Abstract
Normal expression of the human β-globin domain genes is dependent on at least three types of regulatory elements located within the β-globin domain: the locus control region (LCR), globin enhancer elements (3′β and 3′Aγ), and the individual globin gene promoter and upstream regions. It has been postulated that regulation occurs through physical interactions between factors bound to these elements, which are located at considerable distances from each other. To identify the elements required for promoter-enhancer interactions from a distance, we have investigated the expression of the wild-type, truncated, and mutated γ-globin promoters linked to the 5′HS2 enhancer. We show that in K562 cells, 5′HS2 increases activity approximately 20-fold from both a wild-type and truncated (-135 → +25) γ promoter and that truncation or site-directed mutagenesis of the tandem CCAAT boxes eliminated the enhancement by 5′HS2. Mutation of the γ-globin gene promoter GATA-1 binding sites did not decrease either promoter strength or enhancement of activity by 5′HS2. To determine if enhanced expression of γ-globin gene promoters carrying mutations associated with hereditary persistence of fetal hemoglobin (HPFH) was due to greater interactions with enhancers, we linked these HPFH γ-globin gene promoters to 5′HS2 and demonstrated a twofold to threefold higher expression than the corresponding wild-type promoter plus enhancer in MEL cells. Addition of the Aγ-globin gene 3′ enhancer to a plasmid containing the γ-globin gene promoter and 5′HS2 did not further enhance promoter strength. Furthermore, we have demonstrated that the previously identified core 5′HS2 enhancer (46-bp tandem AP-1/NF-E2 sites) increased expression only when located 5′, but not 3′, to the γ-globin-luciferase reporter gene, suggesting that its enhancer effect is not by DNA looping. Our results suggest that CCAAT boxes, but not GATA or CACCC binding sites, are required for interaction between the γ-globin promoter and the LCR/5′HS2 and that regulatory elements in addition to the core enhancer may be required for the enhancer to act from a distance.
THE FIVE FUNCTIONAL human β-globin family genes (ε, Gγ, Aγ, δ, and β) are grouped within a 70-kb domain located on chromosome 11.1 The Gγ and Aγ genes are normally expressed at high levels only during fetal development, followed by switching to adult β-globin around the time of birth. As a result of this switching, Hb F constitutes only 1% to 2% of the total hemoglobin in normal adults. An exception are individuals with hereditary persistence of fetal hemoglobin (HPFH). These individuals have elevated (5- to 80-fold) expression of γ-globin and Hb F levels of up to 20% during adult life. Elevated Hb F levels can be beneficial to patients with β-thalassemias or sickle cell anemia by reducing the severity of their disease.
The β-globin domain genes are regulated by a region containing a series of erythroid-specific DNase I hypersensitivity sites (5′HS1-4) located 6 to 18 kb upstream of the ε-globin gene termed the locus control region (LCR).2-5 The β-globin domain LCR performs at least two important functions: first, it creates an open chromosomal region more accessible to trans-acting transcription factors6,7 and second, it contains strong enhancers required for high-level, developmentally correct expression of the individual β-globin domain genes.8-12 Within the LCR, 5′HS2 behaves as a strong enhancer in both transient expression and transgenic mice experiments.8-10,13 It is sufficient for appropriate developmental expression of a linked globin gene cluster in transgenic mice.14 Analysis of 5′HS2 has identified tandem AP-1/NF-E2 sites, and a 46-bp fragment containing these sites is necessary and sufficient to enhance expression of a linked γ-globin reporter gene.15,16 Although the molecular mechanisms underlying how the LCR directs correct globin gene expression are unknown, physical interactions between multiple protein-DNA complexes bound to individual gene promoters and the LCR are postulated to be involved, perhaps via DNA looping5,17(and references therein). Recently, Cavallesco and Tuan18divided the HS2 enhancer into an enhancer core and five modulatory domains to identify regions necessary for productive interaction with the β-globin gene promoter. They found that different subdomains modulated enhancement of the β-globin gene promoter activity both positively and negatively at different developmental stages. Furthermore, recent work by Ronchi et al19 led them to conclude that multiple functionally redundant promoter elements are involved in γ-globin gene promoter/LCR interactions. These interactions may be required for the formation of an active transcription complex that contains both the LCR and individual gene promoters. In such a model, promoter elements could have dual roles of promoting transcription initiation and aiding promoter/enhancer interactions.
Besides the LCR, the Gγ and Aγ-globin genes are regulated by cis-acting elements found within their promoters. Based on DNA-protein binding experiments, promoter activity experiments, and the identification of γ-globin promoter mutations in patients having nondeletional HPFH, at least nine sites of DNA-protein interaction (−30 ATAAA, −50 GGGGCCGG, −85 and −112 CCAAT, −145 CACCC, −170 and −190 GATA, −180 ATGCAAAT, and −200 CCCGGG elements) have been identified within the −260 → +25 region of the γ-globin promoter.20-26 The functional importance of these binding sites has been examined by numerous investigators using transient and stable expression experiments. Within the γ-globin 5′ upstream region the ATAAA and tandem CCAAT elements constitute a minimal promoter. These sites direct correct, but low level, transcription of the γ-globin gene.27,28 The −145 CACCC site binds Sp1 and is required for high level expression. Deletion or mutation of the CACCC element reduces γ-globin promoter activity to 10% to 20% of the wild-type level.29-31 Further upstream, the −150 → −260 region of the γ-globin promoter is involved in directing correct developmental stage-specific expression. When combined with a minimal β-globin promoter, this region directed expression of the otherwise silent β-globin gene in K562 cells and in transgenic mice.32 33
Not previously studied is how globin gene enhancers such as the LCR/5′HS2 interact with the γ-globin promoter from a distance to increase γ-globin expression. Current models predict that multiple promoter-enhancer interactions will be involved in the correct developmental stage-specific expression. One approach to identify regulatory factors involved in promoter-enhancer interaction is to perform expression assays with normal and mutated promoters in the absence and presence of an enhancer. The introduction ofcis-acting mutations in the γ-globin gene promoter in such assays could identify transcription factor binding sites that are required for physical interaction of the promoter and enhancer because they may have a much greater effect on levels of enhancer-mediated expression relative to basal expression. Studies of human β-globin domain genes using this approach have identified a GATA binding site within the ε-globin promoter and an erythroid Kruppel-like factor (EKLF) binding site within the β-globin gene promoter that are required for interaction between these promoters and erythroid enhancers.34-36
Our goal in these studies was to identify elements required for interaction of the γ-globin promoter with 5′HS2 in a fetal erythroid environment. In this report, we focus on the γ-globin promoter and report a systematic analysis of the regulatory elements within the human Gγ-globin promoter required for the interaction of the promoter with 5′HS2. Although normally expressed only during fetal development, γ-globin is expressed during adult life in patients with HPFH. Therefore, we also examined the effect of three nondeletional HPFH mutations on the interaction between the γ-globin promoter and globin enhancers to investigate whether increased interaction between the HPFH promoters and the LCR might contribute to the continued expression of γ-globin in adult erythroid cells. Because HPFH patients express γ-globin during both fetal and adult life, we performed the assays with the HPFH mutations in both fetal and adult erythroid cells. Finally, we compared the effect of the full-length and core 5′HS2 enhancers on γ-globin gene expression to determine if the core 5′HS2 enhancer was sufficient for interaction with the γ-globin promoter. Our results demonstrate the 5′HS2 enhancer is able to activate a minimal γ-globin gene promoter and suggest that CCAAT boxes, but not GATA or CACCC binding sites, are required for interaction between the γ-globin promoter and the LCR/5′HS2. Also, regulatory elements within the full-length 5′HS2, but not the 46-bp core enhancer, may be required for the enhancer to act from a distance.
MATERIALS AND METHODS
Plasmid DNA constructions.
DNA fragments containing Gγ-globin promoter sequence from −259, −204, −160, −135, and −72 to +25 relative to the start of transcription were generated by polymerase chain reaction and cloned as BamHI-HindIII fragments into the promoterless luciferase reporter plasmid pOLUC.37The 5′HS2 (1 kb, Genebank nucleotide [nt] no. 8260-9260), core HS2 (37 bp, nt no. 8651-8687), and 3′β (250-bpPst I, nt no. 64310-64560) enhancer fragments were first cloned into the multiple cloning site of pUC118, and a PvuII fragment containing the enhancer sequence plus plasmid sequence was then cloned into the unique PvuII site in pOLUC. The PvuII fragment of pUC118 alone was used to create the reporter plasmids lacking an enhancer. When the core HS2 enhancer was located 5′ to the γ-globin promoter, two complementary oligonucleotides withBamHI ends were annealed together and directly cloned into the unique BamHI site of the γ-globin-luciferase plasmid. The correct 5′ → 3′ orientation of enhancer fragments relative to the γ promoter-luciferase reporter gene was determined by restriction enzyme analysis. γ-globin promoter mutations were created using polymerase chain reaction site-directed mutagenesis38and confirmed by DNA sequencing. Plasmids used in the stable transformation colony assays were constructed by replacing the luciferase reporter gene with the neomycin resistance (neoR) gene from pUC9-Aγneo.31The plasmid pCMV-SEAP expresses secreted alkaline phosphatase39 under control of the intermediate early gene promoter of the human cytomegalovirus. The plasmid pRSV-bGAL contains the bacterial lacZ reporter gene under the control of the Rous Sarcoma Virus LTR promoter. Plasmid DNA used in electroporation experiments was purified by CsCl/EtBr ultracentrifugation or Qiagen plasmid kit (Qiagen, Chatsworth, CA).
Cell growth and electroporation conditions.
Human erythroleukemia K562 and murine erythroleukemia MEL cells were grown in RPMI 1640 medium plus 10% fetal bovine serum and penicillin/streptomycin (GIBCO-BRL, Grand Island, NY) in a humidified 5% CO2 environment. The cells were routinely maintained at a density of 1 to 5 × 105/mL. Transient DNA transfections were performed by electroporation using a BIORAD Gene Pulser (Bio-Rad Laboratories, Hercules, CA). Approximately 2.5 × 106 cells were electroporated with 10 μg supercoiled reporter plasmid DNA at 960 μF and 220 mV or 280 mV for K562 and MEL cells, respectively. A total of 5 μg supercoiled pCMV-SEAP plasmid or pRSV-βGAL plasmid DNA was also included to measure electroporation efficiency. The transfected cells were harvested after 16 hours and then assayed for luciferase and secreted alkaline phosphatase enzyme activity or β-galactosidase enzyme activity. At least two different DNA preparations of each reporter plasmid were tested.
Enzyme assays.
Electroporated cells were collected by centrifugation, washed with 1X phosphate-buffered saline (PBS) and resuspended in KPO4 buffer (100 mmol/L KH2PO4 pH, 7.8, 3 mmol/L MgCl2, 1 mmol/L dithiothreitol [DTT]). Extracts were prepared by adding an equal volume of KPO4 + 2% NP-40 buffer, incubation on ice for 10 minutes, centrifugation for 10 minutes at 13,000g at 4°C, and collection of the supernatant. Luciferase enzyme activity was determined as described40using a model 1251 Luminometer (Pharmacia Biotech, Piscataway, NJ). Secreted alkaline phosphatase enzyme activity was determined in the culture supernatant by the method of Berger et al.39 β-Galactosidase enzyme activity was determined on an aliquot of transfected cells or cell extract using the method of Miller.41 Protein concentrations were determined by the method of Bradford42 using bovine serum albumin as the standard. Luciferase activity was corrected for electroporation efficiency using the measured SEAP or β-galactosidase activity and calculated as light units per minute per mg protein per unit SEAP or β-galactosidase activity. Stable transformation colony assays were performed as described.43
Gel retardation assays.
Nuclear extracts were prepared by the method of Dignam et al.44 Gel retardation assays were performed according to the method of Sykes and Kaufman45 with high-performance liquid chromatography (HPLC) purified double-stranded oligonucleotides containing either the wild-type γ-globin promoter sequence from nt no. −165 to −212 5′ATCTTGGGGGCCCCTTCCCCACACTATCTCAATGCAAATATCTGTCT 3′ or the mutated GATA sequence 5′ATCTTGGGGGCCCCTTCCCCACACGCGATCAATGCAAAGCGATGTCT 3′. The rabbit polyclonal GATA-1 antiserum was raised againstEscherichia coli overproduced mouse GATA-1 protein in the following manner. Full-length mouse GATA-1 was overexpressed in E coli using the T7 RNA polymerase system of Studier et al.46 The predominantly insoluble protein was gel purified from a sodium dodecyl sulfate (SDS)-agarose Prosieve gel (FMC Bioproducts, Rockland, ME) and used to immunize rabbits. Positive antiserum was identified by immunoblotting and gel retardation assays using E coli expressed, mouse, and human GATA-1 protein (data not shown).
RESULTS
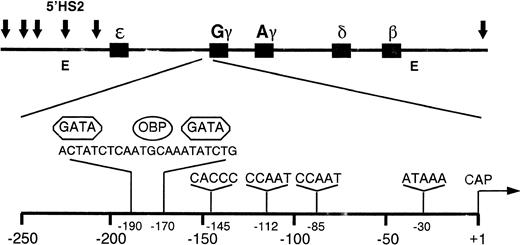
The location of the Gγ-globin gene, 5′HS2, and the Aγ and 3′β enhancers in the β-globin domain are shown in Fig 1. DNA fragments containing wild-type, mutant, and truncated γ-globin gene promoter sequences were cloned upstream of the luciferase reporter gene in the plasmid p0LUC. Enhancer sequences were cloned 150-bp 3′ to the reporter gene and in the normal 5′ → 3′ orientation to minimize any promoter-like activity they might have. In this location, the enhancers are required to work from a distance, rather than have a local effect on the promoter. These reporter plasmids were transfected into K562 cells and the luciferase activity was measured to determine the level of γ-globin gene promoter activity. K562 cells were used as a model of fetal erythroid development, as they express embryonic and fetal, but not adult, globin.47 In some experiments, MEL cells were used as host cells to test the expression of some of the plasmids in an adult erythroid environment because MEL cells express predominately β-globin.48
Diagram of the human β-globin gene domain showing the arrangement of the five β-globin genes, the position of the six identified DNase I hypersensitive (HS) sites (arrows), and the location of the 5′HS2 and 3′β enhancer elements (E). The expanded view of the G γ gene promoter region shows the recognition sites for the DNA-binding proteins GATA-1 and OBP.
Diagram of the human β-globin gene domain showing the arrangement of the five β-globin genes, the position of the six identified DNase I hypersensitive (HS) sites (arrows), and the location of the 5′HS2 and 3′β enhancer elements (E). The expanded view of the G γ gene promoter region shows the recognition sites for the DNA-binding proteins GATA-1 and OBP.
Deletion analysis of the γ-globin promoter.
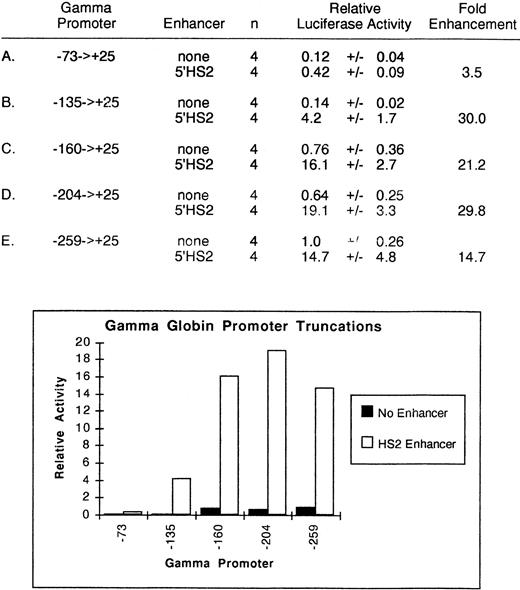
To assess the effect of cis-acting elements on the 5′HS2-mediated enhancement of γ-globin gene expression, deletions of theGγ-globin promoter were created at positions −259, −204, −160, −135, and −73 relative to the start of transcription. Deletion endpoints were selected to sequentially remove known binding sites within the γ-globin promoter (Fig 1). In the first set of experiments, no enhancers were included in the expression vector. The relative promoter activity of these segments of the γ-globin gene was assessed by transfection into K562 cells and measurement of luciferase activity, shown in Fig2. Deletion of γ-globin promoter sequence from −259 to −160 reduced activity approximately 25%, while deletion to −135 further decreased activity 85% over the full-length construct, confirming the importance of the −145 CACCC binding site for full γ-globin promoter activity.30,31 49
Relative activity of truncated γ-globin promoter plus enhancer plasmids in uninduced K562 cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± the standard error of the mean (SEM) and n denotes the number of assays performed.
Relative activity of truncated γ-globin promoter plus enhancer plasmids in uninduced K562 cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± the standard error of the mean (SEM) and n denotes the number of assays performed.
Identical plasmids to which the 5′HS2 enhancer was added were then tested under similar conditions. The addition of 5′HS2 to the expression vector augmented the full-length (−259) γ-globin gene promoter expression approximately 15-fold to 20-fold over the vector without the 5′HS2 element. The promoter strength of the truncation fragments from −204, −160, or −135 showed similar levels of enhancement by the addition of 5′HS2, suggesting that there were no cis-acting elements in that region that were important in 5′HS2 enhancer-promoter interactions. For example, although the −135 γ-globin promoter without the 5′HS2 enhancer had only 14% of the −259 → +25 promoter's activity, the addition of 5′HS2 was still able to increase activity 30-fold. Only the −73 γ-globin promoter did not show a strong enhancement with 5′HS2. These results demonstrate that the 5′HS2 enhancer is able to activate a minimal γ-globin promoter and suggest that cis-acting elements located within the −135 → −73 region are required for γ promoter-5′HS2 enhancer interaction. Two CCAAT boxes and a degenerate GATA binding site have been previously identified in this region.
Effect of CCAAT box mutations on γ-globin expression.
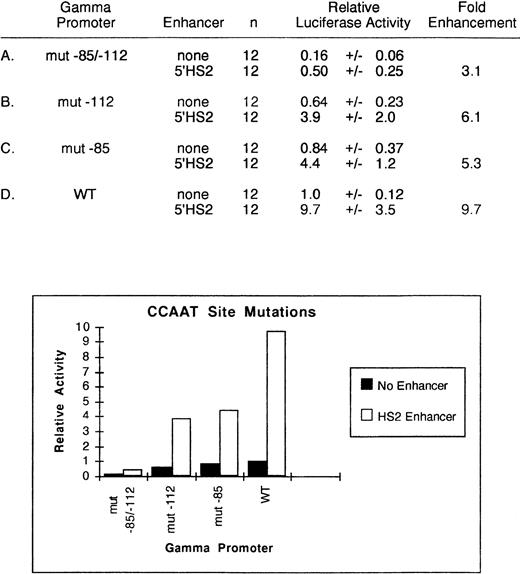
To directly test the importance of the CCAAT boxes, we mutated (CCAAT → CCGTT) the −112 and the −85 CCAAT boxes within the −259 → +25 γ-globin promoter and performed transient assays in K562 cells. As shown in Fig3, in the absence of any enhancer, the mutation of either the upstream or downstream CCAAT box decreased relative promoter activity to .64 ± 0.23 or .84 ± 0.37, respectively. Mutation of both sites reduced activity to 16% of the wild-type level. In these experiments, addition of the 5′HS2 enhancer to the −259 → +25 γ-globin promoter resulted in a 9.7 ± 3.5-fold enhancement. In the presence of the 5′HS2 enhancer, the individual mutation of the −112 and the −85 boxes reduced activity to 40% and 46% of this activity, respectively, while mutation of both sites decreased activity to 5% of the wild-type level. Thus, mutation of both CCAAT boxes reduced activity to the same level as truncation of the γ-globin promoter to −73 → +25. Based on these results, we conclude that at least one CCAAT box is required for γ-globin promoter function and for enhancement of γ-globin promoter activity by 5′HS2.
Relative activity of wild-type and mutated CCAAT γ-globin promoter plus enhancer plasmids in uninduced K562 cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed.
Relative activity of wild-type and mutated CCAAT γ-globin promoter plus enhancer plasmids in uninduced K562 cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed.
Role of γ-globin GATA binding sites.
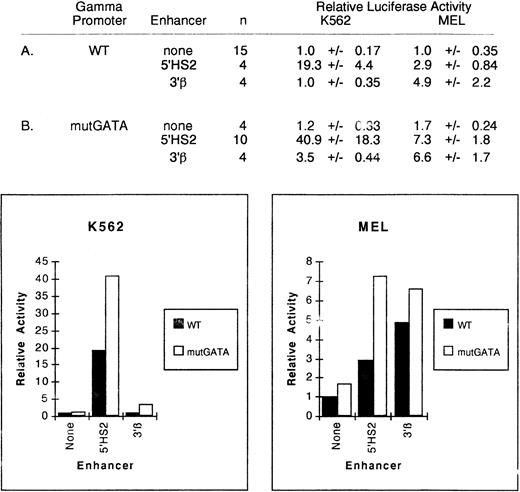
In the human ε-globin promoter, a conserved GATA binding site is required for interaction of the ε promoter with an erythroid-specific enhancer, but loss of the site does not alter expression in the absence of an enhancer.36 In the γ-globin promoter, two GATA binding sites are located at position −169 → −195 separated by an OCT-1 binding site (Fig 1). These binding sites are conserved in humans and simian primates expressing a fetal form of β-globin, implying an important role in γ-globin expression.50 Deletion of the segment of DNA containing these elements had no significant effect on 5′HS2-mediated enhancement of the γ-globin promoter, but did not rule out a specific role for the GATA elements. To determine if these conserved sites are needed for γ-globin promoter function or interaction of the promoter with the 5'HS2 or 3′β enhancers, we mutated the core DNA sequence of both GATA binding sites from GATA to TCGC (mutGATA) by site-directed mutagenesis and again performed transient expression assays in K562 cells. Mutation of both GATA sites did not reduce γ promoter activity (Fig 4). We interpret these results to indicate that an essential interaction between the promoter and 5′HS2 was not eliminated. Although there was an increased expression of the γmutGATA promoter when linked to 5′HS2, these differences in expression were not statistically significant.
Relative activity of wild-type and mutGATA γ-globin promoter plus enhancer plasmids in uninduced K562 and MEL cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed. The wild-type γ-globin promoter data in part A are repeated from Figs 2 and 3.
Relative activity of wild-type and mutGATA γ-globin promoter plus enhancer plasmids in uninduced K562 and MEL cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed. The wild-type γ-globin promoter data in part A are repeated from Figs 2 and 3.
Because mutations that reduce GATA binding to these GATA binding motifs are associated with elevated expression of the human γ-globin gene during adult life, we examined the expression of these promoters in MEL cells, which have an adult developmental pattern of expression of globin genes. Again, the γ-globin gene promoter carrying the GATA mutations had a higher level of expression than the wild-type promoter when linked to 5′HS2 (2.9 ± 0.84 compared with 7.3 ± 1.8, respectively, P < .005).
To determine if this effect was specific for the 5′HS2 enhancer, we linked the 3′β enhancer to the wild-type and GATA mutant γ-globin promoters and assayed their expression in K562 and MEL cells. The 3′β enhancer had no significant enhancing activity when linked to the wild-type promoter in K562 cells. In contrast, it increased the expression of the GATA mutant promoter 3.5-fold in K562 cells (P < .001). In MEL cells, the 3′β enhancer increased expression of the wild-type promoter approximately fivefold and increased the expression of the mutant GATA promoter 6.6-fold, a difference that is not statistically significant. These data indicate that both the enhancer and the environment affect the degree of activation in these experimental systems.
To determine if these effects could be duplicated with vectors that were integrated into the genome, we performed stable transformation colony assays.43 The level of γ-globin expression was measured by the number of productive integration events that resulted in G418-resistant colonies. As shown in Table1, mutGATA γ-globin expression was again not decreased compared with the WT promoter and an increased number of integration events were seen with the GATA mutant promoter.
Activity of Wild-Type and mutGATA γ-Promoters in Stably Transformed K562 Cells
| γ Promoter . | Enhancer . | No. NeoRColonies per 1 × 106 Cells . | Fold Enhancement . | ||
|---|---|---|---|---|---|
| Exp. No. 1 . | Exp. No. 2 . | Exp. No. 1 . | Exp. No. 2 . | ||
| WT | None | 33 | 78 | 1.0 | 1.0 |
| WT | 5′HS2 | 1,650 | 3,225 | 50.0 | 41.3 |
| mutGATA | None | 102 | 219 | 3.1 | 2.8 |
| mutGATA | 5′HS2 | 2,610 | 6,000 | 79.0 | 77.0 |
| γ Promoter . | Enhancer . | No. NeoRColonies per 1 × 106 Cells . | Fold Enhancement . | ||
|---|---|---|---|---|---|
| Exp. No. 1 . | Exp. No. 2 . | Exp. No. 1 . | Exp. No. 2 . | ||
| WT | None | 33 | 78 | 1.0 | 1.0 |
| WT | 5′HS2 | 1,650 | 3,225 | 50.0 | 41.3 |
| mutGATA | None | 102 | 219 | 3.1 | 2.8 |
| mutGATA | 5′HS2 | 2,610 | 6,000 | 79.0 | 77.0 |
Equal numbers of K562 cells were electroporated with plasmids containing wild-type or mutGATA γ promoters ± 5′HS2 enhancer driving the neomycin resistance (neoR) gene as described in Materials and Methods. The number of neoR colonies were counted after plating and growth in complete media plus 1 mg/mL G418 and 0.3% methylcellulose.
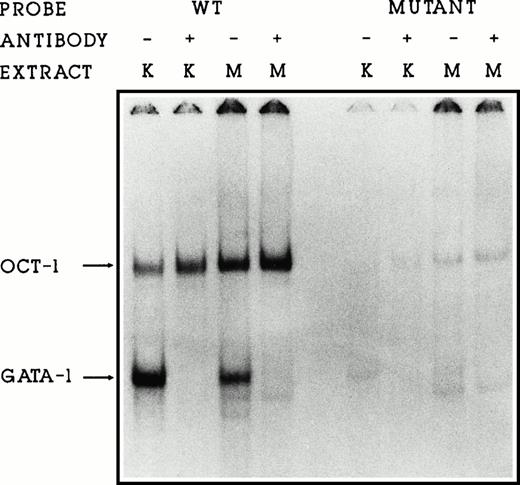
To ensure that these sequence changes abolish GATA-1 protein binding to the DNA, we performed gel retardation assays using DNA fragments containing the wild-type or mutated binding sites. Incubation of nuclear proteins from K562 or MEL cells with the wild-type fragment containing GATA and OCT binding sites resulted in one GATA-1 and one OCT-1 protein/DNA complex being formed (Fig5, lanes 1 to 4). Addition of GATA-1 antiserum in lanes 2 and 4 abolishes the GATA-1/DNA complex. Mutation of the GATA sequences eliminates both GATA-1 and OCT-1 binding (Fig 5, lanes 5 to 8). Because the proximal GATA binding site and the OCT binding sites overlap (nt no. −175, see Fig 1), OCT-1 binding is also lost with these mutations. We conclude that the binding of the transcription factors GATA-1 and OCT-1 to the γ-globin promoter are not required in this system for either promoter function or interaction between the promoter and the 5′HS2 or 3′β enhancers. Furthermore, loss of binding may permit higher levels of expression in this system.
GATA-1 and OCT-1 do not bind to the mutated GATA binding site. Gel retardation assays showing the binding of K562 (K) and MEL (M) nuclear proteins to the wild-type and mutGATA binding sites. GATA-1 antiserum (lanes 2, 4, 6, and 8) disrupts the GATA-1/DNA complex.
GATA-1 and OCT-1 do not bind to the mutated GATA binding site. Gel retardation assays showing the binding of K562 (K) and MEL (M) nuclear proteins to the wild-type and mutGATA binding sites. GATA-1 antiserum (lanes 2, 4, 6, and 8) disrupts the GATA-1/DNA complex.
Do three HPFH point mutations enhance γ-globin/5′HS2 interaction?
Based on these results, we examined the possibility that nondeletional HPFH mutations might alter the interaction of the γ-globin promoter with 5′HS2, especially in an adult erythroid environment. Mutations were made in the −259 → +25 γ-globin promoter to create the −202 C → G, -198 T → C, and −175 T → C mutations associated with HPFH, and these promoters were examined for enhancer-mediated γ-globin expression in both K562 and MEL cells (Figs 6 and7). These mutations were also tested because they lie in previously identified binding sites within the γ-globin promoter.
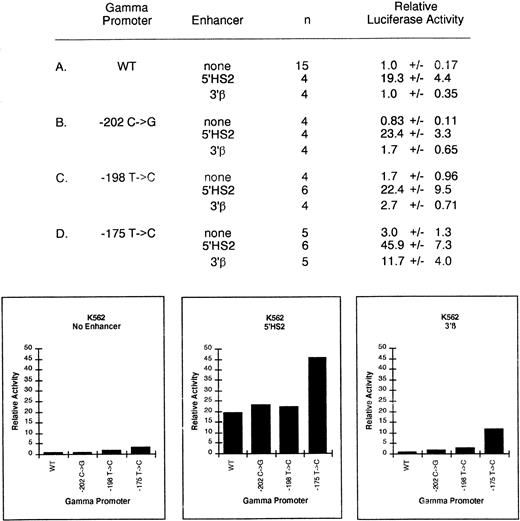
Relative activity of γ-globin HPFH promoter plus enhancer plasmids in uninduced K562 cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed. A value of 1.0x represents 45,947 ± 7,860 light units per minute per mg protein per unit SEAP activity.
Relative activity of γ-globin HPFH promoter plus enhancer plasmids in uninduced K562 cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed. A value of 1.0x represents 45,947 ± 7,860 light units per minute per mg protein per unit SEAP activity.
Relative activity of γ-globin HPFH promoter plus enhancer plasmids in uninduced MEL cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed. A value of 1.0x represents 3,087 ± 1,089 light units per minute per milligram protein per unit SEAP activity.
Relative activity of γ-globin HPFH promoter plus enhancer plasmids in uninduced MEL cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed. A value of 1.0x represents 3,087 ± 1,089 light units per minute per milligram protein per unit SEAP activity.
In the absence of enhancer elements, the activity of the −202 and −198 γ-globin HPFH promoters were unchanged compared with the wild-type promoter. In agreement with previous reports, we found the −175 T → C change increased expression in K562 cells 3.0 ± 1.3-fold (P < .001) (Fig 6D).21,51-53 No other significant increase in expression was observed in K562 or MEL cells with any HPFH mutation. Previous studies have suggested that these mutations do not cause persistence of γ-globin expression merely by increasing promoter strength.31
We next tested the effects of addition of the 5′HS2 enhancer on the HPFH promoters in K562 cells. This enhancer is active in K562 and MEL cells. In addition, we used the vector that substituted the 5′HS2 enhancer with the 3′β enhancer. The use of the 3′β enhancer permitted us to determine if the effect of the 5′HS2 enhancer is specific and allowed us to test for the possibility that an HPFH mutation might permit the γ-globin gene promoter carrying the mutation to have a productive interaction with another globin enhancer, such as 3′β. In K562 cells, only the −175 T → C promoter plus an enhancer had significantly increased activity. The 5′HS2 enhancer (Fig 6D) linked to the –175 T → C promoter produced a level of expression of 45.9 compared with the level of 19.3 seen when 5′HS2 is linked to the wild-type promoter (P < .001). This does not necessarily indicate a greater enhancer/promoter interaction, as the basal expression level of the –175 T → C promoter had a threefold level of expression over the wild-type. Substitution of the 3′β enhancer for 5′HS2 resulted in an 11.7-fold increased expression of the mutant promoter compared with the threefold 3′β enhancement of the wild-type promoter, again producing an effect that can be explained by the basal promoter strength of the −175 mutant promoter and not more productive interactions with the enhancer. The −198 T → C and −202 C → G mutations plus either enhancer did not significantly increase expression compared with the wild-type promoter.
In MEL cells (Fig 7), both the −202 C → G and −175 T → C promoters increased expression approximately twofold (P < .001) higher with the 5′HS2 enhancer than the wild-type promoter. The −198 T → C promoter plus 3′β enhancer increased expression 2.7-fold higher (P < .02) than the wild-type promoter plus 3′β enhancer. Thus, in an adult erythroid environment, each of the three HPFH promoters in combination with a globin enhancer showed twofold to threefold increased enhancement compared with the wild-type promoter plus enhancer, while in fetal erythroid cells, only the −175 T → C promoter showed increased enhancer-mediated expression by globin enhancers.
Relative activity of wild-type γ-globin promoter plus core HS2 enhancer plasmids in uninduced K562 cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed.
Relative activity of wild-type γ-globin promoter plus core HS2 enhancer plasmids in uninduced K562 cells. Activities are calculated relative to the wild-type γ-globin promoter-luciferase plasmid without enhancer. The activity values are presented as the mean ± SEM and n denotes the number of assays performed.
Do other γ-globin gene regulatory elements participate in the interaction with HS2 or the γ-globin gene promoter?
The previous studies have examined the interaction of HS2 with the γ-globin gene promoter. However, these studies do not examine the γ-globin gene promoter in its natural context linked to the 3′γ enhancer. Therefore, we examined the effect of the 5′HS2 enhancer on γ-globin gene promoter function when the 3′γ enhancer was linked in cis. This enhancer was inserted 5′ to the promoter and the 5′HS2 enhancer was left in the 3′ location used for all the previous assays. The expression level of this plasmid was compared with the plasmid that included only 5′HS2 enhancer. The addition of the 3′γ enhancer to the gamma-luciferase expression vector resulted in no significant increase in expression in transient assays (relative activity wild-type + no enhancer = 1.0 v wild-type + Aγ enhancer = 0.9). Therefore, we conclude that in this assay system, the presence of 3′γ had no significant effect on the interaction of 5′HS2 with the γ-globin gene promoter region.
The ultimate goal of these studies is to determine the factors in both the enhancer and promoter that interact. Previous studies of 5′HS2 have identified a tandem AP-1/NF-E2 core that functions as an inducible enhancer in erythroid cells.15,16,54 When linked 5′ to the γ-globin gene promoter, it serves as a strong activator at approximately 40% to 50% level of the full 5′HS2 fragment. To determine if this core 5′HS2 enhancer was responsible for the interaction with the γ-globin promoter, we substituted the 46-bp core enhancer for the full-length 5′HS2 enhancer in either the 5′ or 3′ position of our expression vector (Fig 8). When assayed in K562 cells, as previously reported, the core HS2 enhancer increased expression 11.5-fold when placed immediately upstream of the promoter.54 In contrast to the full-length 5′HS2 enhancer, however, the core HS2 enhancer did not increase expression when placed 150 bp 3′ to the γ-globin promoter-luciferase reporter gene. This suggests that the core HS2 enhancer is unable to activate γ-globin expression from a distance and therefore may not be functioning as a true enhancer. Furthermore, they indicate that the core element is not responsible for the interaction of 5′HS2 with the γ-globin promoter. A more detailed analysis of 5′HS2 will be required.
DISCUSSION
Many models of globin switching propose that selective interaction between individual promoters and the LCR is a central mechanism for regulating globin gene expression.5,17 Using transient expression assays, we have attempted to identify elements in the γ-globin promoter that are essential for productive interactions with globin gene enhancers. These experiments have focused on the 5′HS2 enhancer because it has been demonstrated to be sufficient for normal developmental expression of globin genes in experimental systems.14 As previously reported, the strong 5′HS2 enhancer increased expression from the γ-globin promoter approximately 20-fold in uninduced K562 cells.54 55 In MEL cells, γ-globin promoter activity was low and the globin enhancers had only small effects on expression, reflecting the normally low fetal globin levels found during adult life. Our data support the hypothesis that interaction between the γ promoter and the LCR/5′HS2 element can produce high level γ-globin expression and that this interaction occurs predominately during fetal development.
Within the γ-globin promoter, loss of both CCAAT boxes by either truncation or site-directed mutagenesis eliminated the enhancement by 5′HS2. Mutation of either the proximal or distal CCAAT box, however, resulted in an approximately 50% reduction in activity with 5′HS2. Thus, it appears that at least one CCAAT box is required for promoter-enhancer interaction and that either CCAAT box is functional. The ability of 5′HS2 to enhance a minimal globin promoter containing CCAAT and TATA binding sites has also been reported for both the ε- and β-globin genes. A truncated ε-globin promoter containing only its CCAAT and TATA sites was still activated 33-fold by 5′HS2.56 Also, a truncated human β-globin promoter containing only a TATA and CCAAT element was activated by LCR sequences in MEL cells.34 Therefore, CCAAT boxes may be generally required for the LCR to interact with globin promoters.
In erythroid cells, the factors CP1/NFY, CDP, NF-E3, and GATA-1 have been shown to interact in vitro with the γ-globin promoter CCAAT box region.57-60 Of these factors, GATA-1 is the only factor previously shown to be required for a promoter-enhancer interaction. Several observations make it unlikely, however, that GATA-1 binding to the γ-globin CCAAT box region is playing a similar role. First, GATA-1 binding to the γ-globin CCAAT boxes is not observed in embryonic and fetal cells, but is strong in extracts from adult cells where γ-globin is not normally expressed.59 Second, in adult cell extracts, GATA-1 binds to the wild-type, but not the −117 G → A HPFH CCAAT box.57,59 And finally, our mutation of the CCAAT boxes did not alter the GATA binding site, yet we still observed a decrease in γ-globin enhancement. Thus, the pattern of GATA-1 binding to the γ-globin CCAAT box region is consistent with GATA-1 having a repressive, not positive function. Of the three remaining known binding factors, CP1/NFY and NF-E3 are both positive activators,61,62 and therefore elimination of their binding would be consistent with our mutation results. Recent experiments by Ronchi et al62 with purified NF-E3 did not detect binding to the –85 CCAAT box and led them to conclude that the −112 CCAAT box is predominately occupied by CP1/NFY, rather than NF-E3 in cells expressing γ-globin. Thus, the involvement of CP1/NFY seems most consistent with our findings.
Previous studies of the ε- and β-globin gene promoters have identified GATA and CACCC sites as important for interaction with the LCR. Gong et al36 demonstrated that within the ε promoter the GATA site at −165 is required for interaction of the promoter with erythroid enhancers. In similar experiments, Motamed et al56 reported that the ε promoter CACCC box is an essential component of the 5′HS2 enhancer-ε promoter interaction. In the β-globin promoter, recent studies with the CACCC box binding factor erythroid Kruppel-like factor (EKLF) have shown that EKLF increased expression of a HS2-βCAT reporter plasmid 30-fold in K562 cells and when linked to a γ-LUC reporter HS2-βCAT was activated 1,000-fold.35 EKLF has also been shown to be critical for inducible expression directed by either the α- or β-LCR63 and mutations in the β-globin promoter CACCC box strongly decreased its response to the LCR in induced MEL cells.34 Our finding that 5′HS2 is able to strongly activate a −130 → +25 γ-globin promoter is in agreement with the results of Stamatoyannopoulos et al,64who found that the LCR was able to activate and correctly regulate a −141Aγ transgene in mice. Combined with our finding that mutation of the γ-globin promoter GATA sites resulted in no decrease in promoter activity, these results suggest that the γ-globin promoter does not require GATA-1 or CACCC binding factors to interact with the LCR. This implies that the LCR must be interacting with the γ-globin promoter in a different manner than with the ε- and β-globin promoters.
The role of GATA-1 in the regulation of the human γ-globin genes is still unclear. The fact that the GATA sites are evolutionarily conserved in all globin genes expressed during fetal development and the occurrence of the −175 T → C HPFH mutation suggests that GATA-1 is involved in regulating γ-globin expression. Unlike the human delta and epsilon genes,36,65,66 however, mutation of the γ-globin GATA sites had no effect in this system on activation, repression, or promoter-enhancer interaction. The modest increase in activity we observed with γmutGATA + 5′HS2 may be the result of eliminating OCT-1 binding, as a twofold repressive activity for OCT-1 in K562 cells has been reported.30 67 These results taken together suggest that GATA-1 and OCT-1 binding are not essential for γ-globin expression in fetal cells, although we have not ruled out a repressive effect.
HPFH is a benign disorder where patients express high levels of γ-globin during adult life. In a model of deletional HPFH, the increased proximity of enhancer sequences to the γ-globin genes resulting from inherited deletions of sequences 3′ to the γ-globin genes has been proposed as being responsible for the increased Hb F production.68,69 We examined the possibility that promoter/upstream mutations in nondeletional HPFH may be acting by altering the interaction of the γ-globin promoter with the LCR and either continuing promoter-enhancer interactions in adult cells normally present only in fetal cells or by fostering interaction with a different enhancer element, possibly the 3′β enhancer. The HPFH point mutations (−202 C → G, −198 T → C, −175 T → C) tested in this study fall in previously identified regulatory elements and alter the binding of transcription factors in vitro. The −202 C → G mutation slightly increases Sp1 binding and creates a binding site for the stage selector protein.45,70 The −198 T → C change creates a strong Sp1 binding site28,71 and the −175 T → C mutation alters GATA-1 binding, while increasing promoter strength approximately fourfold in erythroid cell lines.21 51-53 Our finding that in MEL cells all three HPFH promoters plus an enhancer had twofold to threefold higher expression is consistent with the hypothesis that single-base HPFH mutations may act to increase promoter-enhancer interactions. The small increases found in these experiments, however, make determining the biologic significance difficult. Additional studies will be required to answer the question of whether increased interactions between HPFH promoters and globin enhancers may be contributing to elevated Hb F levels in adults.
The finding that the core HS2 element only functioned as an enhancer when located immediately 5′ to the γ-globin gene promoter points out the importance of separating promoter and enhancer elements when studying enhancer function. Experiments performed with enhancer elements located adjacent to a promoter may only be measuring transcriptional activation potential. Our core HS2 data suggests to us a simple model where the tandem AP-1/NF-E2 sites act to bind transcriptional activators such as NF-E2 and behaves as an activator. Additional regulatory elements found within the 1-kb 5′HS2 enhancer, however, are required for interaction from a distance with the γ-globin gene. Several binding sites within the 5′HS2 enhancer element have been previously identified including binding sites for GATA-1, Sp1, USF, and YY1.20,72,73 These binding sites are required for full 5′HS2 activity.74-76 A 280-bp 5′HS2 fragment containing these sites is also able to enhance γ-globin expression from a distance in our assay system (data not shown). Whether one or more of these sites are necessary for the LCR to act from a distance remains to be established.
Supported in part by Grant No. 5P60-HL-2839-1-13 from the National Institutes of Health (NIH). R.E.K. is a member of the DUKE/UNC Comprehensive Sickle Cell Center. S.D.L. was supported by a research fellowship from the Cooley's Anemia Foundation, New York, NY.
Address reprint requests to Russel E. Kaufman, MD, Box 3250, Duke University Medical Center, Durham, NC 27710.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal