Abstract
Expression of neutrophil secondary granule protein (SGP) genes is coordinately regulated at the transcriptional level, and is disrupted in specific granule deficiency and leukemia. We analyzed the regulation of SGP gene expression by luciferase reporter gene assays using the lactoferrin (LF) promoter. Reporter plasmids were transiently transfected into non–LF-expressing hematopoietic cell lines. Luciferase activity was detected from reporter plasmids containing basepair (bp) −387 to bp −726 of the LF promoter, but not in a −916-bp plasmid. Transfection of a −916-bp plasmid into a LF-expressing cell line resulted in abrogation of the silencing effect. Sequence analysis of this region revealed three eight-bp repetitive elements, the deletion of which restored wild-type levels of luciferase activity to the −916-bp reporter plasmid. Electrophoretic mobility shift assay and UV cross-linking analysis identified a protein of approximately 180 kD that binds to this region in non–LF-expressing cells but not in LF-expressing cells. This protein was identified to be the CCAAT displacement protein (CDP/cut). CDP/cut has been shown to downregulate expression of gp91-phox, a gene expressed relatively early in the myeloid lineage. Our observations suggest that the binding of CDP/cut to the LF silencer element serves to suppress basal promoter activity of the LF gene in non–LF-expressing cells. Furthermore, overexpression of CDP/cut in cultured myeloid stem cells blocks LF expression upon granulocyte colony-stimulating factor–induced neutrophil maturation without blocking phenotypic maturation. This block in LF expression may be due, in part, to the persistence of CDP/cut binding to the LF silencer element.
GRANULOPOIESIS involves a carefully regulated series of maturation events wherein the pluripotent stem cell within the bone marrow gives rise to the mature circulating cells of the monocyte and neutrophil lineages.1-3 Progression along the neutrophil maturation pathway involves loss of proliferative potential4 and is marked by a number of morphologically distinct cell intermediates. The maturing neutrophil undergoes a critical commitment step at the transition between the promyelocyte and the myelocyte stage, which is associated with the appearance of the secondary (specific) granules.5 Lactoferrin (LF) along with transcobalamin 1 (TC1), human neutrophil collagenase (HNC), and human neutrophil gelatinase (HNG) comprise the major secondary granule proteins (SGPs) that provide a unique marker of terminal neutrophil maturation. Previous studies have confirmed coordinate, stage-specific expression of these genes that is regulated at the transcriptional level.6-9 Additionally, in acute leukemia and myelodysplastic syndromes, where the regulation of normal myeloid differentiation is disrupted, expression of LF and the other three SGP genes is either aberrant or altogether absent.10 11 Since the specific granule protein (SGP) genes are unlinked and functionally diverse, we hypothesize that the coordinate tissue- and stage-specific expression of these genes is modulated by common factor(s). To identify and define such control elements, we studied the regulation of LF gene expression.
LF is a multifunctional cationic iron-binding glycoprotein that is thought to be responsible for primary defense against microbial infection through its ability to chelate iron, which is required for microbial growth.12 In addition to being expressed in the later stages of neutrophil maturation, LF is expressed in most glandular epithelial tissues, including mammary glands, as well as in the female reproductive organs.13,14 Differential regulation of LF has been reported in these diverse tissues. The recently described overlapping retinoic acid response element (RARE) and an estrogen response element in the 5′ flanking region of the LF gene mediate LF expression in the mammary gland (where estrogen receptors are abundant). This element has differential protein binding in mammary cells and hematopoietic cells, but there are no functional data supporting the significance of the RARE in myeloid-specific LF gene expression.15 The investigators suggest that the LF composite hormone response element may account for differential hormone sensitivities, and predict a role for it as a critical signaling switch in the regulation of differential expression of LF.
The nucleotide sequence of the LF promoter and its 5′ flanking region, which has been previously reported by us7 and by others,16 is known to contain a number of familiar transcription factor binding motifs, which include a TATA-like sequence, two CCAAT boxes, an AP-2 site, PU.1 boxes, GATA-1 sites, and overlapping COUP-TF and RAR elements.7,17 The latter two elements have been shown to influence LF expression in mammary cells.18-20
To define regulatory elements within the LF promoter, varying lengths of the 5′ flanking sequences of the LF gene were tested for promoter activity in reporter gene plasmids transfected into hematopoietic cell lines. We report here the identification of a negative regulatory element within the LF gene promoter that is recognized by the CCAAT displacement protein (CDP/cut), a homeodomain protein with extensive homology to the Drosophila cut protein.21 CDP/cut also regulates expression of the phagocyte-specific cytochrome heavy chain gene (gp91-phox), which is expressed exclusively in differentiating granulocytes.22 Since expression of gp91-phox precedes that of LF in the granulocyte maturation pathway, the role of CDP/cut in the temporal regulation of these myeloid-specific genes remains to be elucidated.
MATERIALS AND METHODS
Plasmid construction.5′ end deletions of the LF promoter were obtained from a previously described LF genomic clone containing 950 basepairs (bps) 5′ of the transcription start site.7 Oligonucleotides were synthesized with a SmaI restriction site at the 5′ end: (−916 to −899) 5′ AATGGCATGCAGTTGTGC 3′, (−726 to −715) 5′ AGGAGATGGAGA 3′, (−649 to −631) 5′ GGCATCTTTCTCATTGGC 3′, and (−384 to −358) 5′ AGGTCATCTGCTGAAGAAGATAGCAG 3′. An antisense oligonucleotide corresponding to the +7 to +22 coordinates of the LF gene having the sequence 5′ CGACTTGGCAAACGAAG 3′ with a synthetic SalI site at its 5′ end was also prepared. Each 5′ oligomer was paired in turn with the 3′ antisense oligomer and used in a standard polymerase chain reaction (PCR) with the genomic LF clone7 as a template. The PCR products were directly cloned into the pCRII vector (Invitrogen, La Jolla, CA) and sequenced. A Sma I-Sal I fragment was isolated and subcloned into a Sma I-Xho I–digested pGL2-basic promoterless vector (Promega Biotechnology, Madison, WI) upstream of the luciferase reporter gene. Construction of the −916 silencer deletion plasmid (−916(d)S) followed the same PCR and cloning strategy, except that the sense oligo contained a 56-bp deletion and had the following sequence: 5′ AATGGCATGCAGTGGTGCCTGGAAACATTTGTACCTGGGGTGCTGTGTGTCATGGGAATTCTGACATTTATGGCGAT 3′.
Large-scale plasmid preparations (Qiagen, Chatsworth, CA) were made, divided into aliquots, and stored at −20°C.
Tissue culture, transient transfection, and luciferase assay.Human K562 and erythroleukemic HEL cells were obtained from the American Type Culture Collection (Rockville, MD) and maintained and grown in RPMI 1640 medium (GIBCO, Grand Island, NY) supplemented with 10% fetal calf serum (GIBCO), 0.2 mmol/L glutamate, 50 U/mL penicillin, and 50 μg/mL streptomycin. 32Dcl3 cells were a kind gift from Dr Giovanni Rovera (Wistar Institute, Philadelphia, PA), and 32Dwt18 cells were a gift from Dr Dan Link (University of Washington, St Louis, MO). Both cell lines were grown in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% fetal calf serum and 10% WEHI-conditioned medium as a source of interleukin-3 (IL-3).23 Murine promyelocytic MPRO cells,24 25 kindly provided by Dr Schickwann Tsai (Fred Hutchinson Cancer Research Institute, Seattle, WA), were grown in IMDM supplemented with 10% fetal calf serum and 10% HM-5–conditioned medium as a source of granulocyte-macrophage colony-stimulating factor (GM-CSF). All cells were maintained at 37°C in a humidified 5% CO2 incubator.
Differentiation of 32Dcl3 cells was performed as described previously.26 Briefly, exponentially growing cells were washed twice with phosphate-buffered saline (PBS) and resuspended in growth medium containing 3 × 103 U/mL G-CSF (Neupogen; Amgen Inc, Thousand Oaks, CA). Maturation was monitored by Wright-Giemsa staining. MPRO cells were induced toward neutrophils by addition of 10 μmol/mL all-trans-retinoic acid (ATRA) in the presence of GM-CSF. Maturation was also monitored by Wright-Giemsa staining.
For transient transfection experiments, approximately 1 × 107 cells were gently pelleted, washed twice with PBS, and resuspended in 180 μL HEPES-buffered saline in electroporation cuvettes. Ten micrograms of each reporter plasmid and 1 μg pCMVβgal (Clontech, Palo Alto, CA), an internal control plasmid used to monitor transfection efficiency, were added to each aliquot of cells. Following a 5-minute incubation period at room temperature, the DNA-cell samples were electroporated using a BioRad Gene Pulser (Melville, NY). K562 and HEL cells were electroporated at 200 mV with 960 microfarads of capacitance. 32Dcl3 and 32Dwt18 cells were electroporated at 400 mV with a capacitance of 250 μF. Transiently transfected cells were incubated in growth medium at 37°C in 5% CO2 for 16 to 20 hours. Luciferase activity was then determined using an assay kit from Promega per the manufacturer's instructions. In all experiments, luciferase expression levels were normalized to the levels of β-galactosidase expression, assayed as previously described.27
Stable transfection and erythropoietin induction of 32Dwt18 cells.Exponentially growing 32Dwt18 cells (1.0 × 107) were washed twice with PBS and resuspended in 180 μL HEPES-buffered saline. Twenty micrograms of each reporter gene plasmid was next cotransfected by electroporation with 1 μg pBabe puro plasmid28 as described earlier. The electroporated cells were resuspended in standard growth medium overnight. The cells were then selected for puromycin resistance in IMDM supplemented with 1 μg/mL puromycin (Sigma). Stable transfection was confirmed by Southern blot analysis.
Stably transfected 32Dwt18 cells were induced toward neutrophil maturation by addition of 1 U/mL erythropoietin (Epo) (Amgen) in medium lacking IL-3. Luciferase measurements were made on alternate days, and viability was monitored by trypan blue exclusion. Neutrophil maturation was monitored by Wright-Giemsa staining and by Northern blot analysis using a mouse LF cDNA probe as previously described.6
Stable transfection of 32Dcl3 cells.CDP/cut-overexpressing 32Dcl3 cells were obtained by cotransfecting the previously described pMT2-CDP/cut plasmid29 and the pSVneo plasmid at a 20:1 ratio as described earlier. Increased CDP/cut levels in the resulting 32Dcl3 subclone were confirmed by Western blot analysis using the anti-CDP/cut antiserum described previously.29 These cells were induced to undergo neutrophil maturation by addition of G-CSF. LF expression was monitored by Northern blot analysis as previously described.6
Preparation of nuclear extracts.Nuclear extracts were prepared essentially as described previously.7 Briefly, 1 × 107 cells were washed twice in ice-cold PBS and once in buffer A containing 10 mmol/L HEPES-KOH (pH 7.9), 1.5 mmol/L MgCl2 , 10 mmol/L KCl, 0.5 mmol/L dithiothreitol (DTT), and 0.5 mmol/L phenylmethylsulfonyl fluoride (PMSF). Cells were then lysed following a 5-minute incubation on ice in buffer A with 0.1% Nonidet P-40. Nuclei were recovered by centrifugation at 4°C for 15 minutes. The nuclei were lysed in high-salt buffer C (20 mmol/L HEPES-KOH (pH 7.9), 10% glycerol, 420 mmol/L NaCl, 10 mmol/L KCl, 0.2 mmol/L EDTA, and 0.5 mmol/L DTT), and nuclear extracts were recovered by centrifugation at 4°C for 15 minutes. Salt concentration in the nuclear extracts was adjusted by addition of buffer D (20 mmol/L HEPES-KOH (pH 7.9), 20% glycerol, 0.05 mmol/L KCl, 0.2 mmol/L EDTA, 0.5 mmol/L DTT, and 0.5 mmol/L PMSF). All buffers used included the following cocktail of protease inhibitors at the final concentration indicated: pepstatin A (10 μg/mL), leupeptin (10 μg/mL), aprotinin (1 μg/mL), and Perfabloc (1 mg/mL) (Boehringer, Indianapolis, IN). Total protein concentration in the nuclear extract preparations was assayed using the BioRad kit per the manufacturer's instructions. In general, most preparations yielded 1.5 to 2 μg/mL protein. Aliquots of nuclear protein extracts were frozen immediately and stored at −70°C until required.
Oligonucleotides and electrophoretic mobility shift assays.Complementary oligonucleotides were annealed and labeled at the 5′ ends using [γ-32P]ATP (6,000 Ci/mmol; Amersham, Arlington Heights, IL) and T4 polynucleotide kinase (NEB, Beverly, MA). Radiolabeled double-stranded oligonucleotides were separated from unincorporated nucleotide by passage through a Sephadex G-25 column (Boehringer). Probes were stored at −20°C.
EMSAs were performed by incubating 15 μg nuclear extracts with 20,000 cpm double-stranded oligonucleotide in a 20-μL reaction mixture containing 10 mmol/L HEPES-KOH buffer (pH 7.9), 50 mmol/L KCl, 2.5 mmol/L MgCl2 , 1 mmol/L DTT, 10% glycerol, 1 μg acetylated bovine serum albumin (NEB), and 0.5 μg poly(dI-dC) at 25°C for 20 minutes. For competition analysis, a 100-fold molar excess of unlabeled oligonucleotides was added to the nuclear extracts before addition of the labeled probe. For the supershift assay, polyclonal anti-CDP/cut antiserum21 was incubated with nuclear extracts for 15 minutes following addition of the radiolabeled probe. Binding reactions were resolved on a 4% nondenaturing polyacrylamide gel containing 1× TBE (0.089 mol/L Tris borate, 0.089 mol/L boric acid, and 0.002 mol/L EDTA) and electrophoresed at 200 V for 3 hours at 4°C. Gels were exposed to x-ray film with an intensifying screen overnight at −80°C. The oligonucleotides used in EMSA analysis were DM2, 5′ AGATGTATTCTAGAAGCAGTATTCTAG 3′; NCAM, 5′ CTTTGAAAATCGAACCGAATCTAAAAT 3′30; and mut NCAM, 5′ CTTGAAACATCGTACCGATGCTAAGAT 3′.30
UV cross-linking analysis.A binding reaction between 32P-labeled oligonucleotide probes and nuclear extracts was performed as described in the previous section. The binding reaction mixtures were irradiated at a wavelength of 254 nm with the source of radiation at 5 cm. After 30 minutes, the covalently linked DNA-protein complexes were resolved on a 7.5% sodium dodecyl sulfate (SDS)-polyacrylamide gel at 200 V for 4 hours. Prestained protein markers (BioRad) were analyzed alongside. Gels were dried and exposed to x-ray film with an intensifying screen at −80°C.
Northern blot analysis.Northern blot analysis was performed as described previously.10 Briefly, 10 μg total RNA was separated on a 1% denaturing gel, transferred to nitrocellulose filters, and hybridized to 32P-labeled cDNA fragments at 42°C in 50% formamide. The filters were washed at high stringency in 0.1% SDS and 0.1× SSC at 55°C and autoradiographed. Blots were probed with a 600-bp mouse LF probe cloned in our laboratory.6
RESULTS
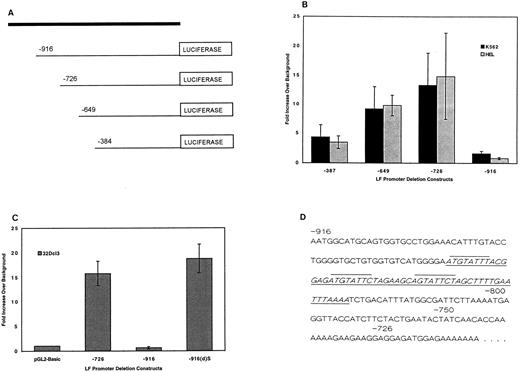
Identification of negative regulatory element within the LF gene promoter.Functional analysis of the LF promoter and its 5′ flanking sequences was initiated by fusing varying lengths of the LF promoter into the promoterless pGL2-basic vector to direct the synthesis of the firefly luciferase reporter gene in transient transfection assays. Initially, four LF promoter segments spanning −384 bp, −649 bp, −726 bp, and −916 bp upstream of the LF transcription start site were cloned into the pGL2-basic vector (Fig 1A). These plasmids were transiently transfected into several hematopoietic cell lines, including K562, a leukemic cell line derived from a patient with CML in blast crisis,31 HEL, a human erythroleukemia cell line32; and 32Dcl3, and IL-3–dependent murine myeloid progenitor cell line.33 The leukemic lines do not express LF, and 32Dcl3 expresses the gene only after G-CSF–induced differentiation. Measurable luciferase activity was detected for the −384 bp, −649 bp, and −726 bp plasmids in leukemic cells (Fig 1B) and uninduced 32Dcl3 cells (Fig 1C). However, the −916 bp plasmid expressed background levels of luciferase activity. This effect was not attributable to a specific DNA preparation or to mutations in the −916 bp LF promoter plasmid.
Transient transfection analysis of LF promoter gene plasmids. (A) Schematic diagram indicating the position of 5′ deletions of the LF promoter relative to the luciferase gene. (B) Luciferase activity of the LF promoter deletion plasmids transfected in K562 and HEL cells expressed as fold increase over luciferase activity measured for the promoterless pGL2-basic vector (background expression, which was consistently 100 to 200 RLU, was given a value of 1). Luciferase activity was normalized to β-galactosidase activity. The data represent 4 to 8 independent experiments each performed in duplicate for each promoter plasmid, and are expressed as the mean with SDs represented as error bars. (C) Luciferase activity of −726 bp, −916 bp, and −916(d)S LF promoter plasmids in uninduced 32Dcl3 cells. The −916(d)S plasmid was created by deletion of the underlined 56-bp fragment in (D). Luciferase activity was normalized to cotransfected β-galactosidase activity (pCMVβgal plasmid). The average of 3 independent experiments, each performed in duplicate, is presented. (D) The nucleotide sequence between the −916-bp and −728-bp coordinates of the LF promoter containing the triad octamers (overlined) and the 56-bp deletion (underlined) of the −916-bp plasmid to yield the −916(d)S plasmid.
Transient transfection analysis of LF promoter gene plasmids. (A) Schematic diagram indicating the position of 5′ deletions of the LF promoter relative to the luciferase gene. (B) Luciferase activity of the LF promoter deletion plasmids transfected in K562 and HEL cells expressed as fold increase over luciferase activity measured for the promoterless pGL2-basic vector (background expression, which was consistently 100 to 200 RLU, was given a value of 1). Luciferase activity was normalized to β-galactosidase activity. The data represent 4 to 8 independent experiments each performed in duplicate for each promoter plasmid, and are expressed as the mean with SDs represented as error bars. (C) Luciferase activity of −726 bp, −916 bp, and −916(d)S LF promoter plasmids in uninduced 32Dcl3 cells. The −916(d)S plasmid was created by deletion of the underlined 56-bp fragment in (D). Luciferase activity was normalized to cotransfected β-galactosidase activity (pCMVβgal plasmid). The average of 3 independent experiments, each performed in duplicate, is presented. (D) The nucleotide sequence between the −916-bp and −728-bp coordinates of the LF promoter containing the triad octamers (overlined) and the 56-bp deletion (underlined) of the −916-bp plasmid to yield the −916(d)S plasmid.
To further investigate this silencing effect, the sequence between bps −916 and −726 was dissected to yield two additional reporter gene plasmids, ie, bps −750 and −800. Both of these plasmids had measurable luciferase activity (data not shown). Similar results were observed following transfection of these LF promoter reporter plasmids in a nonhematopoietic cell line such as HeLa cells (data not shown). These observations suggest the presence of a negative regulatory element between bps 800 and 916 upstream of the LF gene transcription start site.
Deletion of silencer sequences from the −916 bp plasmid.The nucleotide sequence between the −916 bp and −726 bp coordinates of the LF promoter contains a triad of octamers (overlined in Fig 1D). Two of these repeats were found to be identical to the MATα2 protein binding site.34 The MATα2 gene product is part of a well-defined system that governs cell-specific transcription in the Saccharomyces cerevisiae mating types a and α.34 35 The MATα2 protein serves as a repressor of a-specific genes in α-mating–type cells by binding to a conserved DNA motif present in the promoters of a-specific genes. In light of this observation, we hypothesized that these sequences might mediate the silencing effect. A deletion plasmid of the −916 bp plasmid was prepared such that a 56-bp sequence (underlined in Fig 1D) containing the triad of octamers was removed. The resulting promoter plasmid, designated −916(d)S, was tested for promoter activity in transiently transfected K562 cells (data not shown) and in uninduced 32Dcl3 cells (Fig 1C). The LF −916(d)S promoter plasmid expressed wild-type levels (comparable to the −726 bp plasmid) of luciferase activity. A similar pattern of expression was observed in K562 cells. Restoration of wild-type levels of luciferase activity by removal of this 56-bp fragment further suggested that the deleted fragment is a cis-acting silencer of LF expression, as discussed in detail later herein.
Protein-DNA complex formation in LF-expressing and non–LF-expressing cells.To define the nucleotide sequence responsible for protein recognition within the deleted 56-bp LF silencer sequence, three overlapping double-stranded oligonucleotides were synthesized (Fig 2A; DM 1, 2, and 3). Each of the three double-stranded oligonucleotides was used in EMSA analysis using K562 nuclear extracts. The DM2 region (Fig 2A) bound most specifically and avidly to proteins from the K562 nuclear extracts (data not shown) and were used in all subsequent experiments. Oligomers containing only one octamer demonstrated no binding.
(A) Dissection of coordinates −858 bp to −810 bp containing the LF silencer element into oligomers DM (double module) 1, 2, and 3. Each oligomer (except DM3) contains 2 of the 3 octamer repeats (overlined) that comprise the LF silencer element. (B) UV cross-linking analysis of proteins bound to the DM2 oligomers. DM2 oligomers were [γ-32P]dATP-labeled and incubated with equal concentrations of nuclear extracts (N.E.), with (+) or without (−) a 100-fold molar excess of unlabeled DM2 oligos, prepared from K562 cells (lanes 2 and 3), uninduced 32Dcl3 cells (lanes 4 and 5), G-CSF–induced 32Dcl3 cells (lanes 6 and 7), MPRO cells (lanes 8 and 9), and MPRO + ATRA cells (lanes 10 and 11). The protein-DNA complexes formed were subjected to UV light at 254 nm for 30 minutes before electrophoresis in a 7.5% SDS-polyacrylamide gel. The position of prestained protein molecular weight markers is indicated. F, unbound probe. Lane 1, probe without nuclear extract.
(A) Dissection of coordinates −858 bp to −810 bp containing the LF silencer element into oligomers DM (double module) 1, 2, and 3. Each oligomer (except DM3) contains 2 of the 3 octamer repeats (overlined) that comprise the LF silencer element. (B) UV cross-linking analysis of proteins bound to the DM2 oligomers. DM2 oligomers were [γ-32P]dATP-labeled and incubated with equal concentrations of nuclear extracts (N.E.), with (+) or without (−) a 100-fold molar excess of unlabeled DM2 oligos, prepared from K562 cells (lanes 2 and 3), uninduced 32Dcl3 cells (lanes 4 and 5), G-CSF–induced 32Dcl3 cells (lanes 6 and 7), MPRO cells (lanes 8 and 9), and MPRO + ATRA cells (lanes 10 and 11). The protein-DNA complexes formed were subjected to UV light at 254 nm for 30 minutes before electrophoresis in a 7.5% SDS-polyacrylamide gel. The position of prestained protein molecular weight markers is indicated. F, unbound probe. Lane 1, probe without nuclear extract.
EMSA was performed using nuclear extracts from LF-expressing and non–LF-expressing cells. The protein-DNA complexes formed by non–LF-expressing cells, namely K562, uninduced 32Dcl3, and the promyelocytic MPRO cells, were essentially identical. However, the absence of a large protein-DNA complex was noted in nuclear extracts prepared from LF-expressing cells (data not shown). To confirm this differential binding of proteins, UV cross-linking analysis was performed to determine the molecular weight of protein species that specifically recognized the DM2 oligomers (Fig 2B). The DM2 oligos bound several protein species ranging in molecular weight from approximately 55 to 180 kD. However, a protein of approximately 180 kD (indicated by the arrowhead in Fig 2B) bound specifically to the DM2 oligonucleotides when exposed to nuclear extracts prepared from non–LF-expressing cells, ie, K562 cells (lane 2) and uninduced 32Dcl3 cells (lane 4), but not from LF-expressing nuclear extracts, ie, G-CSF–induced 32Dcl3 cells (lane 6), uninduced MPRO cells (lane 8), and ATRA-induced MPRO cells (lane 10). The MPRO cell line is a GM-CSF–dependent cell line derived from mouse bone marrow cells that have been transduced with a dominant negative retinoic acid α-receptor cDNA.24,25 These cells are arrested at the late promyelocyte stage and can be induced to undergo neutrophil maturation following treatment with ATRA. Although uninduced MPRO cells express some LF, maximal expression of this gene comparable to that seen in induced 32D cells occurs 1 to 3 days after ATRA induction.36 The DM2 oligomers formed similar specific complexes using nuclear extracts prepared from uninduced cells (lane 8) and from ATRA-induced MPRO cells (lane 10), presumably because both populations of cells are positive for LF expression. Longer exposures of similar UV cross-linking gels to x-ray film never revealed the presence of the 180-kD protein in LF-expressing nuclear extracts. However, such long exposures did reveal the apparently ubiquitous 130-kD protein in both the uninduced and ATRA-induced MPRO lanes.
EMSA and UV cross-linking data confirm the presence of a large protein-DNA complex (180-kD protein) that is absent from LF-expressing cells. The large molecular weight of this protein suggested that the DNA binding species might be the CCAAT displacement protein (CDP/cut), which has been implicated in expression of the gp91-phox (cytochrome heavy chain) gene that is expressed relatively early in the granulocytic differentiation pathway (Berliner N., unpublished observations).
CDP/cut is a 180- to 200-kD protein that was first described as a DNA binding activity in sea urchins.37 It has been shown to act as a negative regulator of gene expression in a large number of developmentally regulated genes such as the myeloid-specific gp91-phox gene,29,38 the NCAM gene,30 the γ-globin genes,39 and others. Progress in understanding the role of CDP/cut in gene regulation has been hampered by the fact that CDP/cut is ubiquitously expressed and that it lacks a well-defined consensus DNA binding motif29 (and references therein). However, it has been clearly established that in several instances decreased CDP/cut binding activity is associated with increased expression of CDP/cut-regulated genes.
CDP/cut recognizes the LF silencer motif.To determine whether CDP/cut was involved with binding to the LF silencer DM2 oligomers, EMSA was performed using 32P-labeled DM2 oligomers and nuclear extracts from a non–LF-expressing source, ie, K562 cells. The protein-DNA complexes formed were subjected to competition by unlabeled oligomers prepared from the NCAM promoter that had previously been shown to bind CDP/cut specifically.30 Mutant NCAM oligos, which do not bind to CDP/cut,30 served as a control (Fig 3A). A high–molecular-weight protein-DNA complex (indicated by the arrowhead) was specifically competed away by the wild-type NCAM oligomers (lane 4), but not by the mutant NCAM oligomers (lane 5). However, the mutant NCAM oligomers did appear to compete away a relatively faster-migrating complex (indicated by a •). The relevance of this observation, if any, remains unclear. However, it is noteworthy that the NCAM CDP/cut binding oligomers share no homology with the LF DM2 oligomers at the nucleotide level, although both sets of oligomers are recognized by CDP/cut as a binding site. These observations suggest that CDP/cut recognizes and binds specifically to the LF silencer motif.
(A) EMSA analysis of K562 nuclear proteins bound to DM2 oligomers in the presence of NCAM and mutant NCAM cold competitor oligomers. [γ-32P]dATP-labeled DM2 oligomers were incubated with K562 nuclear extracts in the presence of a 100-fold molar excess of cold DM2 oligomers (lane 3), NCAM oligomers (lane 4), and mutant NCAM oligomers (lane 5). Lane 2, K562 nuclear extracts with no added competitor (No comp.). The DM2 probe was run without any nuclear extracts in lane 1 (DM2 probe). (B) Supershift EMSA analysis of K562 nuclear proteins bound to DM2 oligomers in the presence of an anti-CDP/cut antibody. In the left panel, [γ-32P]dATP-labeled DM2 oligomers were complexed with nuclear proteins from K562 cells (lane 2) and the DNA-protein complexes exposed either to preimmune serum (lane 3) or to anti-CDP/cut antibody (lane 4). In the right panel, NCAM oligomers were labeled with [γ-32P]dATP and allowed to bind to nuclear proteins from K562 nuclear extracts. The resulting protein-DNA complexes were exposed either to preimmune serum (lane 6) or to anti-CDP/cut antibody (lane 7). Lane 1, DM2 oligomers without nuclear extract.
(A) EMSA analysis of K562 nuclear proteins bound to DM2 oligomers in the presence of NCAM and mutant NCAM cold competitor oligomers. [γ-32P]dATP-labeled DM2 oligomers were incubated with K562 nuclear extracts in the presence of a 100-fold molar excess of cold DM2 oligomers (lane 3), NCAM oligomers (lane 4), and mutant NCAM oligomers (lane 5). Lane 2, K562 nuclear extracts with no added competitor (No comp.). The DM2 probe was run without any nuclear extracts in lane 1 (DM2 probe). (B) Supershift EMSA analysis of K562 nuclear proteins bound to DM2 oligomers in the presence of an anti-CDP/cut antibody. In the left panel, [γ-32P]dATP-labeled DM2 oligomers were complexed with nuclear proteins from K562 cells (lane 2) and the DNA-protein complexes exposed either to preimmune serum (lane 3) or to anti-CDP/cut antibody (lane 4). In the right panel, NCAM oligomers were labeled with [γ-32P]dATP and allowed to bind to nuclear proteins from K562 nuclear extracts. The resulting protein-DNA complexes were exposed either to preimmune serum (lane 6) or to anti-CDP/cut antibody (lane 7). Lane 1, DM2 oligomers without nuclear extract.
To further confirm binding of CDP/cut to the DM2 oligomers, a supershift EMSA was performed using an anti-CDP/cut antibody and K562 nuclear extracts (Fig 3B). 32P-labeled NCAM oligomers were used as a positive control (right panel). As indicated by arrowheads in both panels, addition of anti-CDP/cut antibody resulted in a supershift of the DM2 oligomers (lane 4) similar to that observed for the NCAM oligomers (lane 7). No supershift was observed by addition of preimmune serum to either the DM2 or the NCAM oligomers (lanes 3 and 6). This experiment confirms that CDP/cut binds to the LF silencer.
Overexpression of CDP/cut in 32Dcl3 cells downregulates LF expression.The role of CDP/cut as a repressor of the myeloid-specific gp91-phox gene has been recently demonstrated by constitutive expression of stably transfected CDP/cut cDNA in HL60 cells, which results in inhibition of expression of the target gp91-phox gene.29 In a similar experiment, we transfected the CDP/cut cDNA into the 32Dcl3 cell line to determine whether constitutive overexpression of CDP/cut influences LF gene expression following G-CSF induction. CDP/cut overexpression before and after G-CSF induction was confirmed by Western blot analysis (Fig 4A). CDP/cut-overexpressing 32Dcl3 cells were induced with G-CSF; morphologic neutrophil maturation was monitored by Wright Giemsa staining, and LF expression was assessed by Northern blot analysis. Morphologic differentiation of the CDP/cut-overexpressing stable transformants was normal (data not shown), but overexpression of CDP/cut in these cells completely blocked LF gene expression at the mRNA level (Fig 4B, lanes 4 and 5). In contrast, 32Dcl3 cells transfected with the pSVneo plasmid alone expressed high levels of LF mRNA following terminal myeloid differentiation with G-CSF (Fig 4, lane 2).
Western and Northern blot analysis of G-CSF–induced 32Dcl3 cells stably transfected with the pMT2-CDP/cut plasmid. (A) Western blot using anti-CDP antiserum confirming overexpression of CDP/cut in CDP-transfected 32Dcl3 cells before (0) and after (4d) G-CSF induction. 32Dwt18/CDP/cut cells were used as a positive (+) control. Each lane contains 100 μg nuclear protein extracts. (B) Ten micrograms each of total RNA isolated from uninduced or 2-day and 4-day G-CSF–induced 32Dcl3 cells stably cotransfected with pMT2-CDP/cut and pSVneo (lanes 3, 4, and 5, respectively) plasmids or pSVneo plasmid alone (uninduced, lane 1; 4-day G-CSF–induced, lane 2) was electrophoresed on a 1% formaldehyde/agarose gel and blotted onto a nitrocellulose filter. The filter was probed with an [α-32P]dCTP–labeled LF cDNA probe. The position of 28S and 18S rRNA markers is indicated. LF mRNA is denoted by the arrowhead. To ensure equal loading of RNA in each lane, the blot was stripped and reprobed with an [α-32P]dCTP–labeled β-actin probe, as indicated in the bottom panel.
Western and Northern blot analysis of G-CSF–induced 32Dcl3 cells stably transfected with the pMT2-CDP/cut plasmid. (A) Western blot using anti-CDP antiserum confirming overexpression of CDP/cut in CDP-transfected 32Dcl3 cells before (0) and after (4d) G-CSF induction. 32Dwt18/CDP/cut cells were used as a positive (+) control. Each lane contains 100 μg nuclear protein extracts. (B) Ten micrograms each of total RNA isolated from uninduced or 2-day and 4-day G-CSF–induced 32Dcl3 cells stably cotransfected with pMT2-CDP/cut and pSVneo (lanes 3, 4, and 5, respectively) plasmids or pSVneo plasmid alone (uninduced, lane 1; 4-day G-CSF–induced, lane 2) was electrophoresed on a 1% formaldehyde/agarose gel and blotted onto a nitrocellulose filter. The filter was probed with an [α-32P]dCTP–labeled LF cDNA probe. The position of 28S and 18S rRNA markers is indicated. LF mRNA is denoted by the arrowhead. To ensure equal loading of RNA in each lane, the blot was stripped and reprobed with an [α-32P]dCTP–labeled β-actin probe, as indicated in the bottom panel.
Because greater than 80% of 32Dcl3 cells die within the first 24 hours of IL-3 removal and G-CSF induction, these results were confirmed using 32Dwt18, an IL-3–dependent subline of 32Dcl3. This cell line carries a chimeric form of the G-CSF receptor containing the extracellular domain of the Epo receptor and the cytoplasmic signaling domain of the G-CSF receptor. These cells differentiate in response to Epo and express LF at a level comparable to that seen with the 32Dcl3/G-CSF system. The main advantage is that constitutive cytokine receptor expression prevents the high rate of cell death encountered upon induction of 32Dcl3 with G-CSF, permitting easier selection and induction of single cell clones. 32Dwt18 cells were transfected with the CDP expression vector and cloned by limiting dilution. Pooled cells showed markedly reduced LF expression upon Epo induction, and five single-cell clones showed a complete block to Epo-induced LF expression (data not shown). It is therefore inferred that overexpression of CDP/cut results in repression of LF expression without any apparent effect on the G-CSF–induced morphologic neutrophil maturation. As previously described by Lievens et al,29 this is an effect specifically seen following induction of cells overexpressing CDP/cut. It is notable that endogenous expression of CDP/cut at the mRNA level remains constant upon G-CSF induction29 (data not shown).
CDP/cut remains bound to the LF silencer motif in 32Dcl3-CDP/cut-overexpressing cells.We next used EMSA analysis to ascertain whether the block to upregulation of LF expression in 32Dcl3/CDP/cut cells is reflected in binding to the LF silencer. 32P-labeled DM2 oligomers were incubated with nuclear extracts prepared by uninduced and G-CSF–induced 32Dcl3Neo+ cells (Fig 5A) and with nuclear extracts prepared from uninduced and G-CSF–induced 32Dcl3/CDP/cut cells (Fig 5B). In contrast to 32Dcl3Neo+ cells, where G-CSF induction led to decreased binding of the CDP/cut protein to the DM2 oligomers, as indicated by the arrow (Fig 7A), the levels of CDP/cut bound to the DM2 oligomers remained high despite G-CSF induction of 32Dcl3 cells overexpressing CDP/cut (Fig 5B). This suggests that CDP/cut remains bound to the LF silencer element in G-CSF–induced 32Dcl3/CDP/cut cells, and this may contribute to the abrogation of G-CSF–induced LF expression in these cells.
EMSA of 32Dcl3Neo+ and 32Dcl3CDP/cut nuclear proteins bound to the DM2 LF silencer oligomers following G-CSF induction. (A) Double-stranded DM2 oligomers were [γ-32P]dATP–labeled and incubated with equal concentrations (15 μg) of nuclear extracts prepared from uninduced (lane 2) and G-CSF–induced day 2 (lane 3) and day 4 (lane 4) 32Dcl3Neo+ cells. Lanes 1 and 5, labeled DM2 oligomers without addition of any nuclear extracts. (B) [γ-32P]dATP–labeled DM2 oligomers were incubated with 15 μg/lane of nuclear extracts prepared from uninduced (lane 6), G-CSF–induced (day 2, lane 7), and G-CSF–induced (day 4, lane 8) 32Dcl3CDP/cut cells. Arrows in A and B represent the CDP/cut protein-DNA complex.
EMSA of 32Dcl3Neo+ and 32Dcl3CDP/cut nuclear proteins bound to the DM2 LF silencer oligomers following G-CSF induction. (A) Double-stranded DM2 oligomers were [γ-32P]dATP–labeled and incubated with equal concentrations (15 μg) of nuclear extracts prepared from uninduced (lane 2) and G-CSF–induced day 2 (lane 3) and day 4 (lane 4) 32Dcl3Neo+ cells. Lanes 1 and 5, labeled DM2 oligomers without addition of any nuclear extracts. (B) [γ-32P]dATP–labeled DM2 oligomers were incubated with 15 μg/lane of nuclear extracts prepared from uninduced (lane 6), G-CSF–induced (day 2, lane 7), and G-CSF–induced (day 4, lane 8) 32Dcl3CDP/cut cells. Arrows in A and B represent the CDP/cut protein-DNA complex.
Stable transfection of reporter plasmids in 32Dwt18.We next performed further reporter gene assays in an effort to correlate the silencing effect of CDP overexpression with a functional effect mediated through the identified CDP binding site. In general, the best in vitro model for the study of granulopoiesis has been the IL-3–dependent myeloid progenitor cell line 32Dcl3. This cell line undergoes terminal granulocytic differentiation upon removal of IL-3 and addition of G-CSF.33 40 However, because of the previously mentioned overwhelming cell death within the first 24 hours of IL-3 removal and G-CSF induction, both transient and stable transfection analysis in the induced 32Dcl3 cells proved difficult.
To overcome these difficulties, 32Dwt18 cells were again used for stable transfection of reporter gene plasmids. 32Dwt18 cells were stably cotransfected with the −916 bp and the −726 bp plasmids in conjunction with an expression vector carrying the gene for puromycin resistance. Stably transfected cells were selected in puromycin. Pools of stable transfectants were then induced with Epo. Measurement of luciferase reporter gene activity and viable cell counts were made following induction. A fivefold increase in luciferase activity was observed for the −916 bp plasmid following Epo induction (Fig 6A), as compared with the near-background levels measured for this plasmid in uninduced cells (data not shown). A corresponding increase in the levels of LF mRNA was detected by Northern blot analysis following induction of these 32Dwt18 cells with Epo (Fig 6B). The −726 bp plasmid was also inducible following Epo induction.
(A) Luciferase activity of stably transfected −916 bp and −916(d)S LF promoter plasmids in uninduced and Epo-induced (48 h) 32Dwt18 cells. The fold increase in reporter gene activity following Epo induction (fold induction) was calculated as luciferase activity + Epo per 1 × 106 viable cells/luciferase activity − Epo per 1 × 106 viable cells. Three independent experiments were performed; standard deviations are indicated by error bars. (B) Northern blot analysis of 32Dwt18 cells induced to undergo neutrophil maturation following Epo induction. Ten micrograms of total RNA from uninduced or 2-day Epo- and 4-day Epo-induced 32Dwt18 cells was electrophoresed on a 1% formaldehyde/agarose gel and blotted onto a nitrocellulose filter. The filter was first probed with [α-32P]dCTP–labeled LF cDNA (top) and later stripped and reprobed with an [α-32P]dCTP–labeled β-actin probe to ensure equal loading of RNA in each lane (bottom).
(A) Luciferase activity of stably transfected −916 bp and −916(d)S LF promoter plasmids in uninduced and Epo-induced (48 h) 32Dwt18 cells. The fold increase in reporter gene activity following Epo induction (fold induction) was calculated as luciferase activity + Epo per 1 × 106 viable cells/luciferase activity − Epo per 1 × 106 viable cells. Three independent experiments were performed; standard deviations are indicated by error bars. (B) Northern blot analysis of 32Dwt18 cells induced to undergo neutrophil maturation following Epo induction. Ten micrograms of total RNA from uninduced or 2-day Epo- and 4-day Epo-induced 32Dwt18 cells was electrophoresed on a 1% formaldehyde/agarose gel and blotted onto a nitrocellulose filter. The filter was first probed with [α-32P]dCTP–labeled LF cDNA (top) and later stripped and reprobed with an [α-32P]dCTP–labeled β-actin probe to ensure equal loading of RNA in each lane (bottom).
Of note, although expression of the −916 bp plasmid increased following G-CSF induction, the level of expression of this plasmid remained more than 10-fold lower than that of the −726 bp plasmid. The implications of this are discussed below. However, G-CSF induction of cells transfected with the −916(d)S plasmid expressed levels of luciferase comparable to those of the induced cells transfected with the −726 bp plasmid, suggesting that the deleted silencer-containing 56-bp element does not contain sequences required for upregulation of LF expression following G-CSF induction in 32Dcl3 cells.
DISCUSSION
We have identified a region within the 5′ regulatory sequences of the human LF gene that confers a silencing effect on the expression of reporter gene plasmids in non–LF-expressing cells. We have demonstrated that the silencing effect is abolished in 32Dwt18 cells upon induction of neutrophil maturation and concomitant LF expression with Epo. Negative regulatory elements (NREs) are often associated with tissue- and stage-specific control of gene expression, thereby ensuring repression of certain genes under inappropriate circumstances.41 A variety of NREs in a host of diverse genes have been described in hematopoietic cells. For example, repression of lysozyme gene expression is controlled by three negative regulatory sites prior to macrophage differentiation42; the silencing of κ gene expression in immature pre-B cells is also attributed to NREs.43 Of note, the overall homology at the nucleotide level between human and mouse LF promoters in this region is 82.5%, with conservation of the octamer repeats, as compared with an overall homology of 40% reported for the two promoters from −950 bp to the transcription start site.44 This high degree of conservation may reflect the functional importance of these sequences in regulating LF gene expression.
We have demonstrated that the LF NRE is recognized by CDP/cut. CDP/cut has been implicated in a number of systems as a transcriptional repressor of developmentally regulated genes29 (and references therein). CDP/cut was first described as a repressor of a sea urchin testis–specific H2B gene promoter.38 CDP/cut and related proteins, namely murine cux-1 and canine clox (cut-like homeobox), are members of a unique homeodomain family that includes the Drosophila cut protein, a regulator of cell-lineage specification.22,30,45,46 Members of this family of DNA binding proteins share both the homeodomain and three other motifs called cut repeats. Cut repeats share about 60% amino acid identity and exhibit specific DNA binding.47-49 Thus, CDP/cut proteins contain four putative DNA binding domains with overlapping but distinct sequence specificities.40 50 This may explain the lack of a single well-defined consensus DNA binding motif for CDP/cut proteins.
The role of CDP/cut has been demonstrated in the downregulation of a wide variety of genes, including the human myeloid-specific gp91-phox gene,29,39 human myc gene,51 human γ-globin gene,40 mouse NCAM gene,30 and mouse myosin heavy chain gene.45 However, the mechanism through which the CDP/cut protein mediates repression remains elusive.
The CDP/cut protein binds to the LF gene NRE only in non–LF-expressing cells. As has been demonstrated with the gp91-phox gene,29 39 we have established an inverse relationship between LF gene expression and CDP/cut binding to the LF NRE. The observation that removal of the CDP/cut binding site from the −916 plasmid does not block induction of expression from the LF reporter plasmid implies that although the binding of CDP/cut to the LF NRE may be necessary for repression of LF, it probably does not function by blocking the binding of positive regulators of LF gene expression.
However, it must be noted that the transfection data do not completely mirror the CDP overexpression data. It remains unclear why the −916 plasmid, although no longer silent, does not gain full expression upon G-CSF induction of transfected 32Dwt18 cells. Whether this reflects a more complex mechanism of regulation involving upstream sequences or represents a failure of the transfection approach itself is unclear. Similarly, although as noted, the results of deleting the silencer element appear to rule out the binding of a positive regulator at that site, it remains possible that deletion of the sequences does not give an accurate reflection of the conformation of the DNA in a chromosomal context. However, it seems clear that a loss of CDP/cut binding to the silencer does not account for high-level tissue-specific expression of LF, and that the primary function of CDP in this context may be to block inappropriate basal promoter activity in nonpermissive cells. With the exception of the NCAM gene, this observation has also been made for other CDP/cut-regulated genes such as gp91-phox and the γ-globin gene.40 In the case of gp91-phox, an increase in expression of this gene occurs independently of the binding of CP1, a member of the CCAAT binding enhancer family of positive transcription factors, whose binding site overlaps that of CDP/cut in the gp91-phox promoter.29 In fact, expression of a gp91-phox promoter-driven transgene lacking a CDP/cut/CP1 site is identical to that of the control plasmid, suggesting that CP1 is not essential for expression of the gp91-phox gene.52
Hence, how CDP/cut mediates repression of the LF gene is unclear. The mechanism of transcriptional repression in eukaryotes has been the subject of several recent reviews.41,53 In general, the mode of action of repressors falls into four broad categories. First, the repressor may prevent an activator protein from binding by competing for binding to the same DNA binding motif. An example of this has been proposed for the AP-1 transcription factor, which represses basal and retinoic acid–induced expression of the human osteocalcin gene by competing for overlapping DNA binding sites with the retinoic acid receptor.54 Second, some transcriptional activators can be “titrated” away from their DNA binding sites by inhibitory proteins with which they can form protein-protein complexes. For example, certain helix-loop-helix (HLH) transcription factors can be negatively regulated by the Id protein.55 Third, repressors can bind DNA near a DNA-bound activator and quench its activating capabilities, as the E2A and Jun gene products do upstream of the insulin gene.56 And fourth, repressors can interact directly with the basal transcriptional machinery and prevent genes from becoming transcriptionally active. For example, the Drosophila even-skipped protein has been shown to directly interact with TATA binding protein, an integral part of the basal transcription complex.57 Our studies make the first and second possibilities unlikely. However, the third and fourth possibilities remain potential mechanisms for LF repression by CDP/cut.
Overexpression of the CDP/cut protein in 32Dcl3 cells inhibits LF expression following G-CSF induction without affecting the morphologic changes associated with normal G-CSF–induced myeloid differentiation in these cells. This observation parallels that observed for the gp91-phox gene, where overexpression of CDP/cut in stably transfected HL60 cells induced toward the myeloid lineage resulted in abolition of gp91-phox expression at the mRNA level.29 The decrease in the level of LF mRNA following CDP/cut overexpression in 32Dcl3/CDP/cut cells may be the result of persistent binding of the CDP/cut protein to the LF silencer element. We hypothesize that increased LF expression following G-CSF induction of 32Dcl3 cells is the result of upregulation of yet-to-be-identified transcriptional activating proteins. Since overexpression of CDP/cut in these cells appears to block G-CSF–induced LF expression, it is suggested that CDP/cut interferes in some way with the positive regulation of LF expression. However, our data show that removal of the CDP/cut binding site in the LF gene promoter does not affect upregulation of LF following G-CSF induction, suggesting that the G-CSF–induced transactivators do not compete directly with CDP/cut for binding to the silencer element. How then does CDP/cut overexpression block induction of LF expression? One possibility is that CDP/cut competes with other sites within the LF promoter that bind the putative LF transactivators; for example, there are two CCAAT boxes in the LF promoter that could represent potential CDP binding sites, although preliminary transfection data do not support this. Binding of CDP to multiple sites has been observed in the gp91-phox gene, where CDP/cut binds four different sites within the promoter with different binding affinities.58 Alternatively, CDP/cut may regulate expression of the putative LF transactivators themselves, thereby indirectly influencing LF expression. Inhibition of LF expression in 32Dcl3 cells overexpressing CDP/cut provides further evidence for its function as a repressor; however, retention of the ability to undergo phenotypic maturation argues that CDP/cut is not affecting neutrophil differentiation itself.
Another question raised by our findings is how CDP/cut controls the repression of two myeloid-specific genes, gp91-phox and LF, that are expressed at markedly different points during neutrophil maturation. The evidence presented here does not directly address this question. However, loss of CDP/cut binding clearly precedes LF expression in developing granulocytes. It is possible that basal LF promoter activity begins before the myelocyte stage during differentiation but results in a level of expression that is below the limits of detectability by Northern blot analysis. This would be consistent with the data reported for deletion of the CDP/cut binding site from mice carrying a transgene in which the growth hormone gene was driven by the gp91-phox promoter. Mice carrying a transgene with a promoter containing a deleted CDP/cut binding site had no detectable ectopic growth hormone expression by immunohistochemistry. However, they were three times the size of control animals in which the transgene contained the CDP/cut binding site. This suggests that inappropriate basal promoter activity resulted from the loss of CDP/cut binding.52
Alternatively, differential timing of expression of gp91-phox and LF during myeloid differentiation could reflect functional heterogeneity of CDP/cut itself. Alternately spliced forms of the CDP/cut protein have been described29 that may result in expression of a family of CDP/cut proteins capable of differentially regulating gene expression. Alternatively, CDP/cut has been described to be a phosphoprotein,48 and its state of phosphorylation may have differential effects on its binding to different promoters, especially in view of the wide sequence variation in different CDP/cut binding sites. It has recently been reported that in vitro phosphorylation of the cut repeats by protein kinase C inhibited the DNA binding ability of the CDP/cut protein.59
In summary, we have identified a NRE within the LF gene promoter that silences expression of transfected reporter plasmids. The LF silencer element is bound by CDP/cut in non–LF-expressing cells. Maturation past the promyelocyte stage results in decreased binding of CDP/cut to the silencer element concomitant with high levels of LF expression. This observation, taken together with the finding that deleting the silencer element has no effect on high levels of LF expression, suggests that binding of CDP/cut to the LF silencer element is involved in repression of LF expression in nonpermissive cells, and that loss of CDP/cut binding may be necessary but not sufficient for high-level LF expression during myelopoiesis. The loss of LF expression as a result of CDP/cut overexpression confirms that CDP/cut functions as a repressor. What factor(s) are necessary to drive high levels of LF gene expression in the maturing neutrophil remains unclear.
ACKNOWLEDGMENT
We are grateful to Dr Archibald Perkins, Dr Bernard Forget, and Dr Daniel Tenen for insightful discussions and provision of reagents during the course of this study. We also thank Dr Fridbjorn Sigurdsson for assistance with the figures.
Supported by Grant No. DK42347 from the National Institutes of Health (to N.B.). N.B. is a Scholar of the Leukemia Society of America.
Address reprint requests to Nancy Berliner, MD, Department of Internal Medicine, Section of Hematology, WWW-428, Yale University School of Medicine, 333 Cedar St, New Haven, CT 06510.

![Fig. 2. (A) Dissection of coordinates −858 bp to −810 bp containing the LF silencer element into oligomers DM (double module) 1, 2, and 3. Each oligomer (except DM3) contains 2 of the 3 octamer repeats (overlined) that comprise the LF silencer element. (B) UV cross-linking analysis of proteins bound to the DM2 oligomers. DM2 oligomers were [γ-32P]dATP-labeled and incubated with equal concentrations of nuclear extracts (N.E.), with (+) or without (−) a 100-fold molar excess of unlabeled DM2 oligos, prepared from K562 cells (lanes 2 and 3), uninduced 32Dcl3 cells (lanes 4 and 5), G-CSF–induced 32Dcl3 cells (lanes 6 and 7), MPRO cells (lanes 8 and 9), and MPRO + ATRA cells (lanes 10 and 11). The protein-DNA complexes formed were subjected to UV light at 254 nm for 30 minutes before electrophoresis in a 7.5% SDS-polyacrylamide gel. The position of prestained protein molecular weight markers is indicated. F, unbound probe. Lane 1, probe without nuclear extract.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/7/10.1182_blood.v90.7.2784/3/m_bl_0022f2.jpeg?Expires=1769090887&Signature=Mx3zBj8eQ6gwdNTdcdBf6lZ-24r5mjewKaJKUNAh2q0YI3gjqZrWtez2IQpI5tONt28eB9TYyB7-RAQO5GpSPLBUcrit0jzyoUkOil38~SDd5mmLxKFPG1ZEh5BfBE3sq6usq9zyUEJsdtgpnTwXjlFD3TuUwVfxhQABR7lGlJtaUR834ww0Vj6~8pXsVTzKKziwScIRqXZrTvUs3scNdvIZFKhNIlk81cHpaw4xJQtkhg3REB88bpT9JdhfZtgzDYWdMmCUcWy9akoFGdmpWd9t0ti1cgJw-91IOIulbOpLSNnYJlXRLU04gPeeMI3F--cB~QBxIgDtnYKn~b~XkQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. (A) EMSA analysis of K562 nuclear proteins bound to DM2 oligomers in the presence of NCAM and mutant NCAM cold competitor oligomers. [γ-32P]dATP-labeled DM2 oligomers were incubated with K562 nuclear extracts in the presence of a 100-fold molar excess of cold DM2 oligomers (lane 3), NCAM oligomers (lane 4), and mutant NCAM oligomers (lane 5). Lane 2, K562 nuclear extracts with no added competitor (No comp.). The DM2 probe was run without any nuclear extracts in lane 1 (DM2 probe). (B) Supershift EMSA analysis of K562 nuclear proteins bound to DM2 oligomers in the presence of an anti-CDP/cut antibody. In the left panel, [γ-32P]dATP-labeled DM2 oligomers were complexed with nuclear proteins from K562 cells (lane 2) and the DNA-protein complexes exposed either to preimmune serum (lane 3) or to anti-CDP/cut antibody (lane 4). In the right panel, NCAM oligomers were labeled with [γ-32P]dATP and allowed to bind to nuclear proteins from K562 nuclear extracts. The resulting protein-DNA complexes were exposed either to preimmune serum (lane 6) or to anti-CDP/cut antibody (lane 7). Lane 1, DM2 oligomers without nuclear extract.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/7/10.1182_blood.v90.7.2784/3/m_bl_0022f3.jpeg?Expires=1769090887&Signature=jZsXMLR3QZneNNjcGOUQOLpCoFXEupqsbOL62VA5bIOJKaVZGDlNWrOkLEpxPE4cMoh0uGg1Gm4DU64qFyjiSzsSzCTo1MY7S93iXtaJiVlXF1yd0dZmgg-PREj6Y2anF063sBOp4elDBPzcYeO~d9rZ6y36rs6FHsX7YL0KhyNT0pWOSk8LJhXMl0ufro-IpHuWwN4SbV5gf-P5v72-sxYAG2P07PLkAVX6GTMy5dHedcKK8esIacfYUZ9djUf2HJnkkYiS~LLB2nYDEJwjPhwItNh1Sl44J9~yDG~ngNeJVXn6BKPNJblaSkilwIFdKIeh4QRVI~ho2-EwWzpBWA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Western and Northern blot analysis of G-CSF–induced 32Dcl3 cells stably transfected with the pMT2-CDP/cut plasmid. (A) Western blot using anti-CDP antiserum confirming overexpression of CDP/cut in CDP-transfected 32Dcl3 cells before (0) and after (4d) G-CSF induction. 32Dwt18/CDP/cut cells were used as a positive (+) control. Each lane contains 100 μg nuclear protein extracts. (B) Ten micrograms each of total RNA isolated from uninduced or 2-day and 4-day G-CSF–induced 32Dcl3 cells stably cotransfected with pMT2-CDP/cut and pSVneo (lanes 3, 4, and 5, respectively) plasmids or pSVneo plasmid alone (uninduced, lane 1; 4-day G-CSF–induced, lane 2) was electrophoresed on a 1% formaldehyde/agarose gel and blotted onto a nitrocellulose filter. The filter was probed with an [α-32P]dCTP–labeled LF cDNA probe. The position of 28S and 18S rRNA markers is indicated. LF mRNA is denoted by the arrowhead. To ensure equal loading of RNA in each lane, the blot was stripped and reprobed with an [α-32P]dCTP–labeled β-actin probe, as indicated in the bottom panel.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/7/10.1182_blood.v90.7.2784/3/m_bl_0022f4.jpeg?Expires=1769090887&Signature=z-pgaipnK0tBNux7NWt2N4MShvXVlJk~stS3Is9wQvJkep6ytf2CotiEmQdaEyH1HDHo6Tbjo5wTAgqHDHDvSYvIbE5xbOVaP8hivAW1zTjjLqoEpwvmITZjw-wo-gZwDnl7HgceOEACHjnPcPpuqAxEPeVhwyC5~ADKgTu05ccZfkCL94AbPjzvmHMVC~AxCEKbW7qg5VodEGVEQcVggTDWrfQMnffKVhtARvhzVsYb0l8klacR4sAy3G8mgxEHdRYddJlw476O9v0W2gyOsmb7mimOjRjyNPlw18wexrr3hU-7rvPT-U6XYrM9T2oycy93VTU6jQqTGry5KUNoLg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. EMSA of 32Dcl3Neo+ and 32Dcl3CDP/cut nuclear proteins bound to the DM2 LF silencer oligomers following G-CSF induction. (A) Double-stranded DM2 oligomers were [γ-32P]dATP–labeled and incubated with equal concentrations (15 μg) of nuclear extracts prepared from uninduced (lane 2) and G-CSF–induced day 2 (lane 3) and day 4 (lane 4) 32Dcl3Neo+ cells. Lanes 1 and 5, labeled DM2 oligomers without addition of any nuclear extracts. (B) [γ-32P]dATP–labeled DM2 oligomers were incubated with 15 μg/lane of nuclear extracts prepared from uninduced (lane 6), G-CSF–induced (day 2, lane 7), and G-CSF–induced (day 4, lane 8) 32Dcl3CDP/cut cells. Arrows in A and B represent the CDP/cut protein-DNA complex.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/7/10.1182_blood.v90.7.2784/3/m_bl_0022f5.jpeg?Expires=1769090887&Signature=Q0WkL4j30dhjDAVyt6fsbAfGkDrs94E6c3NLcWc-ZwgWgAhMf1uCkitBFAGInWiK9ivpcKa9BpZvSYC~PLXCviF3Ce3OnNrFEoWydR8isK55j~m0oMs1ccnW1M2zvLi2ea3IWRpHWoi8ppf4YOF6Qt~6lFA9zWjQh~fCk5~auUzcAXDWYc8kqNObuSVg0JDrYnYoBbz4Ke1wvtxy6Z1PGzPFyrSDwkRLngndFHnh1JVA2X88EF6gx84PuvEgAB2n5ZZN~kf-Ef9EbZfsQ45jvjGynGN~Eppx8VvwfJ9wqntUI5OBtu2A6jJ6hHtsfDFIQh8nf2K-4fKYLMVhBBMwow__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. (A) Luciferase activity of stably transfected −916 bp and −916(d)S LF promoter plasmids in uninduced and Epo-induced (48 h) 32Dwt18 cells. The fold increase in reporter gene activity following Epo induction (fold induction) was calculated as luciferase activity + Epo per 1 × 106 viable cells/luciferase activity − Epo per 1 × 106 viable cells. Three independent experiments were performed; standard deviations are indicated by error bars. (B) Northern blot analysis of 32Dwt18 cells induced to undergo neutrophil maturation following Epo induction. Ten micrograms of total RNA from uninduced or 2-day Epo- and 4-day Epo-induced 32Dwt18 cells was electrophoresed on a 1% formaldehyde/agarose gel and blotted onto a nitrocellulose filter. The filter was first probed with [α-32P]dCTP–labeled LF cDNA (top) and later stripped and reprobed with an [α-32P]dCTP–labeled β-actin probe to ensure equal loading of RNA in each lane (bottom).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/7/10.1182_blood.v90.7.2784/3/m_bl_0022f6.jpeg?Expires=1769090887&Signature=RaAzpb59B1uRWLYWqhx74bF2dbcHPrQNF8aJoGJwNiVgna~ysOowvCMjW8UhNTUu3vnDuq5m6~cgannbPmuOKqB2LC6Gky35t3Bx8nUcVgDJJ2KrwJWkrz-2aRQIQj7H3qyhqOlgic2Dm36~C1fBPeyX3GF8FogyKy92~e4y~LG4jYrXlagwbh3qqgCTI-tz5bVoZ8kulsqLEaXfy3IA83TWEY8GNh6XY~U0VL~hfNTVvYiZsYGfTQoOeV-u3FaT7ofguXyrnZmkz2bSw7Lf7qiUx~UG-2iSi2uu6y3XizjDh~m0acLKschjDjEL-~KtxQXV3Zzh~fNg1oeBvjZ8lg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal