Abstract
One of the diagnostic criteria of essential thrombocythemia (ET) is the absence of the Philadelphia chromosome (Ph-neg). On the molecular level, Ph-neg ET patients may carry BCR-ABL transcript. The natural history of BCR-ABL positive Ph-neg ET patients is undetermined. We examined the BCR-ABL status by reverse transcriptase two-step nested polymerase chain reaction in bone marrow aspirates of 25 Ph-neg ET patients. We found 12 BCR-ABL positive and 13 BCR-ABL negative patients in the study group. The comparison showed that the two groups had similar clinical and laboratory characteristics, except for a significant increased patients' age and decreased polymorphonuclear cell count in the BCR-ABL positive group. During a median follow-up of 20 and 22.5 months for the BCR-ABL negative and positive groups, respectively, there was neither blastic transformation nor unrelated death in both groups. We conclude that it is important to look for BCR-ABL transcript in Ph-neg ET patients and to follow them closely to investigate the nature of this translocation in this group of patients.
ESSENTIAL thrombocythemia (ET) is a clonal disorder that originates from a multipotent stem cell1 and is characterized by extreme thrombocytosis for which neither a primary cause nor diagnostic signs of other myeloproliferative disorders can be found.2 The criteria for this indolent3 disease established by The Polycythemia Vera Study Group (PVSG)2 include the absence of the Philadelphia chromosome, ie, Ph-neg. This chromosome results from the reciprocal translocation between the long arm of chromosomes 9 and 22 t(9:22) (q34:q11).4 This translocation disrupts the normal ABL and BCR genes in chromosomes 9 and 22, respectively, giving rise to a chimeric BCR-ABL transcript mRNA encoding a fusion p210 kD protein with transforming ability.5,6 It has been suggested that the abnormal hybrid protein is involved in the control of cell growth, probably transducing continuous proliferation signals to the cells,7 or by inhibiting programmed cell death.8 The absence of the Ph chromosome predominantly helps to distinguish ET from chronic myeloid leukemia (CML) with thrombocytosis. Recently, a small group of patients with the clinical picture of ET with Ph chromosome in their marrow karyotype has been described.9-11 Their clinical presentation, natural history, and grave prognosis due to progression to blastic crisis just as in CML patients, justifies an increased effort to diagnose these patients as early as possible.
We have initiated a study aimed at further characterization of Ph-neg ET patients. We analyzed the molecular profile of these patients with respect to the BCR-ABL status of bone marrow aspirates in Ph-neg ET patients and tried to establish the relation between positive transcripts and clinical presentation.
MATERIALS AND METHODS
Patients.Twenty-seven patients, 3 newly diagnosed and 24 follow-up patients (range, 8 to 87 months), who met the PVSG criteria for ET were enrolled in the study. On presentation, detailed medical history was taken followed by a complete physical examination. The laboratory assessment on admission included complete blood counts and peripheral blood smears with differential leukocyte count, leukocyte alkaline phosphatase (LAP) score, lactate dehydrogenase (LDH), and vitamin B12 levels were obtained. Bone marrow aspiration and biopsy were performed in all of the patients and evaluated for histologic features and cytogenetics.
During the 10-month study period (June 1995 to March 1996), the patients underwent repeated bone marrow aspiration, which was submitted for cytogenetic examination and molecular analysis. The 24 follow-up patients were treated with hydroxyurea.
Isolation of mononuclear cells and RNA extraction.Mononuclear cells were prepared from 3 mL of bone marrow aspirates layered on Ficoll-Hypaque density gradient (Sigma, St Louis, MO) and centrifuged at 1,600 rpm for 20 minutes. The interface cell layer was collected and washed in RPMI media (Biological Industries, Kibbutz Beit Haemek, Israel). Total cellular RNA was obtained from these cells by modification of the guanidium thiocyanate/phenol/chloroform extraction technique using the TRI-Reagent (Molecular Research Central Inc, Cincinnati, OH) as described by Chomczynski.12 The obtained RNA pellet was dried and dissolved in ribonuclease-free water. RNA was quantified spectrophotometrically at 260 nm. Total RNA integrity was checked by electrophoresis on denaturing agarose.
Reverse transcriptase two-step nested polymerase chain reaction (RT-PCR).Three micrograms of total RNA were reverse transcribed into complementary DNA (cDNA) using 0.5 μg oligo(dT) 12-18 primer and 200 U of Superscript reverse transcriptase II (GIBCO/BRL, Grand Island, NY) in a total volume of 20 μL according to the recommendation of the manufacturer. This served as the starting template for the outer primer polymerase reaction. Each patient's sample was transcribed in two tubes. PCR was performed by using the same sets of primer sequences as described by Hughes et al.13 cDNA was amplified by 2.5 U of Taq Polymerase (Appligene, Strasbourg, France). In one tube, the reaction was performed in the presence of outer primers (A and B), which amplify cDNA derived from b3a2 and b2a2 transcripts of BCR-ABL mRNA. The other tube was used for quality control of RNA and successful PCR amplification and was performed in the presence of primers A and C amplifying the ABL product. One tenth of the first BCR-ABL PCR product was reamplified in a second PCR reaction, which used nested inner primers D and E. A total of 10 μL of this PCR product was then electrophoresed through 2% ethidium bromide agarose gel, visualized, and photographed under ultraviolet light. In some of the cases, DNA fragments were transferred onto a positively charged Nylon membrane (GeneScreen Plus, Dupont, NEN Research Products, Boston, MA) using an alkaline transfer protocol by the method of Southern.14 Membranes were hybridized with 5′-end fluorscein-labeled DNA probes for b3a2 and b2a2, as described by Kawasaki et al.15 A chemiluminescent detection kit (Tropix Inc, Bedford, MA) was used for detection.
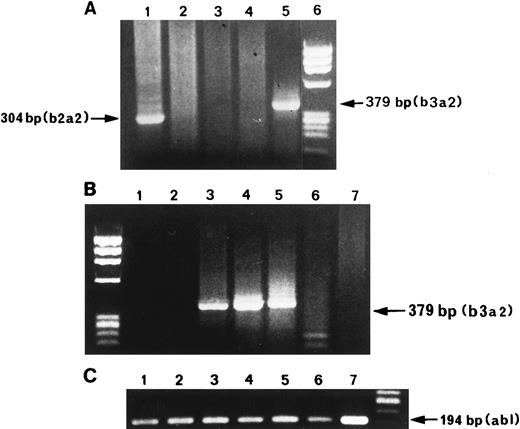
RT-PCR products of BCR-ABL mRNA amplification of ET Ph-neg patients. (A) Lane 1, patient with b2a2 trascript; lanes 2 and 3, patients without BCR-ABL trascripts; lane 4, HL-60 negative control; lane 5, positive control extracted from K562 cells diluted 1:105 with HL-60 negative cells; lane 6, molecular marker. PCR products of b3a2 trascript (B) and ABL (C) of five bone marrow samples. Lanes 1, 2, and 6, patients without BCR-ABL transcript; lane 3, positive control; lanes 4 and 5, positive patients; lane 7, HL-60 negative control.
RT-PCR products of BCR-ABL mRNA amplification of ET Ph-neg patients. (A) Lane 1, patient with b2a2 trascript; lanes 2 and 3, patients without BCR-ABL trascripts; lane 4, HL-60 negative control; lane 5, positive control extracted from K562 cells diluted 1:105 with HL-60 negative cells; lane 6, molecular marker. PCR products of b3a2 trascript (B) and ABL (C) of five bone marrow samples. Lanes 1, 2, and 6, patients without BCR-ABL transcript; lane 3, positive control; lanes 4 and 5, positive patients; lane 7, HL-60 negative control.
Clinical Data at Diagnosis of the Two Groups of Ph-neg ET Patients
| . | Group A . | Group B . | P Value . |
|---|---|---|---|
| . | BCR-ABL Negative . | BCR-ABL Positive . | . |
| . | n = 13 . | n = 12 . | . |
| Male/female | 8/5 | 4/8 | |
| Age (yr) | 52.1 ± 16.8 | 64.5 ± 10.4 | 0.04 |
| Range | 28-81 | 45-82 | |
| Follow-up (mo) | |||
| Median | 20 | 22.5 | |
| Range | 8-87 | 17-63 | |
| Symptoms | |||
| Dizziness | 4 | 5 | |
| Bleeding | 2 | 1 | |
| Thrombosis | 0 | 1 | |
| Tinitus | 1 | 0 | |
| Spleen | |||
| Unpalpable | 8 | 10 | |
| Palpable | 5 | 2 | |
| <3 cm | 2 | 1 | |
| ≥3 cm | 3 | 1 |
| . | Group A . | Group B . | P Value . |
|---|---|---|---|
| . | BCR-ABL Negative . | BCR-ABL Positive . | . |
| . | n = 13 . | n = 12 . | . |
| Male/female | 8/5 | 4/8 | |
| Age (yr) | 52.1 ± 16.8 | 64.5 ± 10.4 | 0.04 |
| Range | 28-81 | 45-82 | |
| Follow-up (mo) | |||
| Median | 20 | 22.5 | |
| Range | 8-87 | 17-63 | |
| Symptoms | |||
| Dizziness | 4 | 5 | |
| Bleeding | 2 | 1 | |
| Thrombosis | 0 | 1 | |
| Tinitus | 1 | 0 | |
| Spleen | |||
| Unpalpable | 8 | 10 | |
| Palpable | 5 | 2 | |
| <3 cm | 2 | 1 | |
| ≥3 cm | 3 | 1 |
Laboratory Data at Diagnosis of the Two Groups of Ph-neg ET Patients
| . | Group A . | Group B . | P Value . | ||
|---|---|---|---|---|---|
| . | BCR-ABL Negative . | BCR-ABL Positive . | . | ||
| . | n = 13 . | n = 12 . | . | ||
| . | Mean ± SD . | Range . | Mean ± SD . | Range . | . |
| Hemoglobin (g/dL) | 13.1 ± 2.1 | 10-15.9 | 13.6 ± 1.9 | 10.4-16.7 | 0.36 |
| Leukocytes (×109/L) | 9.5 ± 2.7 | 6.2-15.7 | 9.90 ± 2.1 | 6.9-14.1 | 0.67 |
| PMN (%) | 71.9 ± 5.3 | 66-86 | 61.0 ± 14.5 | 38-89 | 0.03 |
| Lymph (%) | 19.0 ± 5.8 | 8-25 | 24.9 ± 8.8 | 8-28 | 0.06 |
| Mono (%) | 5.3 ± 2.6 | 2-8 | 8.2 ± 4.8 | 2-19 | 0.08 |
| Eos (%) | 2.8 ± 2.3 | 1-6 | 5.2 ± 4.5 | 1-16 | 0.12 |
| Baso (%) | 0.31 ± 0.5 | 0-1 | 0.33 ± 0.5 | 0-1 | 0.90 |
| Platelets (×109/dL) | 781 ± 267 | 623-1370 | 756 ± 185 | 626-1170 | 0.78 |
| LAP score | 84 ± 58 | 27-207 | 58 ± 31 | 26-112 | 0.19 |
| LDH (U/L) | 484 ± 254 | 172-950 | 406 ± 146 | 163-630 | 0.36 |
| Vit B12 (pmol/L) | 410 ± 116 | 183-555 | 331 ± 119 | 171-580 | 0.13 |
| . | Group A . | Group B . | P Value . | ||
|---|---|---|---|---|---|
| . | BCR-ABL Negative . | BCR-ABL Positive . | . | ||
| . | n = 13 . | n = 12 . | . | ||
| . | Mean ± SD . | Range . | Mean ± SD . | Range . | . |
| Hemoglobin (g/dL) | 13.1 ± 2.1 | 10-15.9 | 13.6 ± 1.9 | 10.4-16.7 | 0.36 |
| Leukocytes (×109/L) | 9.5 ± 2.7 | 6.2-15.7 | 9.90 ± 2.1 | 6.9-14.1 | 0.67 |
| PMN (%) | 71.9 ± 5.3 | 66-86 | 61.0 ± 14.5 | 38-89 | 0.03 |
| Lymph (%) | 19.0 ± 5.8 | 8-25 | 24.9 ± 8.8 | 8-28 | 0.06 |
| Mono (%) | 5.3 ± 2.6 | 2-8 | 8.2 ± 4.8 | 2-19 | 0.08 |
| Eos (%) | 2.8 ± 2.3 | 1-6 | 5.2 ± 4.5 | 1-16 | 0.12 |
| Baso (%) | 0.31 ± 0.5 | 0-1 | 0.33 ± 0.5 | 0-1 | 0.90 |
| Platelets (×109/dL) | 781 ± 267 | 623-1370 | 756 ± 185 | 626-1170 | 0.78 |
| LAP score | 84 ± 58 | 27-207 | 58 ± 31 | 26-112 | 0.19 |
| LDH (U/L) | 484 ± 254 | 172-950 | 406 ± 146 | 163-630 | 0.36 |
| Vit B12 (pmol/L) | 410 ± 116 | 183-555 | 331 ± 119 | 171-580 | 0.13 |
Normal values: LAP score = 15-110; LDH = 230-460 U/L; Vit B12 = 148-700 pmol/L.
Abbreviations: PMN, polymorphonuclear cells; lymph, lymphocytes; mono, monocytes; eos, eosinophils; baso, basophils; LAP, leukocyte alkaline phosphatase; LDH, lactic dehydrogenase; Vit B12, vitamin B12.
Detection limit controls.The detection limit of the method was established by adding 100 K562 cells expressing BCR-ABL transcript (b3a2) to 107 nonexpressing HL-60 cells. Total RNA was extracted and was used as a positive control in all experiments. As a negative control, we used RNA extracted from HL-60 cells or healthy normal volunteers, and a blank control was added to eliminate the possibility of reagent contamination. The recommendations of Kwok and Higushi16 were followed. Each positive result was repeated independently two to three times. Negative results were regarded as true only when a successful ABL PCR product was demonstrated.
Cytogenetic analysis.Mononuclear marrow cells were separated as described above and cultured at a concentration of 1 × 106 /mL in RPMI 1640 medium with 20% fetal calf serum, 10 g/mL L-glutamine, 10 g/mL penicillin and streptomycin. Cultures were incubated for 24 to 72 hours at 37°C. Cells were harvested according to a standard protocol. Metaphase preparations were G-banded using the method of Seabright17 and analyzed according to the international system for human cytogenetic nomenclature (ISCN) guidelines.18 A total of 20 to 30 metaphases were analyzed.
Statistical analysis.We used the two-sided Student's t-test to compare the means of continuous variables examined in our study. P values less than .05 were considered statistically significant.
RESULTS
Twenty-seven patients were enrolled in the study, two of whom were excluded because of failed bone marrow aspiration. All aspirates were negative for the Philadelphia chromosome at the cytogenetic level.
Two groups of Ph-neg ET patients were identified on the basis of RT-PCR for BCR-ABL transcripts. Group A consisted of 13 (52%) patients in whom bone marrow aspirates were negative for rearrangements within the major breakpoint cluster region (M-bcr, ie, BCR-ABL negative); and group B consisted of 12 (48%) patients who were BCR-ABL positive (Fig 1). In group B, a junction between the third exon (b3) of M-bcr and the second exon (a2) of ABL was shown in eight cases (66.6%) and between the second exon (b2) of M-bcr and a2 in four cases (33.3%) (Fig 1). Some of the patients (PCR positive and negative) were tested repeatedly after 6 to 12 months from the first PCR. Specificity of the PCR products was confirmed by Southern blot using b3a2 and b2a2 specific DNA probes.
Table 1 lists the clinical characteristics and Table 2 lists the laboratory data of the two groups of patients at diagnosis. The comparison of the tested variables shows that the two groups had similar clinical characteristics, as well as laboratory results except for a significant increase in the polymorphonuclear (PMN) cell counts accompanied by a decrease in lymphocytes and monocytes in group A. It should be noted that the LAP score of all patients was within the normal range.
During a median follow-up of 20 and 22.5 months for groups A and B, respectively, there were neither blastic transformations nor unrelated deaths in the respective two groups of patients.
DISCUSSION
Stoll et al10 observed that less than 10% of patients with the clinical features of ET may have the Philadelphia chromosome in their myeloid cells, which is associated with a poor prognosis.11 Some of the patients, described in small series during the last few years, had several combinations of CML features including leukocytosis, basophilia, low LAP score, or immature myeloid cells in peripheral blood.9-11
The molecular analysis of the bone marrow aspirates of our ET patients showed that about half of these patients carried the BCR-ABL transcripts in their mononuclear cells. We did not identify which cell lineage expresses BCR-ABL transcripts, but in CML, it is conceived that the translocation originates in a pluripotent stem cell involving both myeloid and lymphoid linages.19 Our BCR-ABL positive group was clinically indistinguishable from the group of BCR-ABL negative ET patients.
The results raise the possibility that BCR-ABL positive ET patients are, in fact, CML patients. We assume that this is not the case because (1) the BCR-ABL positive group had normal LAP scores on admission without any evidence of inflammation, infection or therapy, while 91% of CML patients had low or abscent LAP score20; (2) there was no associated basophilia, in contrast to CML patients in whom basophilia is virtually always present21; (3) the majority had no increase in spleen size, whereas splenomegaly is present in about 90% of CML patients at diagnosis22; (4) there was no blastic transformation or unrelated death in the BCR-ABL positive group during a median follow-up of 22.5 months, which contrasts the onset of blastic crisis in CML patients.23
It was recently reported that 22 of 73 (30.1%) healthy adults carry the BCR-ABL transcript in their peripheral blood,24 which is quite close to the rates (48%) of BCR-ABL transcripts in the bone marrow aspirates of our ET study group. It could be that by adopting the method used by Biernaux et al,24 which is much more sensitive (1:108 cells) than the one used in our laboratory (1:105), in patients with a myeloproliferative disorder, a higher rate of BCR-ABL positive transcripts may be discovered. This would suggest that the incidence of BCR-ABL transcript found in our study is not a reflection of the incidence found in the general population.
Using the fluorescence in situ hybridization (FISH) technique in adjunct to the cytogenetic analysis may detect complex and subtle rearrangements of Ph translocation (“masked” Ph — chromosome) in 3.4% to 15% of CML cases.25 Because our group was small, the FISH technique would have omitted from the data a negligible part of the patients, which would not change the concept presented herein.
It is assumed that random chromosomal aberrations increase with age. However, in our study, the significantly older age of patients in the BCR-ABL positive group was associated with an otherwise normal karyotype except the BCR-ABL fusion gene, which was the only translocation searched for. Thus, the question whether the frequency of BCR-ABL positive status in ET patients is age-dependent remains to be further investigated.
We believe that a longer study follow-up period is needed to observe the sequence of events leading to the appearance of BCR-ABL transcripts and its contribution to the patients' prognosis.
The observation reported herein could have implications on treatment modalities in ET, and the role of interferon treatment in this group should be assessed. In addition, the results may raise the possibility of defining a new variant of ET whose significance is yet to be explored.
In conclusion, we suggest that the BCR-ABL status be examined in ET patients and that they be followed closely to understand the nature of our observation and its importance.
Address reprint requests to Adina Aviram, PhD, Division of Hematology, Rabin Medical Center, Beilinson Campus, 49100 Petah-Tikva, Israel.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal