematopoietic development, rather than ablation of more primitive progenitors that give rise to myeloid or lymphoid lineages. In our current study, we provide evidence that disruption of PU.1 results in the loss of M-, G-, and GM-CSF-mediated proliferation and development. Concomitant with this loss M-CSF receptors were undetectable, whereas G- and GM-CSF receptor expression was substantially reduced. Independent of the loss of M-, G-, and GM-CSF receptor function, PU.1 deficient myeloid progenitor cells have additional defects that alter differentiation and expansion. Our findings demonstrate that PU.1 is not essential for myeloid or neutrophil commitment, but is required for optimal myeloid expansion and necessary for the development of monocytes/macrophages.
It is apparent from our studies that PU.1 is required for normal expression of M-, G-, and GM-CSF receptors. These results are not surprising particularly given that PU.1 cooperates with other transcription factors, such as C/EBPα and AML1, to regulate the promoters of the myeloid growth factor receptors for M-, G-, and GM-CSF and other myeloid specific genes.12 Although hematopoietic cells from PU.1 null mice do not respond to G- or GM-CSF in vitro, a low level of granulopoiesis still occurs in these mice. A number of mechanisms have been proposed as to how G-CSF regulates granulopoiesis in vivo; these include stimulation of primitive progenitors, proliferation of granulocyte progenitors and induction of granulocyte maturation. However, the in vivo importance of these mechanisms is not at all clear.31 The hypothesis that G-CSF receptor engagement17 is required for differentiation of neutrophils from progenitors is controversial. Evidence from the G-CSF ligand-null,33 G-CSF receptor-null mice,31 and G- and GM-CSF cytokine deficient mice33 demonstrates that cytokines other than G- or GM-CSF allow bone marrow granulopoiesis, but it appears that G-CSF might be required for normal neutrophil numbers in the periphery.31 Results obtained from PU.1 null mice, both in vivo and in vitro, would also argue that granulopoiesis occurs in the absence of detectable G- and GM-CSF response. The presence of neutrophils in PU.1 null mice is consistent with a variation of the proposed stochastic developmental model,34,35 where IL-3, possibly IL-6, or other cytokines provide survival signals to developing neutrophils thereby allowing intrinsic developmental programs to commence. That cytokine/cytokine receptor interaction provides a survival signal, allowing cells at a specific stage to exploit intrinsic or external signals, has been demonstrated for monocytes/macrophages,26 T cells,36 and B cells37 as well. Although developing PU.1 null neutrophils appear to be morphologically normal in having segmented nuclei, CAE+ expression and intermediate expression of Gr-1 (Fig4), we found that colony expansion is limited and terminal differentiation and functional maturity does not occur (K.L. Anderson et al, submitted). What still remains to be determined is whether reduced expansion, loss of functional maturity, and reduced Gr-1 expression are due to the postulated role of G-CSF17,38 or other cytokines such as GM-CSF31or are the result of additional defects induced by the absence of PU.1. Studies are under way to address these issues by restoring the expression of G-, GM-CSF receptors or PU.1 directly in PU.1 null progenitor cells.
The absence of monocytes/macrophages in PU.1 null mice would not be predicted from the studies of M-CSF, G-CSF, and GM-CSF cytokine null mice, which develop monocytes/macrophages, albeit at reduced levels.32,33 Recent studies have provided further support that M-CSF functions as a survival factor, rather than as a differentiation factor, for monocytes.26 Given the clear lack of M-, G-, and GM-CSF response in hematopoietic and myeloid cells from PU.1 deficient mice, and the absence of a normal bone marrow microenvironment, the question of whether IL-3 or cytokines other than M-, G-, and GM-CSF are limiting in vivo and thus not supporting monocyte/macrophage development must be considered. However, we show that SCF, IL-3, and IL-6 allow for HPP-CFC generation and neutrophil development, thus arguing for at least intact IL-3 signaling in PU.1 null cells. However, the loss of PU.1 function cannot be supplanted by these cytokines to rescue monocyte/macrophage development in vitro. Based on our results and published reports demonstrating in vitro (and in myeloid growth factor null mice) that other cytokines in the absence of M-, G-, and GM-CSF are sufficient for low levels of monocyte/macrophage generation, we propose that PU.1, in addition to regulating M-, G-, and GM-CSF receptor expression, is necessary for intrinsic programs required for monocyte/macrophage survival or differentiation. Alternatively, the loss of M- and GM-CSF receptors is not equivalent to loss of their respective cytokines during myeloid development, so that monocyte commitment occurs in the PU.1 null mouse, but in the absence of either M- or GM-CSF receptor expression committed cells do not survive.
Whether PU.1 disruption causes intrinsic defects in primitive myeloid cells beyond the loss of a M-, G-, or GM-CSF mediated response is difficult to ascertain in this study. PU.1 deficient hematopoietic cells generated HPP-CFC in the presence of SCF, IL-3, and IL-6, but the frequency of HPP-CFC per hematopoietic compartment (liver or bone marrow) and the number of cells per colony was reduced as compared with age-matched controls. A recent study has suggested the existence of at least four stages in the mouse HPP-CFC hierarchy, from pro-HPP-CFC to HPP-CFC-3, where HPP-CFC are positioned within the hierarchy based on in vitro cell expansion to combinations of cytokines.28 The most primitive HPP-CFC utilizes M-CSF and/or GM-CSF with SCF, IL-1, IL-3, and/or IL-6 in various combinations for maximum colony expansion.28 The decreased number of HPP-CFC from PU.1 deficient mice could be the result of a more generalized in vivo disruption of primitive progenitors that give rise to pro-HPP-CFC. Alternatively, but not mutually exclusive, is the possibility that the reduced HPP-CFC frequency is due to a diminished survival or expansion of selected HPP-CFC progenitors in vivo as the result of the loss of normal expression of M-CSF or GM-CSF receptors. Although a role for PU.1 in hematopoietic progenitor cells is supported by a recent study in which constitutive expression of PU.1 resulted in enhanced size and numbers of colonies in IL-3, G-CSF, or GM-CSF, the mechanism is unknown.39 Since our studies do not distinguish as to what stage the HPP-CFC belong, we cannot conclude precisely as to where the lack of PU.1 affects early myeloid development. Studies are under way to isolate primitive lin−Sca-1+ populations from age-matched PU.1 null and control littermates to address these issues on a per cell basis, both in vitro and in vivo.
The role of PU.1 in hematopoietic development has also been studied in an independently derived PU.1 gene-targeted mouse.19 A comparison of the PU.1 null mouse discussed here and that generated by Scott et al19 reveals similarities but also some major differences. One difference is that the PU.1 null mice reported here are born alive, whereas in the studies by Scott et al19 all mice die in utero by day 18. A second major difference that exists is the block of all myeloid and lymphoid development in the mouse developed by Scott et al.19 Recent studies by Scott et al40 show that PU.1 gene disrupted mouse fetal liver cells failed to reconstitute the myeloid or lymphoid lineages or rescue lethally irradiated mice. These results suggest that the absence of PU.1 results in a cell-autonomous defect that disrupts primitive hematopoietic stem cell commitment to myeloid and lymphoid lineages. The molecular mechanism(s) contributing to the observed hematopoietic defects in and the fetal death of the PU.1 null mice generated by Scott et al19 are still unexplained. Recently, we have proposed possible explanations for the observed differences between the distinct PU.1 null phenotypes generated in these mice.18
We observe hematopoietic cell-autonomous defect(s) in the PU.1 null mice described herein. Rather than a generalized block in all myeloid development, lineage specific effects were observed. As shown here and previously,18 monocyte/macrophage development is disrupted, but commitment and development along the neutrophil lineage occurs. At least part of the mechanism for the abnormal myeloid development in PU.1 null mice appears to be the loss of normal M-, G-, and GM-CSF receptor expression during myeloid development. In addition to the loss of M-, G-, and GM-CSF receptor regulation there appears to be an independent requirement for PU.1 expression during monocyte/macrophage development and possibly during HPP-CFC expansion. It is tempting to speculate that PU.1 plays a role beyond simply regulating growth factor receptor expression during myelopoiesis. Finally, we would like to propose that the cumulative effect of altered or the absent expression of PU.1-regulated genes, a group which is likely to include genes other than those already known, contributes to the observed PU.1 null phenotype.
ACKNOWLEDGMENT
The authors gratefully acknowledge the technical assistance of Michelle Butler and Kari Carver and the secretarial assistance of Bonnie Towle. We would like to thank Laura Crisa, Toñi Ortiz, Greg Henkel, Deborah Vestal, and Donald Mosier for critically reading the manuscript. This is publication IMM 10705 from The Scripps Research Institute.
Supported by National Institutes of Health Grants No. DK49886 (B.E.T.) and AI30656 (R.A.M.).
Address reprint requests to Bruce E. Torbett, PhD, Department of Immunology, IMM-7, The Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal