Abstract
Monoclonal gammopathy of undetermined significance (MGUS) is a frequent condition in patients over 50 years old, that ultimately leads to multiple myeloma (MM) in 20% of patients after 20 to 35 years of follow-up. Little is known about cytogenetic changes associated with this condition. We studied 19 MGUS patients both at diagnosis and after 12 to 35 months of follow-up (mean = 26), using DNA content measurement of bone marrow plasma cells (BMPC), and a new interphase fluorescence in situ hybridization technique (FISH) allowing the simultaneous identification of monotypic BMPC (fluorescent anti light-chain antibodies) and the determination of the number of copies for two different chromosomes within the same PC nucleus (one biotin-labeled probe coupled next to texas red avidin and one FITC-labeled probe). At diagnosis of the MGUS, single interphase FISH showed at least one numeric chromosome change in 13 of 19 patients, after the use of centromeric probes directed against chromosomes no. 3, no. 7, no. 9, and no. 11. At follow-up, abnormalities found at diagnosis in 13 patients were still shown. Moreover, abnormalities occurred in three of the last six patients (trisomy for one to three different chromosomes), although no patient evolved into MM. Dual interphase FISH showed that some BMPC bore numeric changes with both probes tested whereas other BMPC bore abnormality with only one of the probes tested. In patients who showed trisomy for at least three different chromosomes, distribution of numeric changes within BMPC defined significant numbers of up to seven different BMPC clones. All these various clones were shown both at diagnosis and at follow-up. In every patient, these various clones differed only for the number of abnormalities they exhibited, and could be related to each other in a model of gradual acquisition of chromosome changes. Eventually, data reported here show that MGUS patients acquire slowly, gradually, but ineluctably chromosome changes, distributed within several related subclones. However, these changes are not related to transformation into MM: among the various clones coexisting within the same patient, a peculiar change, still to demonstrate, might develop and lead to overt MM.
A MONOCLONAL component (M-component) in serum may be related in some instances to multiple myeloma (MM) in an aggressive stage (stage III according to Durie and Salmon staging system, or plasma cell leukemia) or not (stage I MM, or smoldering MM). However, in most instances, corresponding to 1% of the population over 50 years old and 3% of the population over 70 years old,1 this M-component is related to a quite asymptomatic condition, designated as monoclonal gammopathy of undetermined significance (MGUS). After 20 to 35 years of follow-up, 20 to 30% of MGUS patients will develop overt MM, but no single laboratory test exists so far that can predict changes towards a malignant evolution.1 2
The origin of MM is largely unknown. As for other malignant tumors,3-5 several genetic changes are certainly required to generate monoclonal malignant plasma cells.6 The detection in some MGUS patients of molecular alterations of interleukin-6 receptor (IL-6R), lck and c-myc genes has been shown to precede rapid progression into aggressive MM,7 raising again MGUS as an intermediate condition between normality and malignant myelomatous disease. In MM patients molecular studies have shown that the tumor cell has undergone extensive somatic hypermutation, and homogeneity in the variable region of the heavy gene sequence indicates that this cell might be considered as postfollicular.8 In MGUS patients study of the VH gene sequences in the monoclonal cells showed that in some patients there was no intraclonal variation as in MM, whereas in some other patients tested there was a heterogeneity of clonal sequences. In these latter cases, careful analysis of the sequences in individual patients suggested a clonal history closer to follicular lymphoma than to MM.9
Conventional cytogenetic study shows abnormalities in up to 50% of MM patients, and in nearly all patients when other techniques as DNA content measurement and interphase FISH techniques are used.10 Careful analysis of cytogenetic data shows that no consistent and/or specific cytogenetic abnormality exists in MM,10 in opposition to many leukemias in which one cytogenetic event (for example a specific translocation) may be related to the pathogenesis of the disease. So far, no clear-cut cytogenetic study can be reliably obtained in MGUS using conventional methods, but fluorescence in situ hybridization techniques (FISH) identify numeric chromosome changes within bone marrow (BM) plasma cells (BMPC) in at least 50% of MGUS patients, and chromosomes involved are the same as those found in abnormal numbers in MM patients.11 12
Using DNA content measurement and interphase FISH techniques, our aim in this study was to ascertain more accurately the distribution of the cytogenetic abnormalities associated with MGUS in a large series of patients, both at diagnosis and at follow-up. Some patients had no conspicuous change at diagnosis, whereas others exhibited numeric chromosome changes. Surprisingly these changes were not restricted to one single clone of BMPC, but distributed within several clones bearing either one part or all cytogenetic abnormalities. Also, some of the patients who had no chromosome change at diagnosis acquired a cytogenetic abnormality at follow-up. In each patient, according to an hypothesis of gradual acquisition of chromosome abnormalities, a relationship between the various cytogenetic subclones could be drawn.
DNA Index of BMPC and Results of Single Interphase FISH Study in Patients, at Diagnosis [a] and at Follow-Up [b]
| Pt No. . | Next Sample After (mo) . | Ig Peak . | BMPC (%) . | DNA Index of BMPC . | Single Interphase FISH Two (=D), Three (=T), or Four (=TT) Signals Within BMPC (% affected cells) . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | Chr No. 3 . | Chr No. 7 . | Chr No. 9 . | Chr No. 11 . |
| 1.a | GK | 4 | 0.98 | D | D | D | D | |
| b | 12 | 1 | 1.00 | D | D | D | D | |
| 2.a | GL | 2 | 0.99 | D | D | D | D | |
| b | 23 | 3 | 1.00 | D | D | D | D | |
| 3.a | GK | 6 | 1.02 | D | D | D | D | |
| b | 27 | 3 | 0.99 | D | D | D | D | |
| 4.a | GL | 7 | 0.99 | D | D | D | D | |
| b | 26 | 6 | 0.98 | D | D | D | T (22%) | |
| 5.a | GK + GL | 1 | 1.01 | D | D | D | D | |
| b | 27 | 2 | 0.99 | D | D | T (15%) | D | |
| 6.a | GK | 4 | 0.98 | D | D | D | T (51%) | |
| b | 16 | 1 | 0.96 | D | D | D | T (48%) | |
| 7.a | AL | 2 | 1.01 | T (25%) | T (24%) | D | D | |
| b | 35 | 2 | 1.07 | T (45%) | T (53%) | D | D | |
| 8.a | GK | 3 | 0.97 | T (10%) | T (11%) | T (12%) | D | |
| b | 12 | 2 | 1.02 | T (10%) | T (9%) | T (12%) | D | |
| 9.a | GL | 3 | 1.11 | D | D | T (50%) | T (72%) | |
| b | 26 | 3 | 1.11 | nd | nd | T (62%) | T (73%) | |
| 10.a | GK | 3 | 0.99 | D | D | D | D | |
| b | 30 | 2 | 1.05 | T (14%) | D | T (22%) | T (26%) | |
| 11.a | GK | 4 | 1.17 | T (78%) | D | T (52%) | T (62%) | |
| b | 12 | 5 | 1.15 | T (74%) | D | T (45%) | T (50%) | |
| 12.a | GK | 2 | 1.12 | T (70%) | T (56%) | T (39%) | D | |
| b | 24 | 2 | 1.12 | T (63%) | T (62%) | T (40%) | D | |
| 13.a | AL | 5 | 0.99 | T (17%) | T (15%) | D | D | |
| b | 27 | 8 | 0.98 | T (56%) | T (44%) | T (44%) | T (39%) | |
| 14.a | GK | 1 | 0.99 | D | D | T (17%) | T (25%) | |
| b | 35 | 1 | 1.04 | T (19%) | D | T (21%) | D | |
| 15.a | GK | 5 | 1.07 | T (76%) | T (63%) | T (62%) | D | |
| b | 24 | 1 | 1.13 | T (72%) | T (66%) | T (51%) | D | |
| 16.a | GK | 3 | 1.13 | T (61%) | T (44%) | T (43%) | T (55%) | |
| b | 23 | 1 | 1.12 | T (51%) | T (43%) | T (38%) | T (43%) | |
| 17.a | GK | 1 | 1.13 | T (51%) | T (33%) | T (42%) | T (54%) | |
| b | 32 | 4 | 1.12 | T (65%) | T (53%) | T (57%) | T (61%) | |
| 18. | GK | 5 | 1.13 | T (58%) | T (61%) | D | T (67%) | |
| 19.* | GL | 4 | 1.45 | T + TT (75%) | T + TT (83%) | T + TT (62%) | T + TT (81%) | |
| Pt No. . | Next Sample After (mo) . | Ig Peak . | BMPC (%) . | DNA Index of BMPC . | Single Interphase FISH Two (=D), Three (=T), or Four (=TT) Signals Within BMPC (% affected cells) . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | Chr No. 3 . | Chr No. 7 . | Chr No. 9 . | Chr No. 11 . |
| 1.a | GK | 4 | 0.98 | D | D | D | D | |
| b | 12 | 1 | 1.00 | D | D | D | D | |
| 2.a | GL | 2 | 0.99 | D | D | D | D | |
| b | 23 | 3 | 1.00 | D | D | D | D | |
| 3.a | GK | 6 | 1.02 | D | D | D | D | |
| b | 27 | 3 | 0.99 | D | D | D | D | |
| 4.a | GL | 7 | 0.99 | D | D | D | D | |
| b | 26 | 6 | 0.98 | D | D | D | T (22%) | |
| 5.a | GK + GL | 1 | 1.01 | D | D | D | D | |
| b | 27 | 2 | 0.99 | D | D | T (15%) | D | |
| 6.a | GK | 4 | 0.98 | D | D | D | T (51%) | |
| b | 16 | 1 | 0.96 | D | D | D | T (48%) | |
| 7.a | AL | 2 | 1.01 | T (25%) | T (24%) | D | D | |
| b | 35 | 2 | 1.07 | T (45%) | T (53%) | D | D | |
| 8.a | GK | 3 | 0.97 | T (10%) | T (11%) | T (12%) | D | |
| b | 12 | 2 | 1.02 | T (10%) | T (9%) | T (12%) | D | |
| 9.a | GL | 3 | 1.11 | D | D | T (50%) | T (72%) | |
| b | 26 | 3 | 1.11 | nd | nd | T (62%) | T (73%) | |
| 10.a | GK | 3 | 0.99 | D | D | D | D | |
| b | 30 | 2 | 1.05 | T (14%) | D | T (22%) | T (26%) | |
| 11.a | GK | 4 | 1.17 | T (78%) | D | T (52%) | T (62%) | |
| b | 12 | 5 | 1.15 | T (74%) | D | T (45%) | T (50%) | |
| 12.a | GK | 2 | 1.12 | T (70%) | T (56%) | T (39%) | D | |
| b | 24 | 2 | 1.12 | T (63%) | T (62%) | T (40%) | D | |
| 13.a | AL | 5 | 0.99 | T (17%) | T (15%) | D | D | |
| b | 27 | 8 | 0.98 | T (56%) | T (44%) | T (44%) | T (39%) | |
| 14.a | GK | 1 | 0.99 | D | D | T (17%) | T (25%) | |
| b | 35 | 1 | 1.04 | T (19%) | D | T (21%) | D | |
| 15.a | GK | 5 | 1.07 | T (76%) | T (63%) | T (62%) | D | |
| b | 24 | 1 | 1.13 | T (72%) | T (66%) | T (51%) | D | |
| 16.a | GK | 3 | 1.13 | T (61%) | T (44%) | T (43%) | T (55%) | |
| b | 23 | 1 | 1.12 | T (51%) | T (43%) | T (38%) | T (43%) | |
| 17.a | GK | 1 | 1.13 | T (51%) | T (33%) | T (42%) | T (54%) | |
| b | 32 | 4 | 1.12 | T (65%) | T (53%) | T (57%) | T (61%) | |
| 18. | GK | 5 | 1.13 | T (58%) | T (61%) | D | T (67%) | |
| 19.* | GL | 4 | 1.45 | T + TT (75%) | T + TT (83%) | T + TT (62%) | T + TT (81%) | |
Abbreviation: nd, not done.
For patient no. 19, 10% to 25% BMPC exhibited 4 signals (=TT) after hybridization, and results within brackets include BMPC with 3 + 4 signals.
PATIENTS AND METHODS
Samples from each of the 19 patients from this report were collected in two instances: first at discovery of the M-component, and next after 12 to 35 months (mean = 24) of follow-up. Diagnosis of the MGUS was performed in each patient using criteria previously described.1 Age at diagnosis ranged from 37 to 81 years (mean = 65). Monoclonal peak was Gκ in 12 instances, Gλ in 4 instances, Aλ in 2 instances, and Gκ + Gλ in one instance. In patients with either Gκ or Gλ peak, total amount of IgG ranged from 13.6 to 36.4 g/L (mean = 21.7 g/L) and was 8.8 and 13.0 g/L for the two patients with IgA peak, respectively. Patients no. 1 and no. 16 had IgG more than 30 g/L: levels were 29.8 g/L and 33.1 g/L after 12 and 23 months, respectively, and as clinical and other biological data did not change, both patients were included in this study. Six patients had reduced levels of one or both classes of uninvolved polyclonal Ig. One patient had slight increase of polyclonal IgA. A small amount of Bence Jones proteinuria was found in one patient (not found at follow-up). BMPC (500 cells counted in each instance) was 1% to 7%, according to the patients (Table 1). The number of BMPC expressing Ki 67 antigen ranged from 0% to 5% (median = 1.3; mean = 1.4). C reactive protein (CRP), serum lactate dehydrogenase (LDH), calcemia, and β2 microglobulin were tested in all patients and were always within normal ranges.
Fluorescent green cytoplasm allows identification of a plasma cell (patient no. 6a) and hybridization of DNA probes shows 2 copies for chr no. 9 (2 red signals; biotin-labeled probe) and 2 copies for chr no. 11 (2 green signals; FITC-labeled probe) [dual band-pass filter]. (B) in the same patient (no. 6a), one plasma cell is disomic for chr no. 9 (2 red signals) and trisomic for chr no. 11 (3 green signals) [dual band-pass filter]. (C) Using a dual band-pass filter, one plasma cell from patient no. 15a exhibits trisomy for chr no. 7 (FITC-labeled probe) and disomy for chr no. 9 (biotin-labeled probe): red signals close to the green ones lead to yellow fluorescence, and the use of single band-pass filters shows 2 red signals (D: texas-red filter) and 3 green ones (E: FITC filter). (F ) and (G) Plasma cells exhibiting trisomy for chr no. 7 (FITC-labeled probe) and trisomy for chr no. 9 (biotin-labeled probe), using the triple band-pass filter (patient no. 17b in F and no. 17a in G). (H) Plasma cell exhibiting 3 signals for chr no. 9 (biotin-labeled probe) and 2 signals for chr no. 7 (FITC-labeled probe) (patient no. 17a).
Fluorescent green cytoplasm allows identification of a plasma cell (patient no. 6a) and hybridization of DNA probes shows 2 copies for chr no. 9 (2 red signals; biotin-labeled probe) and 2 copies for chr no. 11 (2 green signals; FITC-labeled probe) [dual band-pass filter]. (B) in the same patient (no. 6a), one plasma cell is disomic for chr no. 9 (2 red signals) and trisomic for chr no. 11 (3 green signals) [dual band-pass filter]. (C) Using a dual band-pass filter, one plasma cell from patient no. 15a exhibits trisomy for chr no. 7 (FITC-labeled probe) and disomy for chr no. 9 (biotin-labeled probe): red signals close to the green ones lead to yellow fluorescence, and the use of single band-pass filters shows 2 red signals (D: texas-red filter) and 3 green ones (E: FITC filter). (F ) and (G) Plasma cells exhibiting trisomy for chr no. 7 (FITC-labeled probe) and trisomy for chr no. 9 (biotin-labeled probe), using the triple band-pass filter (patient no. 17b in F and no. 17a in G). (H) Plasma cell exhibiting 3 signals for chr no. 9 (biotin-labeled probe) and 2 signals for chr no. 7 (FITC-labeled probe) (patient no. 17a).
Specific osteolytic lesions were absent both at diagnosis and at follow-up, and no patient developed MM after a median follow-up of 26 months. Results found at follow-up showed that the amount of M-component was unchanged in each patient. For patients with IgG peak, total IgG ranged from 12.4 to 33.1 g/L (mean = 20.5 g/L). Decrease of polyclonal Ig was found in the same patients as at diagnosis. Percentages of BMPC were within the same ranges at diagnosis and at follow-up (Table 1), as were calcemia, CRP, LDH, and β2 microglobulin (not reported). Ki 67 index ranged from 0% to 7.5% (median = 0.7; mean = 1.2).
Two of the 19 patients (no. 18 and no. 19) had clinical and biological follow-up in the same conditions as the other 17; however, BM sample was diluted at second examination and specific BM studies were performed on the first sample only.
Controls. BM samples from eight healthy volunteers (BM donors) were studied in the same conditions as those applied for MGUS patients.
BM aspirates were obtained from the posterior iliac crest or the sternum and collected on EDTA as anticoagulant. Mononuclear cells were separated by density gradient centrifugation (density = 1.077; MSL; Eurobio, Les Ulis, France). Cytocentrifuge slides were tested either by the next 2 days or stored at −20°C (without specific treatment).
DNA content of plasma cells. Slides were stained according to the Feulgen procedure, and computerized image analysis (Quantimet 600S; Leica,4 Rueil Malmaison, France) was used to ascertain nuclear staining, as previously described.13 Mean nuclear staining of BMPC was divided by mean nuclear staining of surrounding nonplasmacytic cells, to obtain the DNA index (DI).
Interphase FISH studies. Directly labeled fluorescein isothiocyanate (FITC) and biotin-labeled α-satellite or classical satellite probes for chromosomes (chr) no. 3, no. 7, no. 9, no. 11, and no. 15 were used (D3Z1, D7Z1, D9Z1, D11Z1, D15Z1; Oncor, Gaithersburg, MD). One part of the experiments used biotin-labeled probes (single interphase FISH), and another part used a mixture of one biotin-labeled probe and one directly labeled probe, both specific for different chromosomes (dual interphase FISH): these latter mixtures allowed the simultaneous enumeration of copies for 2 different chromosomes within the same nucleus. In each experiment, the couples of probes used were as follows: chr no. 3 and chr no. 7, chr no. 3 and chr no. 9, chr no. 3 and chr no. 11, chr no. 7 and chr no. 9, chr no. 7 and chr no. 11, and chr no. 9 and chr no. 11. In preliminary experiments, a mixture of FITC-labeled and texas red-labeled probes was used for dual hybridization; however, texas red probes failed to give reproducible strong red spots in some instances and we decided to use biotin-labeled probes according to the following protocol.
Cytocentrifuge slides were fixed in cold acetone (5 minutes), in 4% paraformaldehyde in phosphate-buffered saline (PBS; pH = 7.40), rinsed and allowed to dry for 15 minutes. The step corresponding to immunostaining of BMPC was performed as follows. After rehydratation of slides with PBS, diluted (1/100) rabbit polyclonal anti human light chains (either κ or λ) antibody, directly labeled with FITC (Dako, Glostrup, Denmark), was applied for a brief period (5 minutes); next, slides were rinsed with PBS, air dried, and dehydrated in graded alcohols. The step corresponding to the hybridization of DNA probe(s) was done as follows. Slides were denaturated for 5 minutes at 72°C in 70% formamide solution (in 2× sodium saline citrate); 10 μL of hybridization mixture was applied next (0.5 μL of biotin-labeled probe in 9.5 μL of Hybrisol VI (Oncor, Gaithersburg, MD) for single interphase FISH, and mixture of 1 μL of FITC-labeled probe and 0.5 μL of biotin-labeled probe in 8.5 μL of Hybrisol VI for dual interphase FISH; denaturation for 2 minutes at 72°C), coverslipped and sealed with rubber cement. After 1 night of hybridization at 37°C, amplification of the biotinylated probe was performed using successive layers of texas red avidin (Vector, Burlingame, CA), biotinylated anti avidin antibody (Vector), and texas red avidin once again, as previously described.11
Slides were counterstained with 4, 6-diamidino-phenyl indole (DAPI) and cells analyzed under a fluorescence microscope (DMRD microscope; Leica) equipped with various single, double and triple band-pass filters to visualize either 1, 2, or all 3 fluorescent dyes.
RESULTS
After interphase FISH, cytoplasmic green fluorescence allowed easy recognition of the monotypic BMPC (see Fig 1). Hybridization using biotin-labeled probes led to intranuclear fluorescent red signals. Dual interphase FISH hybridization (biotin-labeled and FITC-labeled probes) led to fluorescent red and fluorescent green signals within the same nucleus: the use of optimally diluted anti light-chain antibodies, as mentioned in Patients and Methods, was a crucial step in this study. Various band-pass filters were preconized at times to assert the number of these fluorescent signals (see Fig 1).
Controls and Values Used for Reference
DI of BMPC ranged from 0.98 to 1.00 in normal controls (n = 8).
Single interphase FISH studies. In each control (n = 8) 64 to 112 PC could be evaluated according to the probes tested.
Using biotinylated probes, the number of PC who exhibited three or one red signal(s) was always less than 5% or 8%, respectively. Using the mean plus two standard deviations, trisomy was ascertained when at least 8% PC exhibited 3 red signals, and monosomy was ascertained over 15% PC exhibiting one red signal. However, in this study we did not find monosomy in any patient with the probes tested.
Using directly labeled probes (FITC) background levels were lower (threshold of 5% to define trisomy), but for an easier interpretation of data, we decided to use the same thresholds for both biotin- and FITC-labeled probes.
On each slide enumeration of 200 cells that did not react with anti-Ig antibodies was performed: less than 5% and 10% of cells with 3 or one signal had to be found, respectively, independently of the nature of the probe tested, to assert optimal hybridization.
Dual interphase FISH. Most nuclei from controls showed 2 red and 2 green signals, and we never observed more than 5% BMPC exhibiting both 3 red and 2 green signals or vice versa. Also, no more than 1% BMPC showing both 3 red and 3 green signals could be observed. In this study, we considered results of dual interphase FISH as relevant over the following thresholds: >8% BMPC nuclei exhibiting 3 signals of one color (red or green) and 2 signals of the other one color; >3% BMPC exhibiting 3 red signals and 3 green signals; >3% BMPC exhibiting 4 signals of one color and 3 signals of the other color; >3% BMPC exhibiting 4 red signals and 4 green signals (although these latter two features were never observed on control slides).
Overall efficiency of hybridization was appreciated on Ig− cells, as described above for single FISH.
Analysis of Data Obtained After Single Interphase FISH in MGUS Patients
One hundred to 143 BMPC were enumerated on each slide and results converted next into %. Results are reported in Table 1.
At diagnosis. Six patients (patients no. 1 to no. 5 and patient no. 10) had no abnormality after FISH study, at least for the chromosomes tested here, and DI was within normal values in four patients and slightly elevated in two patients. Thirteen patients showed numeric abnormalities within BMPC: one, four, five, and three patients exhibited trisomy for 1, 2, 3, or 4 different chromosomes, respectively. DI of BMPC was abnormal in 10 patients and within normal range in three patients.
At follow-up. Study was performed in 11 of 13 patients who had chromosome abnormalities at diagnosis, and conspicuous changes were observed in three: increase in the % of abnormal BMPC in patients no. 7 and no. 13 (this latter patient acquiring additional abnormalities), and changes in the nature of the involved chromosomes in patient no. 4 (trisomy for chr 3 appeared whereas trisomy for chr 11 disappeared). DI clearly rose in three patients, remained abnormal in five patients, and inconspicuous changes around normal values were noted in three patients.
Among the six patients without numeric chromosome change at diagnosis, three showed abnormalities within BMPC at follow-up. Trisomy for 1 chromosome (chr no. 11 or chr no. 9) was found in patients no. 4 and no. 5, and trisomy for chr no. 3, no. 9, and no. 11 was found in patient no. 10 (Table 1). DI did not change in patients no. 4 and no. 5 (normal values in both instances), and increased from 0.99 to 1.05 in patient no. 10 (Table 1).
Analysis of Data Obtained After Dual Chromosome Interphase FISH in MGUS Patients
Ninety-one to 165 BMPC were enumerated on each slide, according to the patients. Figure 1 shows various aspects of dual hybridization within BMPC. Intranuclear signals were easy to observe using dual or triple band-pass filters. In some instances green and red signals were close to each other, leading to yellow signal: in those instances the use of single band-pass filters confirmed that yellow spots corresponded to superimposed green and red signals (see Fig 1).
Results of Dual Interphase FISH Performed in Patients No. 11, 12, 15, and 17, Both at Diagnosis [a] and at Follow-Up [b]
| Patient No. . | 11 a (%) . | 11 b (%) . | 12 a (%) . | 12 b (%) . | 15 a (%) . | 15 b (%) . | 17 a (%) . | 17 b (%) . |
|---|---|---|---|---|---|---|---|---|
| Testing probes for chr no. 3 and no. 7: | ||||||||
| 2 sign. no. 3 + 2 sign. no. 7 | 24 | 26 | 15 | 8 | 34 | 21 | ||
| 3 sign. no. 3 + 2 sign. no. 7 | 19 | 27 | 12 | 18 | 16 | 15 | ||
| 2 sign. no. 3 + 3 sign. no. 7 | 10 | 12 | 10 | |||||
| 3 sign. no. 3 + 3 sign. no. 7 | 49 | 29 | 56 | 58 | 30 | 48 | ||
| 4 sign. no. 3 + 3 sign. no. 7 | 6 | |||||||
| 3 sign. no. 3 + 4 sign. no. 7 | 5 | |||||||
| Testing probes for chr no. 3 and no. 9: | ||||||||
| 2 sign. no. 3 + 2 sign. no. 9 | 9 | 24 | 18 | 25 | 10 | 19 | 30 | 8 |
| 3 sign. no. 3 + 2 sign. no. 9 | 29 | 40 | 35 | 29 | 19 | 20 | 23 | 19 |
| 2 sign. no. 3 + 3 sign. no. 9 | 18 | |||||||
| 3 sign. no. 3 + 3 sign. no. 9 | 47 | 33 | 40 | 33 | 59 | 47 | 29 | 44 |
| 4 sign. no. 3 + 3 sign. no. 9 | 4 | 16 | ||||||
| Testing probes for chr no. 3 and no. 11: | ||||||||
| 2 sign. no. 3 + 2 sign. no. 11 | 14 | 24 | ||||||
| 3 sign. no. 3 + 2 sign. no. 11 | 19 | 33 | ||||||
| 3 sign. no. 3 + 3 sign. no. 11 | 60 | 39 | ||||||
| Testing probes for chr no. 7 and no. 9: | ||||||||
| 2 sign. no. 7 + 2 sign. no. 9 | 31 | 30 | 16 | 15 | 56 | 20 | ||
| 3 sign. no. 7 + 2 sign. no. 9 | 23 | 24 | 11 | 15 | 11 | |||
| 2 sign. no. 7 + 3 sign. no. 9 | 12 | 14 | 17 | 15 | 17 | 23 | ||
| 3 sign. no. 7 + 3 sign. no. 9 | 31 | 32 | 46 | 42 | 16 | 28 | ||
| 3 sign. no. 7 + 4 sign. no. 9 | 5 | 3 | ||||||
| 4 sign. no. 7 + 3 sign. no. 9 | 7 | |||||||
| Testing probes for chr no. 7 and no. 11: | ||||||||
| 2 sign. no. 7 + 2 sign. no. 11 | 40 | 38 | 45 | 22 | ||||
| 3 sign. no. 7 + 2 sign. no. 11 | 12 | |||||||
| 2 sign. no. 7 + 3 sign. no. 11 | 56 | 48 | 16 | 17 | ||||
| 3 sign. no. 7 + 3 sign. no. 11 | 18 | 47 | ||||||
| Testing probes for chr no. 9 and no. 11: | ||||||||
| 2 sign. no. 9 + 2 sign. no. 11 | 17 | 23 | 50 | 23 | 20 | |||
| 3 sign. no. 9 + 2 sign. no. 11 | 17 | 22 | 39 | 18 | 13 | |||
| 2 sign. no. 9 + 3 sign. no. 11 | 10 | 12 | 27 | 28 | ||||
| 3 sign. no. 9 + 3 sign. no. 11 | 36 | 36 | 27 | 33 | ||||
| 4 sign. no. 9 + 2 sign. no. 11 | 7 | |||||||
| 4 sign. no. 9 + 3 sign. no. 11 | 11 | 5 |
| Patient No. . | 11 a (%) . | 11 b (%) . | 12 a (%) . | 12 b (%) . | 15 a (%) . | 15 b (%) . | 17 a (%) . | 17 b (%) . |
|---|---|---|---|---|---|---|---|---|
| Testing probes for chr no. 3 and no. 7: | ||||||||
| 2 sign. no. 3 + 2 sign. no. 7 | 24 | 26 | 15 | 8 | 34 | 21 | ||
| 3 sign. no. 3 + 2 sign. no. 7 | 19 | 27 | 12 | 18 | 16 | 15 | ||
| 2 sign. no. 3 + 3 sign. no. 7 | 10 | 12 | 10 | |||||
| 3 sign. no. 3 + 3 sign. no. 7 | 49 | 29 | 56 | 58 | 30 | 48 | ||
| 4 sign. no. 3 + 3 sign. no. 7 | 6 | |||||||
| 3 sign. no. 3 + 4 sign. no. 7 | 5 | |||||||
| Testing probes for chr no. 3 and no. 9: | ||||||||
| 2 sign. no. 3 + 2 sign. no. 9 | 9 | 24 | 18 | 25 | 10 | 19 | 30 | 8 |
| 3 sign. no. 3 + 2 sign. no. 9 | 29 | 40 | 35 | 29 | 19 | 20 | 23 | 19 |
| 2 sign. no. 3 + 3 sign. no. 9 | 18 | |||||||
| 3 sign. no. 3 + 3 sign. no. 9 | 47 | 33 | 40 | 33 | 59 | 47 | 29 | 44 |
| 4 sign. no. 3 + 3 sign. no. 9 | 4 | 16 | ||||||
| Testing probes for chr no. 3 and no. 11: | ||||||||
| 2 sign. no. 3 + 2 sign. no. 11 | 14 | 24 | ||||||
| 3 sign. no. 3 + 2 sign. no. 11 | 19 | 33 | ||||||
| 3 sign. no. 3 + 3 sign. no. 11 | 60 | 39 | ||||||
| Testing probes for chr no. 7 and no. 9: | ||||||||
| 2 sign. no. 7 + 2 sign. no. 9 | 31 | 30 | 16 | 15 | 56 | 20 | ||
| 3 sign. no. 7 + 2 sign. no. 9 | 23 | 24 | 11 | 15 | 11 | |||
| 2 sign. no. 7 + 3 sign. no. 9 | 12 | 14 | 17 | 15 | 17 | 23 | ||
| 3 sign. no. 7 + 3 sign. no. 9 | 31 | 32 | 46 | 42 | 16 | 28 | ||
| 3 sign. no. 7 + 4 sign. no. 9 | 5 | 3 | ||||||
| 4 sign. no. 7 + 3 sign. no. 9 | 7 | |||||||
| Testing probes for chr no. 7 and no. 11: | ||||||||
| 2 sign. no. 7 + 2 sign. no. 11 | 40 | 38 | 45 | 22 | ||||
| 3 sign. no. 7 + 2 sign. no. 11 | 12 | |||||||
| 2 sign. no. 7 + 3 sign. no. 11 | 56 | 48 | 16 | 17 | ||||
| 3 sign. no. 7 + 3 sign. no. 11 | 18 | 47 | ||||||
| Testing probes for chr no. 9 and no. 11: | ||||||||
| 2 sign. no. 9 + 2 sign. no. 11 | 17 | 23 | 50 | 23 | 20 | |||
| 3 sign. no. 9 + 2 sign. no. 11 | 17 | 22 | 39 | 18 | 13 | |||
| 2 sign. no. 9 + 3 sign. no. 11 | 10 | 12 | 27 | 28 | ||||
| 3 sign. no. 9 + 3 sign. no. 11 | 36 | 36 | 27 | 33 | ||||
| 4 sign. no. 9 + 2 sign. no. 11 | 7 | |||||||
| 4 sign. no. 9 + 3 sign. no. 11 | 11 | 5 |
After hybridization with every couple of probes, each plasma cell showed a variable number of red and green signals (2, 3, or 4 signals with each fluorescence). Ninety-one to 165 PC were enumerated on each slide, and data were converted next in % and reported in the table (only data over the thresholds [see text] are reported). In each patient, chromosomic numeric changes are distributed in several different patterns. These patterns correspond to subpopulations of plasma cells showing disomy, trisomy, or even tetrasomy for one chromosome tested and either disomy, trisomy, or tetrasomy for the other one chromosome tested, in each couple of probes used in the study.
For each couple of probes tested, the respective numbers of BMPC showing 2 red signals and 2 green signals, or 3 red and 2 green, or 2 red and 3 green, or 3 red and 3 green, or 4 red and 3 green, or 3 red and 4 green, or 4 red and 4 green signals were counted on each slide and converted into percentages. Full results from 4 patients, studied both at diagnosis and at follow-up, are reported in Table 2 (only data over thresholds are reported, as defined above).
At diagnosis. In patients no. 11 to no. 19, various patterns of distribution of the signals could be observed within the same dual interphase FISH test, including at least part of the following: trisomy for both chromosomes, trisomy for only one of the chromosomes tested, trisomy for only the other one chromosome tested, and finally disomy for both chromosomes. As an example, testing both probes for chr no. 3 and chr no. 7 in patient no. 15a showed that 56% BMPC were trisomic for both chr no. 3 and chr no. 7 (noted as 3 signals no. 3 + 3 signals no. 7 in Table 2), 12% BMPC were trisomic for chr no. 3 but disomic for chr 7 (noted as 3 signals no. 3 + 2 signals no. 7), and 15% BMPC were disomic for both chr no. 3 and no. 7 (noted 2 signals no. 3 + 2 signals no. 7). In this patient no. 15a, no significant number of BMPC had 2 signals no. 3 + 3 signals no. 7 (no result reported on the relevant line in Table 2). In some patients, namely patients no. 17 (Table 2), no. 18 and no. 19 (not reported), 4 signals with the same color were found for some of the probes tested, defining subclones bearing tetrasomy for the relevant chromosome.
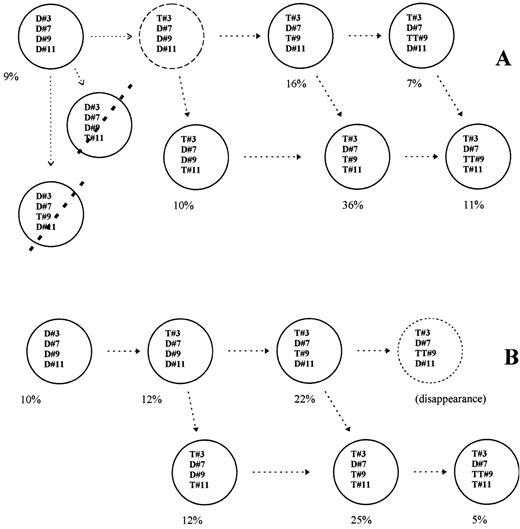
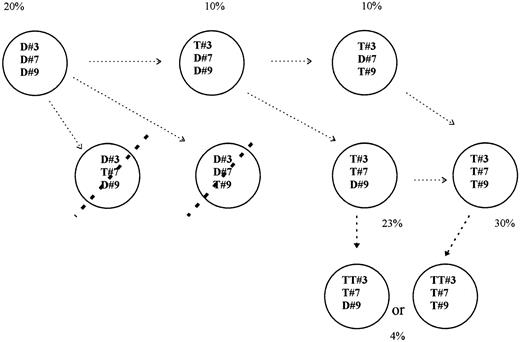
In every patient who exhibited trisomy for at least 3 different chromosomes, one part of BMPC had to or might bear all changes, whereas another part could not bear all the abnormalities. Up to 8 clones could be ascertained in some patients, corresponding to: BMPC without abnormality, BMPC with abnormality either for one (3 different combinations), or for 2 (3 different combinations), or for all chromosomes tested. In each patient, the respective % of BMPC displaying each of these various combinations could be ascertained. In patient no. 11a (Table 2), 47% BMPC had 3 signals for chr no. 3 + 3 signals for chr no. 9, 60% BMPC had 3 signals for chr no. 3 + 3 signals for chr no. 11, and 36% BMPC had both 3 signals for chr no. 9 +3 signals for chr no. 11. So, in this latter patient 36% BMPC could exhibit trisomy for chr no. 3, no. 9, and chr no. 11, but as 60% BMPC were found trisomic for both chr no. 3 and chr no. 11, one part of these latter BMPC had to be disomic for no. 9. Using this strategy it was possible to determine approximate percentages of subclones that bore extra copy(ies) for either all 4, or 3, 2, 1, or no one of the chromosomes tested, in every patient. Full examples are given in Fig 2 for patient no. 11a and in Fig 3 for patient no. 12. As shown in Figs 2 and 3, results of dual interphase FISH ruled out some patterns of chromosome changes (striped subclones in figs). Extra subclones could be also shown when BMPC exhibited tetrasomy for one chromosome (see Figs 2 and 3). In patient no. 11 a part of BMPC that bore 3 signals for chr no. 3 + 3 signals for chr no. 11 but that could not bear 3 signals for chr no. 9, were in fact trisomic for chr no. 3 and no. 11 and tetrasomic for chr no. 9. The same approach was performed in all patients. This led to the definition of up to seven different cytogenetic subclones coexisting together within BM from each MGUS patient.
Results from patient no. 11. Various plasmacytic subclones coexist, at diagnosis (A) and 1 year later. (B) Closed circles correspond to subclones that could be shown within BM after dual interphase FISH; dotted lines striping subclones mean that the corresponding subclones could not be shown. Open circles are either hypothesis or a clone that disappeared at follow-up. Percentages are the estimated amounts of each subclone. A hypothetic link between the various subclones is indicated with the open arrows. D or T followed by one number means Disomy or Trisomy for the corresponding chromosome (#).
Results from patient no. 11. Various plasmacytic subclones coexist, at diagnosis (A) and 1 year later. (B) Closed circles correspond to subclones that could be shown within BM after dual interphase FISH; dotted lines striping subclones mean that the corresponding subclones could not be shown. Open circles are either hypothesis or a clone that disappeared at follow-up. Percentages are the estimated amounts of each subclone. A hypothetic link between the various subclones is indicated with the open arrows. D or T followed by one number means Disomy or Trisomy for the corresponding chromosome (#).
Several cytogenetic subclones may be shown or not for patient no. 12, according to the results from dual interphase FISH (for details, see legend of Fig 2).
Several cytogenetic subclones may be shown or not for patient no. 12, according to the results from dual interphase FISH (for details, see legend of Fig 2).
At follow-up. Patients no. 9 to no. 17 were tested again after 12 to 35 months. Various clones were shown in these instances, and although some changes could be found within the respective percentages, most clones found at diagnosis were still exhibited at follow-up (Table 2 and Fig 2B). In patient no. 14, the clone exhibiting trisomy for chr 11 disappeared, as previously shown using single FISH study (data not reported).
Eventually, abnormalities were distributed within several different cytogenetic clones in most patients, and although some changes occurred, these various subclones were still found at follow-up. A possible link between these various subclones, related to a gradual acquisition of chromosome abnormalities is proposed for patients no. 11 and no. 12 in Figs 2 and 3 (arrows).
DISCUSSION
Chromosomes changes are a common finding within BMPC from MGUS patients,11,12 and results reported here confirm these data, after testing DNA satellite probes corresponding to the chromosomes mostly found in abnormal numbers in MM and in MGUS.14 15 After a median follow-up of 26 months, clinical and biological status of all patients was unchanged: another interphase FISH study performed at this moment demonstrated again cytogenetic abnormalities in all patients who had an abnormality at first examination. Moreover, among the 6 patients who had no chromosome change at diagnosis three acquired an extra copy for 1 to 3 different chromosomes at follow-up. No patient from this study returned to full cytogenetic normality, hypothesizing that appearance of chromosome changes was an irrevocable situation.
Abnormalities demonstrated within BMPC were not focused into one clone but distributed within several clones that differed in the number of abnormalities they displayed (up to 7 subclones could be demonstrated in some patients). Drach et al,12 using single interphase FISH to study BM cells from MGUS patients, also mentioned that in some cases chromosome abnormalities were certainly not restricted to one single clone. A possible explanation is that gradual acquisition of chromosomes occurred within BMPC, and that each subclone might correspond to the remnant of this multistep process. One numeric change occurring into one BMPC would generate one cytogenetically abnormal clone: another chromosome change occurring later would generate another clone exhibiting both abnormalities, and so on. These various and gradual changes either fit the multistep process that might ultimately lead to cancer,3-6,16 or correspond to an incapacity of PC to control their number of chromosomes.10 Clones that were found at diagnosis were also found at follow-up in comparable or in higher percentages in some patients, whereas in other patients some clones disappeared and were replaced by new ones, the latter displaying another change. Maybe the acquisition of peculiar cytogenetic change(s) generated a growth advantage in some instances but led in other instances to the disappearance of the relevant cells.
After the study of Ig variable gene sequences in monoclonal cells of MGUS patients, Sahota et al9 showed that some patients had homogeneous sequences, indicating no continuing exposure to the somatic hypermutation mechanism (as in MM), whereas some other patients had heterogeneous gene sequences, consistent with tumor cells remaining under the influence of the mutator. In those latter instances, the several subclones could be rearranged in a genealogical tree according to the mutational patterns. Our study shows that the various cytogenetic changes we observed might also correspond to the ordered acquisition of chromosome abnormalities. What is the relationship between results drawn from molecular9 and from cytogenetic (this report) techniques? In the case of childhood myelodysplastic syndromes, several findings show that monosomy for chr 7 is a secondary event, constituting a “common final pathway” to different conditions predisposing to myelodysplasia.17 In MGUS, chromosome changes might occur within one cell from an already monoclonal clone (defined according to the Ig gene sequences), or alternately one cell acquiring chromosome change(s) should generate an abnormal clone exhibiting per se an homogeneous Ig gene sequence. Whatever the mechanism, it does not lead to MM in all instances, and at least another step is needed to transform to a malignant disease. Recent data7 showed that molecular alterations of IL-6R, lck, and c-myc genes in MGUS patients resulted in rapid progression into aggressive MM. In that latter study, clonal size of myc rearrangement appeared to be different from clonal size containing lck rearrangement, suggesting the presence of more than one clone in the MGUS patients exhibiting the molecular alterations tested.
Follow-up was for a limited period, ranging from 12 to 35 months, and no patient evolved to MM. This period of time is short if we consider that 15% of patients developed MM within a median time of 9.6 years in a recent study.1 18 Some patients from this study had cytogenetic changes that were not identical at diagnosis and at follow-up, but in other patients abnormalities were in common in both circumstances. So, if a chromosome change may be associated with a growth advantage generating a clone, it is not sufficient enough, at least in our patients, to generate one clone overloading BM and per se all other subclones.
This study, restricted to numeric chromosome changes, raises the question about the existence or not of structural abnormalities associated to numeric ones in BMPC from MGUS patients. Numeric changes are classically considered as a secondary event, and we cannot rule out that one structural chromosome change occurred first, “sensitizing” one cell and generating next a clone in which loss of control for the number of chromosomes should be a secondary event. The various subclones defined according to the numeric changes in this study should all bear one structural abnormality. In MM, careful study of abnormal karyotypes shows that structural changes are quite common, but there is no consistent or specific structural change reported so far.10,14,15 In MGUS, the first structural change should also differ according to the patients, or should be indistinguishable after the use of classical cytogenetic study, as recently described for some translocations in acute leukemia.19 20
BMPC from MGUS patients exhibit various numeric chromosome abnormalities: distribution of changes within BMPC led to defining several cytogenetic clones, and both quantitative and qualitative changes were observed within these clones at follow-up in some patients. Distribution of chromosome abnormalities within the clones favors the hypothesis of gradual acquisition of cytogenetic changes within BMPC, in agreement with the concept of a multistep process for tumorigenesis in monoclonal gammopathies.3-6 16
ACKNOWLEDGMENT
We thank Dr Bataille and Dr Avet-Loiseau for their helpful comments.
Supported by grants from the Comité du Nord de la Ligue Nationale contre le Cancer and the Centre Hospitalier Régional de Lille, Lille, France.
Address reprint requests to Marc Zandecki, PhD, Laboratoire d'Hématologie Biologique, Centre hospital-Universitaire d'Angers, 4800 Angers, France.

![Fig. 1. Fluorescent green cytoplasm allows identification of a plasma cell (patient no. 6a) and hybridization of DNA probes shows 2 copies for chr no. 9 (2 red signals; biotin-labeled probe) and 2 copies for chr no. 11 (2 green signals; FITC-labeled probe) [dual band-pass filter]. (B) in the same patient (no. 6a), one plasma cell is disomic for chr no. 9 (2 red signals) and trisomic for chr no. 11 (3 green signals) [dual band-pass filter]. (C) Using a dual band-pass filter, one plasma cell from patient no. 15a exhibits trisomy for chr no. 7 (FITC-labeled probe) and disomy for chr no. 9 (biotin-labeled probe): red signals close to the green ones lead to yellow fluorescence, and the use of single band-pass filters shows 2 red signals (D: texas-red filter) and 3 green ones (E: FITC filter). (F ) and (G) Plasma cells exhibiting trisomy for chr no. 7 (FITC-labeled probe) and trisomy for chr no. 9 (biotin-labeled probe), using the triple band-pass filter (patient no. 17b in F and no. 17a in G). (H) Plasma cell exhibiting 3 signals for chr no. 9 (biotin-labeled probe) and 2 signals for chr no. 7 (FITC-labeled probe) (patient no. 17a).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/9/10.1182_blood.v90.9.3682/4/m_bl_0041f1.jpeg?Expires=1767784319&Signature=PBuxxmouCMudMVt-3RTdgvOTzK0YDpmwoo0tr1KT38-Rq7BlgI59D7PKRCXjYXNbHVuSRzmCN05oJINRolr~zmAOexBTfCu6wAq5wzXhnhFUfB5DYooHlc7KlI0ipRJ6JtE-hqYQjUWTpZv-hYdEtSQB1RjpNukpP4pa67fO3BQo7-yez25QNv-ISQMaX4YDI7lg0E7OOEPK34ShwUTSvB5iStWLlUixiAEHU9UdVoAPmpLRPBMFD6ZZJRfp-VFo822fGFkYqOfQIQ-zrXq8VIVwSPGtfOInRMrvnq71lrW2aA-vQPLpVHwb6RhUbeKwZixtDxL59g8pT5gN6pQLDA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal