Abstract
The X-linked form of chronic granulomatous disease (CGD) is caused by mutations in the CYBB gene, which encodes the 91-kD subunit of the flavocytochrome b558, a component of the superoxide-generating nicotinamide adenine dinucleotide phosphate (NADPH) oxidase in phagocytic leukocytes. Mutations in this gene are very heterogeneous and often unique for one family. Here we report on a family with two patients (brothers), one with a 3-kb deletion comprising exon 5 and the other with a 3.5-kb deletion comprising exons 6 and 7 of the CYBB gene. Sequence analysis of polymerase chain reaction (PCR)-amplified genomic DNA proved these deletions to be overlapping for 35 bp. Analysis by restriction fragment length polymorphism of genomic DNA from the mother's leukocytes showed her to be a carrier of both deletions in addition to the normal CYBB sequence. This triple somatic mosaicism was confirmed with PCR-amplified genomic and complementary DNA. The presence of the normal CYBB gene in the mother was also proven by the finding of normal superoxide-generating neutrophils in addition to cells lacking this ability. Triple X syndrome was excluded. These findings suggest that the mutations are the result of an event in early embryogenesis of the mother, possibly involving a mechanism like sister chromatid exchange.
CHRONIC granulomatous disease (CGD) is a severe clinical syndrome characterized by recurrent, life-threatening bacterial and fungal infections.1 The disease is caused by failure of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase enzyme in phagocytic leukocytes (neutrophilic and eosinophilic granulocytes, monocytes and macrophages) to generate superoxide, a precursor of microbicidal oxygen metabolites.2 This enzyme is composed of several constituents. The flavocytochromeb558 is located in the plasma membrane of the phagocytes and consists of two subunits, a 91-kD glycoprotein called gp91phox (from phagocyte oxidase) and a 22-kD protein called p22phox.3-5 This flavocytochrome is the actual enzyme that accepts electrons from NADPH in the cytosol and transmits these to oxygen at the other side of the membrane, ie, in the phagosome that contains the microorganisms ingested by the phagocytes. However, under resting conditions, the oxidase is not active. Only when the phagocytes have bound microorganisms or soluble stimuli, such as high concentrations of chemotaxins, the flavocytochrome is converted into an active enzyme. This activation is brought about by translocation of two additional oxidase components from the cytosol to the flavocytochrome, probably by inducing a conformational change in the flavocytochrome.6These cytosolic oxidase components are a 47-kD protein called p47phox and a 67-kD protein called p67phox. A third cytosolic p40phox protein probably stabilizes p47phox and p67phox in resting phagocytes.7 Finally, a membrane-bound and a cytosolic guanosine triphosphate (GTP)-binding protein and several regulating proteins are involved in fine-tuning the activation process.8-10
CGD is caused by mutations in gp91phox, p22phox, p47phox, or p67phox.11 The CYBB gene encoding gp91phox is located at Xp21 and contains 13 exons.12 The genes for the other oxidase components are located on autosomes. Mutations in the CYBB gene, which are found in about two thirds of all CGD patients, are very heterogeneous, comprising deletions from one to several million base pairs, insertions up to 2 kb, splice site mutations, missense mutations, and nonsense mutations.11 Usually, these mutations are family-specific, but now, after analysis of about 250 X-CGD families, a few ‘hot spots’ of point mutations can be recognized, most of them in CpG dinucleotides.13 In contrast, the origin of the deletions in the CYBB gene is not clear, but occasionally a family presents with a defect that gives some insight into this phenomenon. We report here on a family in which two sons suffer from X-linked CGD due to two different deletions in the CYBB gene, while their mother carries the same deletions in her somatic (hair root and white blood) cells in addition to the normal CYBB sequence. This indicates that the mother is a triple mosaic in her somatic cells, possibly due to breakage and reunion of single strands of the sister chromatids early in her embryonic development.
PATIENT HISTORIES
Patient 1 is the oldest of two brothers, born in 1979 and presenting at the age of 2 years with a disseminated Salmonella infection. CGD was diagnosed with a stimulated nitroblue tetrazolium (NBT) dye test showing 0% formazan-positive granulocytes. This result was confirmed by subsequent NBT tests and by chemiluminescence tests.
The boy was otherwise well until the age of 5 when he had a cellulitis of the left foot with associated lymphangitis and inguinal lymphadenitis. At 11 years of age, he developed a first submandibular lymphadenitis that needed to be drained. The patient was subsequently given cotrimoxazole as prophylaxis.
When he was 13, tetracycline was given for severe acne. Two years later, the boy developed a left upper lobe pneumonia, which was succesfully treated with antibiotics. A few months thereafter, he had a second episode of submandibular lymphadenitis. The lymph node was excised, and the culture was positive for Aspergillus fumigatus. Treatment with amphotericin B followed, and the patient recovered. Itraconazole was given as prophylaxis.
Because of uncontrolled severe acne, isotretinoin was given at the age of 16. This drug was discontinued after a few months because of presumed drug-induced fever and other side effects. That same year, the patient developed rectal bleeding due to ulcerative lesions in the left colon. Pathologic examination showed granulomatous inflammation, but no specific pathogen was recovered from cultures. The patient was treated with mesalazin and steroids. This treatment was successful for the colitis. However, after 6 weeks, a nodular pneumonia appeared. Broncho-alveolar lavage showed no pathogens, but Aspergilluswas felt to be the most likely cause. The patient was successfully treated for 6 weeks with amphotericin B, while tapering off the steroids.
As of this writing, the patient is 17 years old and in good general health. He takes cotrimoxazole and itraconazole as prophylaxis for bacterial and fungal infections.
Patient 2 is the youngest brother, born in 1981. CGD was diagnosed in the first weeks of life by means of an NBT dye test. The diagnosis was confirmed by the absence of chemiluminescence after neutrophil stimulation in vitro.
At the age of 2 months, the patient developed a submandibular lymphadenitis, which was successfully treated with parenterally administered antibiotics. At 6 years of age, he had a pneumonitis with associated hilar lymphadenopathy, from which the boy recovered after a 10-day course of intravenously administered antibiotics. Three years later he was hospitalized for a period of 3 months because of a liver abscess, caused by Staphylococcus aureus. The course was protracted and complicated with ulcerative duodenitis and subsequent duodenal substenosis, drug-induced renal salt wasting with hyponatremia and convulsions, and drug-induced leukopenia. The boy finally recovered after surgical drainage of the abscess, prolonged courses of multiple antibiotics, and the administration of γ-interferon. While on cotrimoxazole, rifampin, and fluconazole prophylaxis, he was readmitted 14 months later with a pneumonitis and pericarditis caused byAspergillus. Despite treatment with amphotericin B, the patient deteriorated and developed a constrictive pericarditis, for which he underwent pericardectomy 2 months after admission. Cardiac output did not improve, and the patient died of circulatory failure at the age of 10 years and 9 months. The autopsy showed a large thrombus in the jugular veins and in the left atrium. The epicardium was filled withAspergillus hyphi. Both parents had an NBT dye test. It was normal in the father, but gave intermediate results in the mother who showed only about 40% of NBT-positive cells after neutrophil stimulation. Both the father and the mother are in good health. In the mother's family, no one has a clinical history of CGD.
MATERIALS AND METHODS
This study was conducted after appropriate informed consent had been obtained from all human subjects involved.
Granulocyte function assays.
Neutrophils were purified from venous blood as described earlier.14 NADPH oxidase activity of intact cells was measured by oxygen consumption, lucigenin-amplified chemiluminescence, or NBT dye reduction14 after stimulation of the cells with phorbol-myristate acetate (PMA), formyl-methionyl-leucyl-phenylalanine (fMLP) or serum-treated zymosan (STZ) particles.
Western blot analysis.
Neutrophil membrane and cytosolic fractions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electroblotted onto nitrocellulose paper and probed with mouse monoclonal antibodies against gp91phox or p22phox15 or with specific rabbit antisera against p47phox or p67phox.16 Binding of the antibodies was detected with alkaline phosphatase-conjugated antimouse-Ig and antirabbit-Ig, respectively.
Preparation of RNA and DNA and analysis of DNA.
RNA was isolated from blood mononuclear leukocytes as described previously.17 First-strand cDNA was synthesized from RNA by reverse transcription. Part of the gp91phox-coding region (CYBB exon 4-10) was amplified from cDNA by PCR with primers specified in Table 1, under conditions described previously.17 The PCR products were separated on Nusieve 2:1 agarose gel and stained with 0.05% (wt/vol) ethidium bromide. Genomic DNA was isolated from total blood leukocytes.18Southern blot analysis of genomic DNA was performed by digestion with the restriction enzyme EcoRI and probing with a cDNA CYBB fragment that recognizes the entire coding sequence (code pSV-CGD, kindly provided by Dr M. Dinauer, Indiana University School of Medicine, Indianapolis). Fragments of the CYBB gene were amplified from genomic DNA by PCR with the primers specified in Table 1, under conditions defined previously.17 For nucleotide sequencing, the direct sequencing method was used.19
Primer Sequences
| Fragment Amplified . | Primers . |
|---|---|
| CYBB cDNA exons 4-10 | S 5′-AAGAGTTCGAAGACAACTGGACAGG-3′ |
| AS 5′-GAACACATCTTCACTGGCAGTGCCAAAGGGCCCATC-3′ | |
| CYBB gDNA exon 4exon 5 | S 5′-AAGAGTTCGAAGACAACTGGACAGG-3′ |
| AS 5′-GGATCAGAATTATTGACTCGGGCATTC-3′ | |
| CYBB gDNA exon 5exon 6 | S 5′-AGTAGCACTCTCTGAACTTG-3′ |
| AS 5′-GTACCAAAAGACTTCAAAGTAAGACCTCCGGATG-3′ | |
| CYBB gDNA intron 5intron 5 | S 5′-AAAAGTCAATCCTAGTGTC-3′ |
| AS 5′-GTGTCCTATGTATAGAACAATAG-3′ | |
| CYBB gDNA exon 6exon 7 | S 5′-TTTGTGATCTTCTTCATTGGCCTTGCC-3′ |
| AS 5′-GCACAGCCAAACTCTCTGCGGTCTGC-3′ | |
| CYBB gDNA exon 7exon 8 | S 5′-CTCAGTTTGCTGGAAACCCT-3′ |
| AS 5′-CTGTTGAGATCGCCAAAACCG-3′ | |
| CYBB gDNA exon 7intron 7 | S 5′-CTCAGTTTGCTGCAAACCCT-3′ |
| AS 5′-CACTGTGACAATTGCAAGATGAC-3′ |
| Fragment Amplified . | Primers . |
|---|---|
| CYBB cDNA exons 4-10 | S 5′-AAGAGTTCGAAGACAACTGGACAGG-3′ |
| AS 5′-GAACACATCTTCACTGGCAGTGCCAAAGGGCCCATC-3′ | |
| CYBB gDNA exon 4exon 5 | S 5′-AAGAGTTCGAAGACAACTGGACAGG-3′ |
| AS 5′-GGATCAGAATTATTGACTCGGGCATTC-3′ | |
| CYBB gDNA exon 5exon 6 | S 5′-AGTAGCACTCTCTGAACTTG-3′ |
| AS 5′-GTACCAAAAGACTTCAAAGTAAGACCTCCGGATG-3′ | |
| CYBB gDNA intron 5intron 5 | S 5′-AAAAGTCAATCCTAGTGTC-3′ |
| AS 5′-GTGTCCTATGTATAGAACAATAG-3′ | |
| CYBB gDNA exon 6exon 7 | S 5′-TTTGTGATCTTCTTCATTGGCCTTGCC-3′ |
| AS 5′-GCACAGCCAAACTCTCTGCGGTCTGC-3′ | |
| CYBB gDNA exon 7exon 8 | S 5′-CTCAGTTTGCTGGAAACCCT-3′ |
| AS 5′-CTGTTGAGATCGCCAAAACCG-3′ | |
| CYBB gDNA exon 7intron 7 | S 5′-CTCAGTTTGCTGCAAACCCT-3′ |
| AS 5′-CACTGTGACAATTGCAAGATGAC-3′ |
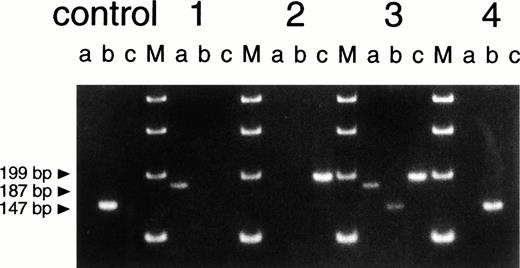
Primers for allele-specific PCR (Fig 4). Primer 1 S 5′-AAAAGTCAATCCTAGTGTC-3′; primer 2 AS 5′-GTGTCCTATGTATAGAACAATAG-3′; primer 3 S 5′-CAGTTCACATTTCCTATGACATC-3′; primer 4 AS 5′-CACTGTGACAATTGCAAGATGAC-3′.
X-chromosome marker analysis was performed as previously described.20 21 Polymorphisms in the ornithine transcarbamylase (OTC) gene, located at the proximal side of the CYBB gene, and in the Duchenne muscular dystrophy (DMD) gene, located distally of the CYBB gene, were used to show the inheritance of the CYBB mutations in the investigated family.
RESULTS
Identification of the CGD subtype.
The patients were identified as CGD patients by complete lack of NBT reduction by their neutrophils with PMA as the stimulus. Moreover, lucigenin-amplified chemiluminescence of the patients' neutrophils stimulated with either PMA or PMA followed by STZ was totally absent. On Western blots of the patients' neutrophil membranes after SDS-PAGE, gp91phox was not detectable, and p22phox was only faintly visible. In the patients' neutrophil cytosols, p47phox and p67phox were normally present. These results suggest that the patients suffer either from the X-linked form of CGD (gp91phox deficiency) or from the autosomal p22phox-deficient form of CGD. These forms of CGD cannot easily be discriminated from each other, because the gp91phox and the p22phox subunits of flavocytochrome b558 need each other for stable expression in the cell. Both forms of CGD therefore lead to lack of expression of both subunits.15 We studied also the neutrophils of the patients' mother. These cells showed a partial lack of NADPH oxidase activity in the oxygen consumption assay: with PMA 2.3 nmol of O2 were consumed per 106 cells per minute (control 8.8) and with fMLP 3.1 (control 8.4). In the NBT slide test, 42% of her PMA-activated neutrophils reduced the NBT dye to formazan (control 98%). This proves that the mother has a mixture of normal and oxidase-deficient neutrophils in her circulation, and therefore is a carrier of the X-linked form of CGD. Thus, her sons were expected to be hemizygote patients with this disease.
Identification of the mutations.
Southern blot analysis of genomic DNA was performed after treatment of the DNA with EcoRI and hybridization with a CYBB cDNA probe. The results (Fig 1, lanes 3 and 4) show that the two brothers have different deletions in the CYBB gene. In the eldest boy, the band at 7 kb has disappeared and a new band at about 3.5 kb is visible. In the youngest boy, the bands at 7 kb and at 4.0 kb have disappeared and a new band at about 8 kb is visible. Most surprisingly, the mother of these two patients (Fig 1, lane 2) was found to carry both mutations in her DNA, in addition to the normal sequence (Fig 1, lane 1).
Southern blot RFLP assay of genomic DNA after digestion with EcoRI. (A) Shows the EcoRI restriction map of the CYBB gene. Hybridization was performed with a 32P-labeled pSV-CGD cDNA probe, containing the entire coding region of CYBB. The length of the expected restriction fragments is indicated. (B) Shows the results with the DNA from a healthy control donor (lane C), from the mother of the patients (lane M), from the eldest patient (lane 1), and from the youngest patient (lane 2). The size of the indicated fragments was determined relative to HindIII digested phage-lambda DNA as size markers. Arrows at right point at differences between lane C and the other lanes.
Southern blot RFLP assay of genomic DNA after digestion with EcoRI. (A) Shows the EcoRI restriction map of the CYBB gene. Hybridization was performed with a 32P-labeled pSV-CGD cDNA probe, containing the entire coding region of CYBB. The length of the expected restriction fragments is indicated. (B) Shows the results with the DNA from a healthy control donor (lane C), from the mother of the patients (lane M), from the eldest patient (lane 1), and from the youngest patient (lane 2). The size of the indicated fragments was determined relative to HindIII digested phage-lambda DNA as size markers. Arrows at right point at differences between lane C and the other lanes.
The existence of two different deletions was also proven by analysis of PCR-amplified cDNA. Fragments comprising CYBB exon 4 through 10 were amplified, separated in agarose and visualized in ethidium bromide. Figure 2 shows that the deletions in the patients are different in size and shows again that the mother carries both deletions and the normal composition. From the maternal grandmother's cDNA a fragment of normal size was obtained. Sequencing of these PCR products showed that the eldest boy lacked exons 6 and 7 and the youngest boy lacked exon 5 (not shown).
Size analysis of PCR products from cDNA. Primers were chosen on exon 4 and exon 10 (Table 1). The PCR products were separated in agarose and visualized with ethidium bromide. Lane 1 contains material from a healthy control donor, lane 2 from the youngest patient, lane 3 from the eldest patient, lane 4 from the mother of the patients, and lane 5 from the maternal grandmother of the patients. Lane M contains size markers of 100 bp.
Size analysis of PCR products from cDNA. Primers were chosen on exon 4 and exon 10 (Table 1). The PCR products were separated in agarose and visualized with ethidium bromide. Lane 1 contains material from a healthy control donor, lane 2 from the youngest patient, lane 3 from the eldest patient, lane 4 from the mother of the patients, and lane 5 from the maternal grandmother of the patients. Lane M contains size markers of 100 bp.
To characterize the deletions in detail, we amplified genomic DNA by PCR in overlapping fragments (for primers, see Table 1) and determined the entire nucleotide sequence of intron 4 through intron 7 of the CYBB gene in the two brothers. In the eldest boy, we found a deletion starting in intron 5 at about 0.2 kb 5′ of the intron-5/exon-6 boundary and ending in intron 7 at about 0.9 kb 3′ of the exon-7/intron-7 boundary (Fig 3). Thus, this deletion removes about 3.5 kb of the CYBB gene, including exons 6 and 7. In the youngest boy, we found a deletion starting in intron 4 at about 1.2 kb 5′ of the intron-4/exon-5 boundary and ending in intron 5 at about 1.7 kb 3′ of the exon-5/intron-5 boundary. As shown in Fig 3, this deletion removes about 3 kb of the CYBB gene, including exon 5. Figure 3 also shows that both deletions are adjacent to each other, overlapping for 35 bp.
Localization of deletions in the CYBB gene of the two patients. The shaded area indicates the deletions, comprising a 3.5-kb nucleotide stretch from intron 5 to intron 7 in patient 1 (the eldest brother) and a 3.0-kb nucleotide stretch from intron 4 to intron 5 in patient 2 (the youngest brother). The deletions overlap for 35 bp.
Localization of deletions in the CYBB gene of the two patients. The shaded area indicates the deletions, comprising a 3.5-kb nucleotide stretch from intron 5 to intron 7 in patient 1 (the eldest brother) and a 3.0-kb nucleotide stretch from intron 4 to intron 5 in patient 2 (the youngest brother). The deletions overlap for 35 bp.
To verify in yet another way that the leukocyte DNA of the mother contained three different compositions of the CYBB gene, we devised an allele-specific PCR to differentiate between these three forms. Primers were chosen around the two deletions, thus leading to product formation only when the complementary sequence was present in the genomic DNA and the distance between the primers was not more than about 3 kb. Figure4 confirms once more that the mother's DNA contains both the normal CYBB sequence and the two different deleted compositions. The maternal grandmother's DNA again showed only the normal CYBB composition. The same test was also applied to DNA from the mother's hair roots. Again, the triple somatic mosaicism was found (not shown).
Size analysis of PCR products from genomic DNA. Primer 1 (sense) was chosen on intron 4, primer 2 (antisense) on intron 5, primer 3 (sense) on intron 5 and primer 4 (antisense) on intron 7 (for positions, see Fig 3; for compositions, see Table 1). Primers 2 and 3 hybridize with DNA sequences within the deletions of patient 1 and patient 2, respectively (Fig 3). Therefore, PCR (a), with primers 3 and 4, only leads to product formation with DNA from patient 1 (not with DNA from a healthy control donor because these primers then hybridize too far apart to lead to product formation under the PCR conditions used). PCR (b), with primers 2 and 3, only leads to product formation with DNA from a healthy control donor. PCR (c), with primers 1 and 2, only leads to product formation with DNA from patient 2. The figure shows the results of PCR (a), (b), and (c) obtained with DNA from a healthy control donor, patient 1 (the eldest brother), patient 2 (the youngest brother), the mother of the patients (3), and the maternal grandmother of the patients (4). The four lanes marked M contain size markers of 100 bp.
Size analysis of PCR products from genomic DNA. Primer 1 (sense) was chosen on intron 4, primer 2 (antisense) on intron 5, primer 3 (sense) on intron 5 and primer 4 (antisense) on intron 7 (for positions, see Fig 3; for compositions, see Table 1). Primers 2 and 3 hybridize with DNA sequences within the deletions of patient 1 and patient 2, respectively (Fig 3). Therefore, PCR (a), with primers 3 and 4, only leads to product formation with DNA from patient 1 (not with DNA from a healthy control donor because these primers then hybridize too far apart to lead to product formation under the PCR conditions used). PCR (b), with primers 2 and 3, only leads to product formation with DNA from a healthy control donor. PCR (c), with primers 1 and 2, only leads to product formation with DNA from patient 2. The figure shows the results of PCR (a), (b), and (c) obtained with DNA from a healthy control donor, patient 1 (the eldest brother), patient 2 (the youngest brother), the mother of the patients (3), and the maternal grandmother of the patients (4). The four lanes marked M contain size markers of 100 bp.
Chromosomal localization of the deletions.
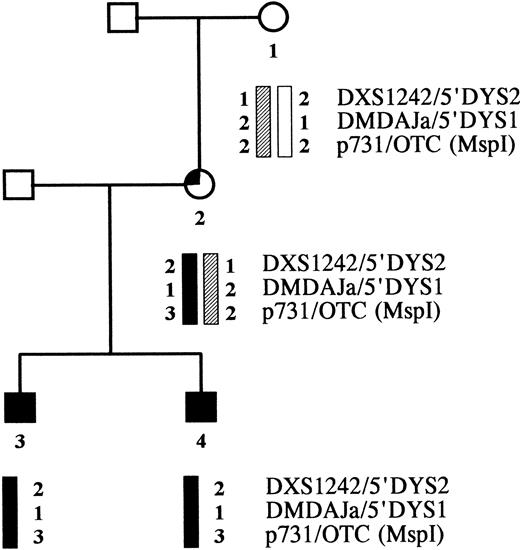
Karyotyping of the mother's lymphocytes in mitosis showed no abnormalities, ie, no X-chromosome trisomy. X-chromosome marker analysis in the family indicated that the two deletions of the brothers originated in the same chromosome. The CYBB-containing part of the brothers' X chromosome proved to be identical to one of their mother's X-chromosome regions, but was not found in their maternal grandmother (Fig 5). Hence, the mutations took place in the CYBB allele that the mother received from her father.
Pedigree of the CGD family. Haplotype analysis was performed with X-chromosome-specific markers flanking the CYBB gene, ie, two CA repeats in the DMD gene and one Msp I polymorphism in the OTC gene.
Pedigree of the CGD family. Haplotype analysis was performed with X-chromosome-specific markers flanking the CYBB gene, ie, two CA repeats in the DMD gene and one Msp I polymorphism in the OTC gene.
DISCUSSION
X-linked CGD is caused by a large variety of mutations in the CYBB gene.11 Usually, only one or two patients are found in small families, due to the severity of the disease. As in other X-linked diseases, patients are sometimes recognized without apparent carriership in the mother or female relatives. This is usually considered to result from a unique meiotic event, but a mitotic origin in early embryogenesis cannot be excluded. Indeed, in some cases of Duchenne muscular dystrophy,22 ornithine transcarbamylase deficiency,23 and hemophilia A24 clear indications for a mitotic origin have been obtained by recurrence of new mutations in a sibship or by somatic mosaicism in the mother or in the patient. If a mutation event occurs during mitosis before the germ-line segregates from other cell lines, the germ-line and somatic tissues can both be expected to be involved.
One family with hemophilia A deficiency has been reported25in which one male patient had a partial factor VIII gene deletion, one sister of the patient was a carrier of this deletion, and one other sister was a carrier of a partial factor VIII gene duplication. Other sisters and brothers of the patient were normal. Thus, the mother had a germ-line mosaicism for two abnormal and the normal factor VIII gene configuration. In the mother's leukocyte DNA, indications for the same three configuration were found by restriction fragment length polymorphism (RFLP) analysis. Therefore, the situation in this family resembles the situation in the family with CGD studied by us.
With three different methods, we have shown that the mother of the two male patients with CGD, each with a different deletion in the CYBB gene, carries both deletions, as well as the normal sequence in the DNA of her somatic cells (leukocytes and hair roots). Because she had no other children, it cannot be established whether the triple mosaicism is also present in her germ-line cells. In the DNA of her own mother, only the normal CYBB sequence was found. This concurs with the lack of the affected allele in her mother. Because the deletions are overlapping, they probably originate from one simultaneous event. Hence, inheritance of one of the mutations from her father or mother is unlikely. Inheritance of both mutations from one parent (ie, from her father) was ruled out by her normal XX karyotype. Most likely, therefore, the mutations originated from a single mitotic event in the early embryogenesis of the patients' mother. The nature of this event is not known, but may involve a mechanism like sister chromatid exchange.
Breaking and reunion of chromatids usually occur without causing any damage. However, when a genetic defect causes genome instability with a high incidence of sister chromatid exchange, loss of DNA may take place.26 If, under such conditions, during late replication of the methylated “inactive” X chromosome a single strand break occurs in each of the newly formed chromatids at a relatively short distance (eg, at 35 bp distance, as in our patients), these ends may hybridize and form a stable double helix. Before repair can take place, separation of the two chromosome halves to daughter cells may occur. Opposite parts of DNA may be torn away and become lost during the next round of replication.
At present it is unknown how the mutated X chromosomes are divided over the mother's somatic cells, ie, whether they are together in one cell (with other cells containing two normal X chromosomes) or whether they are in separate cells (each accompanied by a normal X chromosome). In the former case, one would expect the two mutated X chromosomes to be present in the mother's leukocytes in equal number, whereas in the latter case, one would expect the normal X chromosomes to make up 50% of all X chromosomes. An indication for the existence of the second possibility was obtained from a density scan of Fig 1B. The densities of the 2.7/2.8-kb bands were normalized to correct for the different amounts of DNA applied to the four lanes in the gel and the presence of two X chromosomes in the mother's cells, but only one X chromosome in the cells from her sons and from the (male) control donor. We then found that the (normal) 7-kb band was present for 53% in the mother as compared with the control, the (abnormal) 3.5-kb band for 11% in the mother as compared with the eldest boy, and the (abnormal) 8-kb band for 38% in the mother as compared with the youngest son. This shows that the mother's normal CYBB allele is present in about 50% of her X chromosomes, and that the two mutated forms of the CYBB gene are not present in about equal amounts in her genome. These findings indicate that each of her leukocytes carries one X chromosome with a normal CYBB gene and another X chromosome with either of the two mutated forms of this gene. The 3-kb deletion is present in about 75% of her leukocytes, the 3.5-kb deletion in about 25% of her leukocytes.
Occurrence of germ-line and/or somatic mosaicism may be more common than previously thought. Until now, we have characterized about 110 mutations in the CYBB gene. In 10 of the families involved, two or more patients were identified. In two of these last 10 families, we found different mutations in the patients within one family. One of these families is reported in this article. In the other family, we found two male patients with different deletions in the CYBB gene, ie, one that removes the first 10 exons and intronic sequences and another that encompasses the entire CYBB gene. Again, marker analysis indicated that the two deletions occurred in the same allele. The mother of these patients was found to carry a somatic mosaicism for the normal sequence and for the first-mentioned partial CYBB deletion, but the presence of the second, total CYBB deletion in her DNA must be studied by other methods than PCR and Southern blotting (De Boer, Bakker, Nemet and Roos, unpublished). Thus, the possibility of complicated mosaicisms must be kept in mind when performing genetic studies for, eg, prenatal diagnosis.
Supported by Grant No. 28-2167 from The Netherlands Foundation for Preventive Medicine (Praeventiefonds), The Hague, The Netherlands.
Address reprint requests to Dirk Roos, PhD, CLB, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal