Abstract
We have recently shown that the multiple tumor suppressor gene 1 (MTS1 ) encoding the p16INK4a and p19ARF cell-cycle inhibitors is inactivated by deletion or disruption in most human T-cell acute lymphoblastic leukemias (T-ALLs), representing the most frequent genetic event thus far described in this disease. To analyze the mechanism of these chromosomal events, we used cloning, sequencing, and/or polymerase chain reaction mapping to study 15 rearrangements occurring in the MTS1 locus. We found that these breakpoints occur in two clusters (MTS1bcrα and MTS1bcrβ ). The three rearrangements occurring in MTS1bcrα correspond to a recurrent recombination juxtaposing 5′ MTS2 exon 1 and 5′ MTS1 exon 1α sequences. Breakpoints for 10 of 12 rearrangements within MTS1bcrβ are located at a polymorphic (CA) repeat, suggesting that this sequence might play a role in the clustering. For both MTS1bcrα and MTS1bcrβ, sequence analyses and PCR mapping experiments show that the tightly clustered breakpoints are located in the vicinity of heptamers whose sequence is similar to those involved in the V(D)J recombination. Moreover, short deletions, GC-rich random nucleotide additions, and clone-specific junctional sequences are present in all cases, further suggesting that the rearrangements are due to illegitimate V(D)J recombinase activity. These data are the first demonstration that a tumor suppressor gene can be inactivated by the V(D)J recombinational mechanism. Moreover, they reinforce the view that this process, normally required for T-cell differentiation, plays a crucial role in the pathogenesis of T-ALL.
MULTIPLE TUMOR suppressor gene 1 (MTS1/p16INK4a/CDKN2 ), which is located at chromosome 9p21 and encodes the two distinct cell-cycle inhibitors p16INK4a 1 and p19ARF,2 has been found to be inactivated by mutations and/or deletions in a number of transformed cell lines3,4 as well as in various types of primary tumors (reviewed in Sherr5 ). These data suggested that this gene is a major tumor suppressor gene, a conclusion that has recently been reinforced by analysis of knockout mice lacking functional MTS1 genes.6 Another putative tumor suppressor gene, MTS2,3 is located 12 kb centromeric to MTS1 exon 1β (Fig 1A). This gene encodes the p15INK4b protein, which shares with p16INK4a the ability to inhibit cdk4 and cdk6 kinase activities.7
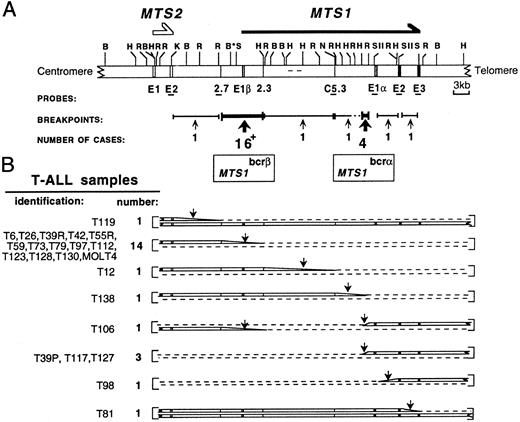
Restriction map of the MTS locus and schematic representation of its configuration in T-ALL samples. (A) An updated MTS locus map is shown. The probes used and the 2 breakpoint clusters MTS1bcrα and MTS1bcrβ are indicated. C5.3, 2.7, and 2.3, sequence tagged sites (STSs); E, exon; *, polymorphic BamHI site; +, the 16 breakpoints include that of the T84 sample, for which the MTS1 configuration was not fully characterized. (B) Schematic representation of the 2 chromosome 9s in cases with rearrangements occurring in the locus (22 T-ALL samples and the MOLT4 cell line).21 In 1 sample, T106, 2 breakpoints occurring on each chromosome 9 were detected. In T39, breakpoints were different at presentation (T39p) and relapse (T39r). Dashed lines indicate deletions; vertical arrows indicate breakpoint-containing regions. The T84 configuration is not shown. All rearrangements within MTS1bcrβ suppress the possibility to express p16INK4a (encoded by exon 1α, exon 2, and exon 3) and p19ARF (encoded by exon 1β, exon 2, and exon 3). In rearrangements within MTS1bcrα, MTS1 exon 1β is deleted and p16INK4a encoding exons remain unchanged. Transcripts initiated from the promoter located 5′ to MTS1 exon 1α and the p16INK4a protein are expressed (data not shown). These data suggest that p19ARF and/or p15INK4b and not p16INK4a could be the functional target(s) of the rearrangements in these cases (Gordie et al, submitted for publication).
Restriction map of the MTS locus and schematic representation of its configuration in T-ALL samples. (A) An updated MTS locus map is shown. The probes used and the 2 breakpoint clusters MTS1bcrα and MTS1bcrβ are indicated. C5.3, 2.7, and 2.3, sequence tagged sites (STSs); E, exon; *, polymorphic BamHI site; +, the 16 breakpoints include that of the T84 sample, for which the MTS1 configuration was not fully characterized. (B) Schematic representation of the 2 chromosome 9s in cases with rearrangements occurring in the locus (22 T-ALL samples and the MOLT4 cell line).21 In 1 sample, T106, 2 breakpoints occurring on each chromosome 9 were detected. In T39, breakpoints were different at presentation (T39p) and relapse (T39r). Dashed lines indicate deletions; vertical arrows indicate breakpoint-containing regions. The T84 configuration is not shown. All rearrangements within MTS1bcrβ suppress the possibility to express p16INK4a (encoded by exon 1α, exon 2, and exon 3) and p19ARF (encoded by exon 1β, exon 2, and exon 3). In rearrangements within MTS1bcrα, MTS1 exon 1β is deleted and p16INK4a encoding exons remain unchanged. Transcripts initiated from the promoter located 5′ to MTS1 exon 1α and the p16INK4a protein are expressed (data not shown). These data suggest that p19ARF and/or p15INK4b and not p16INK4a could be the functional target(s) of the rearrangements in these cases (Gordie et al, submitted for publication).
Recently, our group and subsequent groups showed that inactivation by chromosomal rearrangements of MTS1 (and to a lesser degree, MTS2 ) occurs in the majority of T-cell acute lymphoblastic leukemias (T-ALLs), suggesting an important role in the development of this disease.8-11 We also showed that MTS1 inactivation essentially involved chromosomal deletions, with breakpoints located in the MTS1 locus in approximately one third of the cases.12
To shed light on the mechanism of these rearrangements in T-ALL, we analyzed 15 representative breakpoints by cloning, sequencing, and/or polymerase chain reaction (PCR) mapping. We demonstrate that most breakpoints are tightly clustered, and show evidence that these rearrangements involve an illegitimate V(D)J recombinase activity.
These results show for the first time that a tumor suppressor gene can be inactivated by the V(D)J recombinational mechanism.
MATERIALS AND METHODS
Cells. Bone marrow or peripheral blood samples were obtained from 79 patients with T-ALL. Mononuclear cells obtained from bone marrow or peripheral blood samples were isolated by Ficoll-Hypaque (Eurobio, Les Ulis, France) centrifugation and viably frozen in liquid nitrogen until use. The diagnosis was established according to standard French-American-British criteria.13 Tumor cell phenotype was determined by flow cytometric procedures as previously described.14 A high percentage (usually >95%) of tumor cells were present in all samples. Each patient was identified by a unique number used in all reports from our group. Preliminary Southern blot data from 59 cases have been published previously.12 The T-ALL–derived MOLT4 cell line was also studied.
Southern blot analysis of MTS1 rearrangements.MTS1 and MTS2 configurations were analyzed by Southern blotting15 using a set of probes and various digests as described previously.12 Additional probes were also used. MTS1 5′ exon 1α and MTS1 exon 1β probes were obtained by PCR using OL13/OL17 and OL2/OL4 oligonucleotides, respectively.
Data were analyzed according to a restriction map12 that was updated and corrected with respect to the localization of MTS1 exon 1β. This exon was localized by Southern blot analyses of a P1 phage (P1 2264; Genome Systems, St Louis, MO) containing the MTS1 and MTS2 genes and of an 18-kb subcloned HindIII-HindIII fragment of this phage containing the centromeric part of 2.3, MTS1 exon 1β, 2.7, and MTS2 exon 2.
PCR analysis of chromosomal breakpoints. Standard methods were used.15 Dilutions of DNA extracted from the peripheral blood lymphocytes of a healthy donor in DNA extracted from the pre-B cell line REH, in which both MTS1 and MTS2 genes are deleted, were included in each experiment.
Inverse PCR and breakpoint cloning strategy. To perform the inverse PCR,16 genomic DNA was digested by the relevant restriction endonuclease (described later), and size-selected restriction fragments were ligated at low concentration using T4 DNA ligase (New England Biolabs Ltd, Hertfordshire, UK). DNA circles were PCR-amplified using divergent primers recognizing sequences located on the known part of the rearranged fragment. PCR products were cloned in a PCR Script (SK+ ) vector (Stratagene, La Jolla, CA) and sequenced. To verify that chromosomal rearrangements had been cloned, direct PCR amplification of junctional sequences was performed on genomic DNA using sequence information obtained by the inverse PCR procedure. Moreover, in the case of MTS1bcrβ, Southern blot experiments performed with probes recognizing sequences located on each side of the breakpoints and with relevant restriction enzymes were compared, showing identical data (experiments not shown).
The T117 breakpoint was cloned by inverse PCR using restriction endonuclease Mbo I and primers OL18 and OL16. Cloning of MOLT4, T42, and T123 breakpoints was performed by inverse PCR after BglII digestion using primers OL5 and OL3. The germline sequence containing the sequence fused to MTS1bcrβ in T42 and T123 was characterized by subcloning and sequencing after PCR screening of a P1 library (Genome Systems, St Louis, MO) with OL19 and OL21 oligonucleotides.
Southern blot analysis of PCR fragments. PCR products were electrophoresed in a 2% agarose, 2% NuSieve (FMC BioProducts, Rockland, ME) gel, transferred to a hybond N+ (Amersham International, Amersham, UK) membrane, and hybridized with 32P-radiolabeled oligonucleotides.
Nucleotide sequence analysis. Cloned PCR fragments were sequenced using the T7 Sequencing Kit (Pharmacia, Orsay, France). The oligonucleotide sequences are as follows: OL1, 5′GAAAGACACATCCAAGAGAA3′; OL2, 5′GGGGTGGGGGTGAAGGTG3′; OL3, 5′CGCCCCCTGCCCATCTCC3′; OL4, 5′CGATTGAGGGCTGTGTGAAG3′; OL13, 5′ATTCAAGAGCTAACAGGTATTAG3′; OL16, 5′GGGAGGGAGTCATTGGAAGG3′; OL18, 5′GCCCAACGCACCGAATAGT3′; OL19, 5′AAGAGAGTGAAAGGAAGAACA3′; OL20, 5′GTGCCAGTGTTCTTCCTTTCA3′; OL21, 5′GTTTGATTCTTGGGATATATTT3′; OL22, 5′GTGAACTTTTGGGGGTGTTATT3′; AJT39p, 5′GAGCCACCATCCCTGAGCA3′; AJT117, 5′GTCTCTACGCGGTTGCTTCT3′; AJT127, 5′GTCTCCCAGGGACCTGCTT3′; AJMOLT4, 5′ACACTCTTGACAATAACACC3′; AJT42, 5′TTCCTGCCACTTAGTGCCTG3′; and AJT123, 5′CGGTCGCGGCCTTGGGG3′. Sequences of the other oligonucleotides are shown in the figures.
RESULTS AND DISCUSSION
Clustering of MTS1 breakpoints in T-ALL. Eighty T-ALL samples and the MOLT4 cell line were studied by Southern blot analysis using a set of MTS locus probes12 and EcoRI, HindIII, and, in selected cases, BglII digests (Fig 1). No MTS locus alteration was found in 11% (nine of 80) of the samples. Biallelic and monoallelic deletions of both MTS1 and MTS2 genes were found in 58% (46 of 80) and 4% (three of 80) of the cases, respectively, extending our previous results.12 Twenty-five breakpoints were detected corresponding to 22 T-ALL cases and the MOLT4 cell line.
Apart from five breakpoints scattered over a 42-kb region extending from MTS2 exon 2 to MTS1 exon 3, the other 20 breakpoints were clustered in two regions (Fig 1B). Sixteen of them (64%) are between the 2.7 and 2.3 sequence-tagged sites, thus confirming12 the existence of a major cluster (MTS1bcrβ ). The four remaining breakpoints were located in a 1.1-kb EcoRI-HindIII fragment situated 5′ to the MTS1 exon 1α, thus defining a new minor cluster (MTS1bcrα).
A recurrent rearrangement due to illegitimate V(D)J recombinase activity and juxtaposing 5′ MTS2 exon 1 and 5′ MTS1 exon 1α sequences. To localize the breakpoints within the minor cluster, the three cases with available material (T39p, T117, and T127) were further analyzed by Southern blotting using a 5′ MTS1 exon 1α probe (Fig 2A and B). These experiments suggested that a common rearrangement had occurred, and localized the T117 and T127 breakpoints to a Xba I-Mbo I fragment of 20 nucleotides and T39p breakpoint to a EcoRI-Mbo I fragment of 123 nucleotides (Fig 2A). The T39p breakpoint was more precisely localized by PCR (Fig 2C) to a 50-bp fragment that includes the three breakpoints. Because the sequence of this fragment (Fig 2D) contains a CACTGTG heptamer identical to that found in recombination recognition sequences targeting Ig and T-cell receptor (TCR) gene rearrangements,17 18 we hypothesized that illegitimate V(D)J recombinase activity could be involved in these rearrangements.
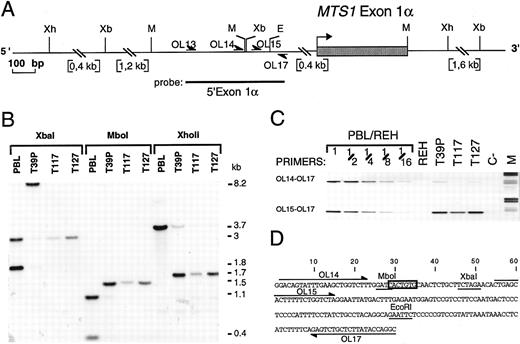
Characterization of the MTS1bcrα breakpoint cluster. (A) Restriction map of the MTS1bcrα region. Only relevant restriction sites are shown. Xb, Xba I; Xh, XhoII; M, Mbo I; E, EcoRI; OL, oligonucleotide. (B) Southern blot analysis of T39p, T117, and T127 rearrangements using the 5′ exon1α probe and Mbo I, XhoII, and Xba I digests. The T39p sample contains approximately 15% nontumoral cells. (C) Reverse view of ethidium bromide–stained gel showing PCR experiments using primers OL17 v OL14 or OL15. Dilutions of DNA from peripheral blood lymphocytes (PBL) from a healthy donor in DNA from the pre-B cell line REH, in which both MTS1 and MTS2 genes are deleted, were included. M, molecular weight marker V (Boehringer Mannheim, Meylan, France); C-, negative control (no DNA). (D) Nucleotide sequence of the MTS1bcrα breakpoint cluster. Mbo I, Xba I, and EcoRI restriction sites are underlined, and the candidate heptamer targeted by the recombinase activity is boxed. Positions of oligonucleotides used to localize the breakpoints are shown.
Characterization of the MTS1bcrα breakpoint cluster. (A) Restriction map of the MTS1bcrα region. Only relevant restriction sites are shown. Xb, Xba I; Xh, XhoII; M, Mbo I; E, EcoRI; OL, oligonucleotide. (B) Southern blot analysis of T39p, T117, and T127 rearrangements using the 5′ exon1α probe and Mbo I, XhoII, and Xba I digests. The T39p sample contains approximately 15% nontumoral cells. (C) Reverse view of ethidium bromide–stained gel showing PCR experiments using primers OL17 v OL14 or OL15. Dilutions of DNA from peripheral blood lymphocytes (PBL) from a healthy donor in DNA from the pre-B cell line REH, in which both MTS1 and MTS2 genes are deleted, were included. M, molecular weight marker V (Boehringer Mannheim, Meylan, France); C-, negative control (no DNA). (D) Nucleotide sequence of the MTS1bcrα breakpoint cluster. Mbo I, Xba I, and EcoRI restriction sites are underlined, and the candidate heptamer targeted by the recombinase activity is boxed. Positions of oligonucleotides used to localize the breakpoints are shown.
The T117 MTS1bcrα breakpoint was first cloned by inverse PCR, and germline chromosome 9 sequences were identified by a search of data banks (Burian DM, Mitchell N, Roe BA, unpublished, 1996; Sveen L, Olopade FI, Rowley JD, unpublished, 1996; accession no. AC000049). Primers recognizing sequences located on each side of the breakpoint (OL16 and OL1) were then designed and used to directly amplify T39p, T117, and T127 breakpoints. The three breakpoints were cloned and sequenced (Fig 3).
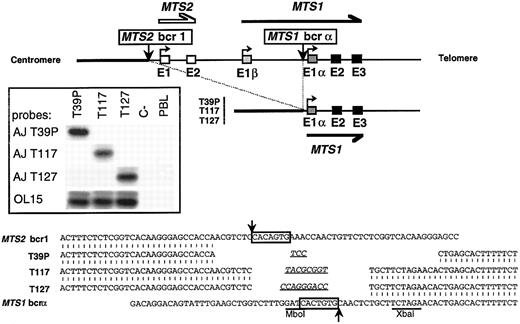
Cloning and sequencing of 3 breakpoints occurring in MTS1bcrα. Top: Schematic representation of the 36-kb deletion that brings sequences 5′ to MTS2 exon 1 (MTS2bcr1 ) upstream to MTS1 exon 1α. (▪) MTS1 exons; (□) MTS2 exons. Vertical arrows represent breakpoints. Insert: Hybridization with junction-specific oligonucleotides after direct PCR amplification of T39p, T117, and T127 breakpoints with OL16 and OL1. PCR products were analyzed in a 2% agarose, 2% NuSieve gel, transferred on a hybond N+ membrane, and successively hybridized with 32P-radiolabeled oligonucleotide specific for junctional sequences of T39p (AJ T39p), T117 (AJ T117), and T127 (AJ T127) and with a common internal oligoprobe, OL15. C-, negative control (no DNA). Bottom: Nucleotide sequence alignment of T39p, T117, and T127 rearrangements with germline chromosome 9 sequences. Canonic heptamers are boxed, and putative N regions are underlined. No consensus nonamer was found. However, highly degenerated nonamers may be present at 23 bp from the MTS2bcr1 heptamer and at 12 bp from the MTS1bcrα heptamer. Localization of breakpoints on germline sequences is indicated by arrows.
Cloning and sequencing of 3 breakpoints occurring in MTS1bcrα. Top: Schematic representation of the 36-kb deletion that brings sequences 5′ to MTS2 exon 1 (MTS2bcr1 ) upstream to MTS1 exon 1α. (▪) MTS1 exons; (□) MTS2 exons. Vertical arrows represent breakpoints. Insert: Hybridization with junction-specific oligonucleotides after direct PCR amplification of T39p, T117, and T127 breakpoints with OL16 and OL1. PCR products were analyzed in a 2% agarose, 2% NuSieve gel, transferred on a hybond N+ membrane, and successively hybridized with 32P-radiolabeled oligonucleotide specific for junctional sequences of T39p (AJ T39p), T117 (AJ T117), and T127 (AJ T127) and with a common internal oligoprobe, OL15. C-, negative control (no DNA). Bottom: Nucleotide sequence alignment of T39p, T117, and T127 rearrangements with germline chromosome 9 sequences. Canonic heptamers are boxed, and putative N regions are underlined. No consensus nonamer was found. However, highly degenerated nonamers may be present at 23 bp from the MTS2bcr1 heptamer and at 12 bp from the MTS1bcrα heptamer. Localization of breakpoints on germline sequences is indicated by arrows.
The sequences located downstream of the breakpoints were identical. A 36-kb interstitial deletion had occurred, juxtaposing sequences located 0.4 kb upstream of MTS2 exon 1 with sequences located 0.7 kb upstream of MTS1 exon 1α (Fig 3, top). On both sides of the deletions, the tightly clustered breakpoints occurred in the vicinity of heptamers with sequences similar to those involved in the V(D)J recombination and that had been deleted by the recombinations. Short deletions and GC-rich N regions were found in all cases (Fig 3, bottom). Clone-specific junctional sequences were generated and shown to be specific by hybridization experiments performed after direct PCR on genomic DNA (Fig 3, insert). All of these data are the hallmark of V(D)J recombinase activity.17 18
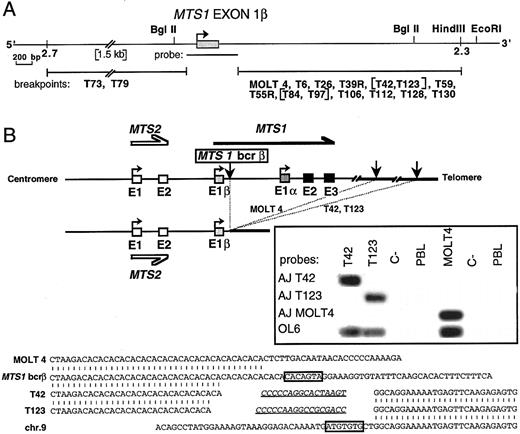
Molecular characterization of rearrangements occurring in MTS1bcrβ. We then investigated if a similar mechanism could be involved in rearrangements occurring in MTS1bcrβ by refining the mapping of breakpoints by Southern blot analysis (not shown) for the 16 cases, using a MTS1 exon 1β probe and various digests. In two cases (T73 and T79), no signal was observed, thus localizing the breakpoints 5′ to MTS1 exon 1β (Fig 4A). In the other 14 cases, rearranged fragments were observed, showing that the corresponding breakpoints were within a 3-kb region located 3′ to MTS1 exon 1β (Fig 4A).
Characterization of the MTS1bcrβ cluster and cloning of 3 representative rearrangements. (A) Partial map of the DNA region flanked by STS2.3 and STS2.7. Localization of the 2 breakpoints occurring 5′ to MTS1 exon 1β (T73 and T79) and of the 14 breakpoints occurring 3′ to exon 1β is shown. Breakpoints were localized by Southern blot experiments using the MTS1 exon 1β probe and BglII digests, and in certain cases, Sal I, BamHI, EcoRI, and HindIII digests. Bracketed cases show rearranged fragments of identical length in various digests. (B) Top: Schematic representation of MOLT4, T42, and T123 rearrangements. Vertical arrows represent breakpoints. Insert: Hybridization with junction-specific oligonucleotides after direct PCR amplification of the MOLT4 breakpoint using primers OL5 and OL22 and the T42 and T123 breakpoints using primers OL5 and OL20. PCR products were successively hybridized with 32P-radiolabeled oligonucleotides specific for junctional sequences of MOLT4 (AJ MOLT4), T42 (AJ T42), and T123 (AJ T123) and with a common internal oligoprobe, OL6. Bottom: Nucleotide sequence alignment of MOLT4, T42, and T123 rearrangements with chromosome 9 germline sequences. Heptamers are boxed, and putative N regions are underlined. No definite nonamer was found.
Characterization of the MTS1bcrβ cluster and cloning of 3 representative rearrangements. (A) Partial map of the DNA region flanked by STS2.3 and STS2.7. Localization of the 2 breakpoints occurring 5′ to MTS1 exon 1β (T73 and T79) and of the 14 breakpoints occurring 3′ to exon 1β is shown. Breakpoints were localized by Southern blot experiments using the MTS1 exon 1β probe and BglII digests, and in certain cases, Sal I, BamHI, EcoRI, and HindIII digests. Bracketed cases show rearranged fragments of identical length in various digests. (B) Top: Schematic representation of MOLT4, T42, and T123 rearrangements. Vertical arrows represent breakpoints. Insert: Hybridization with junction-specific oligonucleotides after direct PCR amplification of the MOLT4 breakpoint using primers OL5 and OL22 and the T42 and T123 breakpoints using primers OL5 and OL20. PCR products were successively hybridized with 32P-radiolabeled oligonucleotides specific for junctional sequences of MOLT4 (AJ MOLT4), T42 (AJ T42), and T123 (AJ T123) and with a common internal oligoprobe, OL6. Bottom: Nucleotide sequence alignment of MOLT4, T42, and T123 rearrangements with chromosome 9 germline sequences. Heptamers are boxed, and putative N regions are underlined. No definite nonamer was found.
Three breakpoints located in this region (MOLT4, T42, and T123) were then cloned and sequenced (Fig 4B). Germline sequences corresponding to the identical sequences rearranged to the MTS1bcrβ in T42 and T123 were characterized after PCR screening of a P1 library. Southern blot experiments on somatic hamster-human hybrids demonstrated that the translocated sequences in T42, T123, and MOLT4 DNA were located on chromosome 9, and deletion analysis indicated that the sequence translocated to the MTS1 locus in MOLT4 was centromeric to the sequences translocated in the T42 and T123 cases (data not shown). Analysis of the nucleotide sequences (Fig 4B, bottom) shows the hallmark of V(D)J recombinase activity. Clone-specific junctional sequences had been generated by the recombinase machinery (Fig 4B, insert).
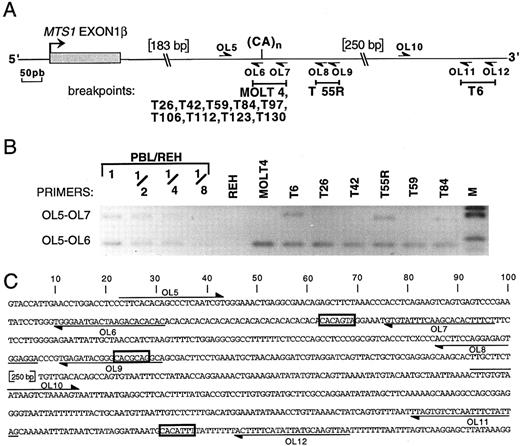
To analyze whether other rearrangements occurring in the corresponding part of MTS1bcrβ (ie, 3′ to MTS1 exon 1β) are generated by the same mechanism, their breakpoints were defined by PCR (Fig 5). In addition to T42, T123, and MOLT4, seven other rearrangements were located in a region of approximately 70 nucleotides that includes a polymorphic (CA)n repeat19 and the CACAGTA heptamer (nucleotides identical to those of the consensus V(D)J recognition signal sequence are underlined). The (CA)n repeat contains CACACAC heptamers that are also potential targets for the recombinase since, as for the CACAGTA heptamer, a CA dinucleotide is present in the sequence adjacent to the heptamer, theoretically contributing to the efficacy of the recombinase process.20 21 Breakpoints of two other cases (T55r and T6) were identified downstream of the (CA) repeat near heptamer-like sequences, CACGCAG and CACATTT, respectively (Fig 5).
Localization of the breakpoints occurring 3′ to MTS1 exon 1β by PCR. (A) Partial map of the region located 3′ to MTS1 exon 1β. The polymorphic dinucleotide CA repeat and the primers OL5 to OL12 used to localize the breakpoints are indicated. Breakpoint-containing regions determined by PCR are represented above by solid bars. (B) Reverse view of an ethidium bromide–stained gel showing representative PCR experiments using primers OL5 v OL7 or OL6. Dilutions of DNA from PBL from a healthy donor in DNA from pre-B cell line REH were included in the experiment. M, molecular weight marker V. One undeleted allele is present in case T84. T128 and T39R could not be studied by PCR, because of the presence of nontumoral cells in the T128 sample and the absence of available material for T39R. (C) Nucleotide sequence of the region located immediately 3′ to MTS1 exon 1β. The position of the relevant oligonucleotides is shown, and candidate heptamers are boxed.
Localization of the breakpoints occurring 3′ to MTS1 exon 1β by PCR. (A) Partial map of the region located 3′ to MTS1 exon 1β. The polymorphic dinucleotide CA repeat and the primers OL5 to OL12 used to localize the breakpoints are indicated. Breakpoint-containing regions determined by PCR are represented above by solid bars. (B) Reverse view of an ethidium bromide–stained gel showing representative PCR experiments using primers OL5 v OL7 or OL6. Dilutions of DNA from PBL from a healthy donor in DNA from pre-B cell line REH were included in the experiment. M, molecular weight marker V. One undeleted allele is present in case T84. T128 and T39R could not be studied by PCR, because of the presence of nontumoral cells in the T128 sample and the absence of available material for T39R. (C) Nucleotide sequence of the region located immediately 3′ to MTS1 exon 1β. The position of the relevant oligonucleotides is shown, and candidate heptamers are boxed.
All of these data suggest that the V(D)J recombinational mechanism is involved in rearrangements occurring in MTS1bcrβ.
Breakpoint clustering at a (CA) polymorphic repeat. Breakpoints of 10 rearrangements, all leading to inactivation of both p16INK4a and p19ARF encoding sequences, are thus clustered at a (CA) repeat, suggesting that this motif may play a role in the clustering. In certain conditions, (CA) repeats adopt a Z-DNA structure and bind nuclear nonhistone high-mobility group proteins 1 and 2.22 Moreover, it has been recently shown by in vitro experiments that these DNA-binding proteins stimulate V(D)J cleavage.23 Purine-pyrimidine tracts (potential Z-DNA) have been described near (but not at) certain translocation breakpoints that occur in T-ALL cases, suggesting a role in targeting the recombination.24 In the case of MTS1 rearrangements, the repeat is precisely located at the breakpoint and may improve the efficacy of V(D)J recombinase–mediated recombination. During the physiologic V(D)J process,25-28 a nick is introduced at the exact border between the recombination signal sequence heptamer and the coding sequence, followed by creation of a hairpin at the coding end and of a blunt signal end. Ligation of coding ends occurs after opening of the hairpin. The particular structure of the CACA portion of the heptamer29 may facilitate the initial cleavage reaction,20 21 especially in cases where the adjacent sequence adopts a similar structure, as is the case for the (CA) repeat.
MTS1, a tumor suppressor gene inactivated by “illegitimate” V(D)J recombinase activity in immature T-cell proliferation. Illegitimate V(D)J recombinase activity has been suggested to be involved in the mechanism of recurrent translocations involving TCR genes as one of the two partners, found in rare T-ALL cases.17,18 Although a V(D)J recombinase–mediated process is clearly involved in the TCR gene breakpoint as an initial or secondary event, whether the same process is used for the non–TCR gene breakpoint remains an open question.18 To date, the best demonstration involving putative or proven oncogenes has been provided by the study of SIL-TAL1 deletions, which activate the proto-oncogene TAL1 in approximately 20% of T-ALL cases.30,31MTS1 disruption is therefore the second example of a V(D)J recombinase–targeted event involving two non-TCR partners in T-ALL, and the first example of tumor suppressor gene inactivation by such a process. The V(D)J process, indispensable to the generation of clone-specific T-cell antigen receptors, therefore has the drawback of being mutagenic and probably plays a crucial role in the development of this aggressive form of human leukemia.
ACKNOWLEDGMENT
We thank S. Genyk for technical assistance and M.H. Stern, J.C. Bories, C. Gazin, and E. Macintyre for very helpful comments. We also thank D.M. Willerford for a critical reading of the manuscript and all G.B.B.M. members for discussion.
Supported by grants from the Fondation Saint Louis, Délégation à la Recherche Clinique AP-HP, and Ligue Nationale contre le Cancer (Comité de Paris).
Address reprint requests to François Sigaux, MD, Laboratory of Molecular Hematology, Centre Hayem, Hôpital Saint-Louis, 1 av Claude Vellefaux, 75475, Paris, Cedex 10, France.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal