Abstract
Human C/EBPε is a newly cloned CCAAT/enhancer-binding transcription factor. Initial studies indicated it may be an important regulator of human myelopoiesis. To elucidate the range of expression of C/EBPε, we used reverse transcription-polymerase chain reaction (RT-PCR) analysis and examined its expression in 28 hematopoietic and 14 nonhematopoietic cell lines, 16 fresh myeloid leukemia samples, and normal human hematopoietic stem cells and their mature progeny. Prominent expression of C/EBPε mRNA occurred in the late myeloblastic and promyelocytic cell lines (NB4, HL60, GFD8), the myelomonoblastic cell lines (U937 and THP-1), the early myeloblast cell lines (ML1, KCL22, MDS92), and the T-cell lymphoblastic leukemia cell lines CEM and HSB-2. For the acute promyelocytic leukemia cell line NB4, C/EBPε was the only C/EBP family member that was easily detected by RT-PCR. No C/EBPε mRNA was found in erythroid, megakaryocyte, basophil, B lymphoid, or nonhematopoietic cell lines. Most acute myeloid leukemia samples (11 of 12) from patients expressed C/EBPε. Northern blot and RT-PCR analyses showed that C/EBPε mRNA decreased when the HL60 and KG-1 myeloblast cell lines were induced to differentiate toward macrophages. Similarly, Western blot analysis showed that expression of C/EBPε protein was either unchanged or decreased slightly as the promyelocytic cell line NB4 differentiated down the macrophage-like pathway after treatment with a potent vitamin D3 analog (KH1060). In contrast, C/EBPε protein levels increased dramatically as NB4 cells were induced to differentiate down the granulocytic pathway after exposure to 9-cis retinoic acid. Furthermore, very early, normal hematopoietic stem cells (CD34+/CD38−), purified from humans had very weak expression of C/EBPε mRNA, but levels increased as these cells differentiated towards granulocytes. Likewise, purified granulocytes appeared to express higher levels of C/EBPε mRNA than purified macrophages. Addition of phosphothiolated antisense, but not sense oligonucleotides to C/EBPε, decreased clonal growth of HL-60 and NB4 cells by about 50% compared with control cultures. Taken together, our results indicate that expression of C/EBPε is restricted to hematopoietic tissues, especially myeloid cells as they differentiate towards granulocytes and inhibition of its expression in HL-60 and NB4 myeloblasts and promyelocytes decreased their proliferative capacity. Therefore, this transcriptional factor may play an important role in the process of normal myeloid development.
WE1 AND OTHERS2 have cloned a novel human gene called C/EBPε, which belongs to the CCAAT/enhancer-binding protein gene family. This family includes C/EBP-α,C/EBP-β (NFIL6, LAP, IL6-DBP, AGP/EBP-β), C/EBP-δ (NFIL6B, CRP3), C/EBP-γ (IG/EBP-1), GADD153 (CHOP) and two homologous genes, CRP1 and HS8, whose expression has not been detected.3-11 These proteins have a bipartite DNA-binding domain composed of a positively charged basic region that contacts the DNA and an amphipathic helix in the conserved C-terminus that mediates dimerization through formation of a “leucine zipper.”12 The less well-conserved amino-terminal portion of these proteins contains regulatory and transactivation domains. The initial studies indicate that C/EBPε is a strong candidate for regulating human myelopoiesis.1 2
The C/EBP proteins are implicated in the differentiation process of a variety of mammalian cells, especially adipocytes and hepatocytes. They may also play important roles in myeloid cell differentiation. The C/EBP-α protein is absent in 3T3-L1 preadipocytes, but its expression increases during terminal differentiation of the cells into nondividing adipocytes.4,13 In addition, C/EBP-α is expressed at low levels in hepatoma cell lines, but is highly expressed in mitotically quiescent hepatocytes.14 On the other hand, C/EBP-β and -δ are similarly expressed in many tissues, including macrophages, especially after their stimulation with a variety of agents including lipopolysaccharides (LPS).3,4,15-22 In contrast to C/EBP-α, both C/EBP-β and -δ are highly expressed in early stages of adipocyte development and this expression decreases with differentiation of these cells.4 The expression patterns for these genes are reversed in myeloid cell differentiation. Some myeloid progenitors have high levels of C/EBP-α, which decrease during granulocytic differentiation23; the levels of C/EBP-β and -δ are low in early myeloid stem cells and increase during granulocytic differentiation.23
C/EBPε is highly homologous to the predicted amino acid sequence of rat CRP1 for which no transcripts have been found in any cell type,11 and is 54% homologous to the myeloid-specific protein, NFM, which is a component of the combinatorial switch involved in the regulation of myeloid genes in cooperation with the c-Myb protein.23-25 NFM is the avian homolog of the human C/EBP-β protein whose expression is not limited to cells of the myelomonocytic lineage. C/EBP-β is dramatically induced in all tissues after stimulation with either LPS or other inflammatory signals.3,4 15-22 This suggests that although C/EBP-β is the structural homolog of NFM, it may not be the functional homolog.
C/EBP-like proteins do not have clear differences in their DNA-binding preferences; therefore, selective expression may play a fundamental role in determining which C/EBP protein is important at a particular stage of hematopoietic cell development. To elucidate the role of C/EBPε in hematopoiesis, we carefully examined the range of its mRNA expression in 28 hematopoietic and 14 nonhematopoietic cell lines, and 17 fresh leukemia samples. Furthermore, we examined mRNA expression of additional C/EBP family members (-α, -β, -δ) and other transcriptional factors that regulate hematopoiesis (AML-1, PU.1, and GATA1). Regulation of C/EBPε expression was also examined in normal human CD34+ CD38−, and CD34+ CD33+ fractionated hematopoietic stem cells subjected to myeloid differentiation in culture, as well as in the myeloid cell lines induced to differentiate toward macrophage-like granulocyte-like cells. Likewise, alterations in clonal growth of HL-60 and NB4 cells were examined in soft-agar culture in the presence of C/EBPε antisense oligonucleotides.
MATERIALS AND METHODS
Samples for analysis and cell culture studies.Twenty-eight hematopoietic and 14 nonhematopoietic cell lines were cultured in RPMI 1640 and Dulbecco's modified Eagle's medium (DMEM) media, respectively, plus 10% fetal calf serum (FCS) and antibiotics (Tables 1 and 2). Cell lines were either established by ourselves26 or obtained from American Type Culture Collection (ATCC; Rockville, MD). Bone marrow samples were obtained after informed consent from 12 acute myelogenous leukemia (AML), two chronic myelogenous leukemia (CML) patients, and one individual with biphenotypic leukemia. Each sample had more than 80% leukemic cells. Peripheral blood granulocytes (polymorphonuclear cells [PMN]) and mononuclear cells were purified from heparinized peripheral blood of healthy volunteers after their informed consent, by density gradient centrifugation as described.27 28 Purity was determined by morphology (>97% granulocytes). Mononuclear cells were cultured in 20 ng/mL GM-CSF for 10 days to induce macrophage differentiation and then harvested. These macrophages were adherent and greater than 95% of the cells had the morphology of macrophages and stained for nonspecific acid esterase, which is characteristic for monocytes/macrophages. To induce monocytic differentiation of HL60 and KG1 cell lines, 12-0-tetradecanoylphorbol diester (TPA, 2 × 10−8 mol/L) was added to the culture medium; cells were harvested at 0, 0.5, 2, 6, 24, and 48 hours after addition. RNA was prepared from all of the above samples using Trizol as described by the manufacturer (GIBCO/BRL, Gaithersburg, MD). Normal human myoblasts were kindly provided by Dr Valerie Askanas (University of Southern California, Los Angeles).
Densitometric Analysis of c/EBPε and MPO in Human Hematopoietic Cell Lines
| Cell Type . | Cell Line . | C/EBPε/ . | MPO/ . |
|---|---|---|---|
| . | . | β-Actin . | β-Actin . |
| Promyelocyte | NB4 | 2.5 | 1.3 |
| Myeloblast* | GFD8 | 2.26 | 0.16 |
| Late myeloblast | HL-60 | 2.24 | 1.39 |
| Myelomonoblast | THP1 | 0.42 | 0.2 |
| Myeloblast* | MDS92 | 0.33 | 0.87 |
| Early myeloblast | KCL22 | 0.25 | 0 |
| Myelomonoblast | U937 | 0.24 | 0 |
| Nl granulocyte | — | 0.19 | 0.54 |
| Early myeloblast | ML-1 | 0.18 | 1.48 |
| Early myeloblast* | TF-1 | 0.06 | 0 |
| Early myeloblast* | GM150 | 0.05 | 0 |
| Early myeloblast* | AML1 | 0.05 | 1.15 |
| Early myeloblast | KG1 | 0.04 | 0 |
| Nl macrophage | — | 0.04 | 0 |
| Erythroblast | HEL | 0 | 0 |
| Erythroblast | K562 | 0 | 0 |
| Megakaryoblast | MEGA1 | 0 | 0 |
| Basophil precursor | KU812 | 0 | 0 |
| Cell Type . | Cell Line . | C/EBPε/ . | MPO/ . |
|---|---|---|---|
| . | . | β-Actin . | β-Actin . |
| Promyelocyte | NB4 | 2.5 | 1.3 |
| Myeloblast* | GFD8 | 2.26 | 0.16 |
| Late myeloblast | HL-60 | 2.24 | 1.39 |
| Myelomonoblast | THP1 | 0.42 | 0.2 |
| Myeloblast* | MDS92 | 0.33 | 0.87 |
| Early myeloblast | KCL22 | 0.25 | 0 |
| Myelomonoblast | U937 | 0.24 | 0 |
| Nl granulocyte | — | 0.19 | 0.54 |
| Early myeloblast | ML-1 | 0.18 | 1.48 |
| Early myeloblast* | TF-1 | 0.06 | 0 |
| Early myeloblast* | GM150 | 0.05 | 0 |
| Early myeloblast* | AML1 | 0.05 | 1.15 |
| Early myeloblast | KG1 | 0.04 | 0 |
| Nl macrophage | — | 0.04 | 0 |
| Erythroblast | HEL | 0 | 0 |
| Erythroblast | K562 | 0 | 0 |
| Megakaryoblast | MEGA1 | 0 | 0 |
| Basophil precursor | KU812 | 0 | 0 |
Densitometry was performed for all the samples and ratios were calculated between hybridization intensity of β-actin and either C/EBPε or MPO. Most samples were analyzed several times with very similar results.
Abbreviation: Nl, normal.
Growth factor-dependent cell lines.
C/EBPε mRNA Expression in Various Nonmyeloid Cell Lines
| Cell Line . | Tissue/Blood Cell . | C/EBPε . | MPO . |
|---|---|---|---|
| CEM | T-lymphocyte | Weak | — |
| TALL-1 | T-lymphocyte | — | — |
| DL-30 | T-cell lymphoma | — | — |
| HSB-2 | T-lymphocyte | Weak | — |
| B1 | B-lymphocyte | — | — |
| BALL-11 | B-lymphocyte | — | — |
| BALL-2 | B-lymphocyte | — | — |
| PALL-1* | B-lymphocyte | — | — |
| PALL-2* | B-lymphocyte | — | — |
| M* | B-lymphocyte | — | — |
| Lu-CSF-1 | Lung | — | — |
| Calu 3 | Lung | — | — |
| DU145 | Prostate | — | — |
| LnCap | Prostate | — | — |
| HOS | Bone | — | — |
| U20S | Bone | — | — |
| SKOV-3 | Ovary | — | — |
| IMR-32 | Neuroblastoma | — | — |
| SKN-SH | Neuroblastoma | — | — |
| HeLa | Cervix | — | — |
| MCF-7 | Breast | — | — |
| HT-29 | Colon | — | — |
| Normal human myoblasts | — | — | |
| W138 | Normal fibroblasts | — | — |
| Cell Line . | Tissue/Blood Cell . | C/EBPε . | MPO . |
|---|---|---|---|
| CEM | T-lymphocyte | Weak | — |
| TALL-1 | T-lymphocyte | — | — |
| DL-30 | T-cell lymphoma | — | — |
| HSB-2 | T-lymphocyte | Weak | — |
| B1 | B-lymphocyte | — | — |
| BALL-11 | B-lymphocyte | — | — |
| BALL-2 | B-lymphocyte | — | — |
| PALL-1* | B-lymphocyte | — | — |
| PALL-2* | B-lymphocyte | — | — |
| M* | B-lymphocyte | — | — |
| Lu-CSF-1 | Lung | — | — |
| Calu 3 | Lung | — | — |
| DU145 | Prostate | — | — |
| LnCap | Prostate | — | — |
| HOS | Bone | — | — |
| U20S | Bone | — | — |
| SKOV-3 | Ovary | — | — |
| IMR-32 | Neuroblastoma | — | — |
| SKN-SH | Neuroblastoma | — | — |
| HeLa | Cervix | — | — |
| MCF-7 | Breast | — | — |
| HT-29 | Colon | — | — |
| Normal human myoblasts | — | — | |
| W138 | Normal fibroblasts | — | — |
Expression determined by RT-PCR; all samples were strongly positive for β-actin transcripts; and all samples were examined at least twice.
Growth factor-dependent cell lines.
The NB4 and HL-60 cell lines were induced to differentiate using 9-cis retinoic acid (9-cis-RA; 10−7 mol/L), all-trans-RA (10−7 mol/L), the potent vitamin D3 analog KH1060 (10−7 mol/L), or a combination of both. Differentiation was assessed by morphology of Giesma-stained cytopreparations, percent cells adherent to plastic, expression of nonspecific acid esterase (NAE) and expression of CD11b and CD14, and reduction of nitroblue tetrozalium (NBT), as previously described.29
For the two-layer soft-agar clonogenic studies of HL-60 and NB4 cells, either the sense or antisense phosphothiolated oligonucleotides, or diluent control was added to the lower layer of soft agar, and HL-60 or NB4 cells (1 × 103) were added to the upper layer of soft-agar as previously described.26 The phosphothiolated oligonucleotides were direct to the beginning of translation and had the sequences 5′-CCC-TGG-GAC-ATG-GCC-GGC-3′; the sense oligonucleotides were 5′-GCC-GGC-CAT-GTC-CCA-CGG-3′ (Research Genetics, Inc, Huntsville, AL).
Stem cell separation and induction of myeloid differentiation.Peripheral blood mobilized mononuclear cells were obtained by leukopheresis and the CD34+ cell population was isolated using a magnetic cell sorting system (MiniMACS;Myltenyi Biotec, Inc, Auburn, CA). CD34+ cells were further purified into CD34+CD38− and CD34+CD33+ cell fractions by fluoresence-activated cell sorter (FACS). Total RNA was extracted from the CD34+CD38− and CD34+CD33+ stem cells before and after culture with stem cell factor (50 ng/mL) and PIXY321 (interleukin-3 [IL-3] and granulocyte-macrophage colony-stimulating factor [GM-CSF ], 20 ng/mL) in RMPI medium containing 30% FCS. Cells were harvested on days 0, 2, 4, 6, 8, and 11 for RNA extraction. Cytospins were prepared concurrently, and slides were stained with Wright-Giemsa for the analysis of cell morphology. For each slide, 200 cells were counted, and the numbers of cells at different stages of differentiation were plotted on a graph.
Reverse transcriptase-polymerase chain reaction (RT-PCR) and Northern blot analyses.The RNA samples were treated with RQ1 RNase-free DNase (1 U/μL Promega; Madison, WI), for 30 minutes at 37°C followed by phenol-chloroform extraction and ethanol precipitation. One microgram of total RNA was heated at 65°C for 10 minutes, and then used as a template for first-strand complementary DNA (cDNA) synthesis using Moloney murine leukemia virus reverse transcriptase and random hexamers as primers (GIBCO/BRL). To control for genomic DNA contamination, a RT reaction was performed without the addition of the enzyme; and one reaction using this sample was included for each set of primers. Furthermore, the primers for C/EBPε spanned an intron resulting in a different size for the cDNA and DNA products. The cDNAs were amplified by 25 cycles of 94°C for 40 seconds, 55°C for 30 seconds, and 72°C for 40 seconds. Each 30-μL PCR reaction contained 1 μL of the cDNA, 200 μmol/L deoxynucleotide triphosphates (dNTPs), and 0.5 U Taq DNA polymerase (GIBCO/BRL), and 1.5 mmol/L MgCl2 . Two sets of primers were used for each sample, including primers specific for the gene of interest and primers for β-actin as an internal control (Table 3). Initial experiments determined the linear range of C/EBPε mRNA expression in the late myeloblast HL60 cell line using RT-PCR and Southern blotting. A total of 20, 23, 25, 27, 30, and 35 cycles were examined using different dilutions of the cDNA; the optimal dilution and 25 cycles of PCR provided a product below the β-actin saturation. All the PCR products were electrophoresed on a 3% agarose gel and transferred to nylon filters (Hybond N+) as described by the manufacturer (Amersham; Arlington Heights, IL). Hybridization of these blots was performed according to the membrane manufacturer's recommendations (Amersham). DNA-probes for C/EBPε, MPO, CD34, C/EBP-α, C/EBP-β, C/EBP-δ, PU.1, AML1, and β-actin were labeled with [α-32P]deoxycytidine triphosphate (dCTP) (3,000 μCi/mmol; NEN/Dupont, Boston, MA) using a random primer labeling kit (GIBCO/BRL).1,7,23 30 Internal oligonucleotides for GATA-1 (5′-TAC TGT GGT GGC TCC GCT CAG CTC ATG AGG CAC AGA GCA-3′) and C/EBPβ (5′-GCA GCA CCA CGA CTT CCT CT-3′) were end-labeled using [γ- 32P]-adenosine triphosphate (ATP) and T4 kinase (GIBCO/BRL). Relative intensities of the signals on the blots were deduced on the basis of densitometric scanning relative to the β-actin signal using a GS-250 molecular imager (Bio-Rad, Hercules, CA) (Table 1).
Sequences of the Primers for RT-PCR
| C/EBPε | 5′-CAGACAGGAAGGCGCTGGG-3′ |
| 5′-CGGCAGTGGCCAAAGGGGCCTT-3′ | |
| C/EBPα | 5′-GAATCTCCTAGTCCTGGCTC-3′ |
| 5′-GATGAGAACAGCAACGAGTAC-3′ | |
| C/EBPβ | 5′-GCCACGGACACCTTCGAGG-3′ |
| 5′-CGGCTCCGCCTTGAGCTG-3′ | |
| C/EBPγ | 5′-CAGCGCCTACATACGACTCC-3′ |
| 5′-GTCTGCCTGGGGCTGCTG-3′ | |
| PU.1 | 5′-TGGAAGGGTTTCCCCTCGTC-3′ |
| 5′-TGCTGTCCTTCATGTCGCCC-3′ | |
| AML-1 | 5′-GTTGAGAGTCGACTGGAAAG-3′ |
| 5′-GAGGGAAAAGCTTCACTCGA-3′ | |
| GATA-1 | 5′-TTAGCCACCTCATGCCTT-3′ |
| 5′-ACATCGGTCTTAAGACCT-3′ | |
| MPO | 5′-AGCGAGGAGCCCCTGGCCAGGAACCTG-3′ |
| 5-GAGCTCGGGCATCTCACTGGAACGG-3′ | |
| CD34 | 5′-TCCAGCTGTTGCGGAGTTTAAGAAG-3′ |
| 5′-TGCTGAATGGCCGTTTCTGGAGGT-3′ | |
| β-Actin | 5′-TACATGGGTGGGGTGTTGAA-3′ |
| 5′-AAGAGAGGCATCCTCACCCT-3′ |
| C/EBPε | 5′-CAGACAGGAAGGCGCTGGG-3′ |
| 5′-CGGCAGTGGCCAAAGGGGCCTT-3′ | |
| C/EBPα | 5′-GAATCTCCTAGTCCTGGCTC-3′ |
| 5′-GATGAGAACAGCAACGAGTAC-3′ | |
| C/EBPβ | 5′-GCCACGGACACCTTCGAGG-3′ |
| 5′-CGGCTCCGCCTTGAGCTG-3′ | |
| C/EBPγ | 5′-CAGCGCCTACATACGACTCC-3′ |
| 5′-GTCTGCCTGGGGCTGCTG-3′ | |
| PU.1 | 5′-TGGAAGGGTTTCCCCTCGTC-3′ |
| 5′-TGCTGTCCTTCATGTCGCCC-3′ | |
| AML-1 | 5′-GTTGAGAGTCGACTGGAAAG-3′ |
| 5′-GAGGGAAAAGCTTCACTCGA-3′ | |
| GATA-1 | 5′-TTAGCCACCTCATGCCTT-3′ |
| 5′-ACATCGGTCTTAAGACCT-3′ | |
| MPO | 5′-AGCGAGGAGCCCCTGGCCAGGAACCTG-3′ |
| 5-GAGCTCGGGCATCTCACTGGAACGG-3′ | |
| CD34 | 5′-TCCAGCTGTTGCGGAGTTTAAGAAG-3′ |
| 5′-TGCTGAATGGCCGTTTCTGGAGGT-3′ | |
| β-Actin | 5′-TACATGGGTGGGGTGTTGAA-3′ |
| 5′-AAGAGAGGCATCCTCACCCT-3′ |
For Northern blot analysis, total RNA (20 μg/lane) was electrophoresed on 1% agarose gels containing 660 mmol/L formaldehyde and transferred to positively-charged nylon membranes (Hybond N+) using the alkali blotting technique as described by the manufacturer (Amersham). Hybridization was performed using a C/EBPε cDNA-probe labeled with α-32P-dCTP as described above.
Western blot analysis.For Western blot analysis, cells were lysed in radio-immunoprecipitation assay (RIPA) buffer (150 mmol/L NaCl, 50 mmol/L Tris-HCl [pH 8.0], 0.5% deoxycholate, 1% NP-40, 1% sodium dodecyl sulfate [SDS]) containing protease inhibitors (1 mmol/L phenylmethylsolfonyl fluoride [PMSF ], 10 μg/mL leupeptin and pepstatin, and 100 μg/mL aprotinin) for protein extraction. Protein concentrations were determined using the Bradford assay as described by the manufacturer (Bio-Rad).
Cell lysate samples (30 μg) were denatured by addition of 2× electrophoresis sample buffer (120 mmol/L Tris-Cl [pH 6.8], 4% SDS, 25% glycerol, 3% β-mercaptoethanol), incubated for 10 minutes at 90°C, and separated on a 12.5% polyacrylamide gel containing SDS.30 The proteins were electroblotted overnight onto Immobilon-P membrane as described by the manufacturer (Millipore, Bedford, MA). Purified rabbit polyclonal anti-C/EBPε antibody (18 μg/mL)1 was incubated with the previously blocked membrane for 1 hour at room temperature, and bound antibody was detected using antirabbit horseradish peroxidase (HRP) antibody conjugate and the enhanced chemiluminescence (ECL) substrate (Amersham). Lysates from Rat-1 cells transfected with either a cytomegalovirus (CMV)-driven, human C/EBPε cDNA expression plasmid1 or empty vector were used as positive and negative controls, respectively.
RESULTS
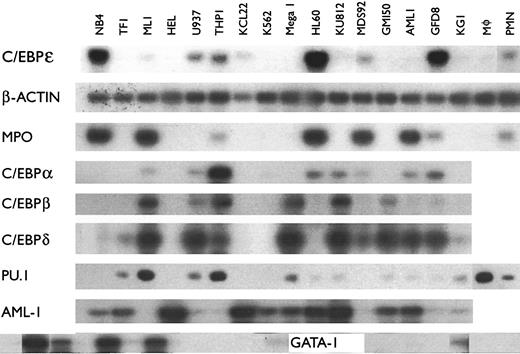
Expression of C/EBPε mRNA in cell lines.A large array of human leukemia cell lines blocked at various stages of differentiation may allow the identification of lineage and differentiation specific proteins. Furthermore, several of these lines can undergo terminal myeloid differentiation when exposed to various inducers of differentiation. We used these lines to help determine the lineage and differentiation specificity of the new transcriptional activator known as C/EBPε. Because detection of the C/EBPε transcripts by Northern blot analysis was difficult in most cells lines except HL-60, NB4, KCL22, SKM1, and PLB9851 (data not shown), we used a semiquantitative RT-PCR approach to analyze expression. We detected the highest levels of C/EBPε mRNA expression in the late myeloblast and promyelocytic cell lines HL60, NB4, and GFD8; the myeloid leukemia cell lines THP1, U937, MDS92, ML-1 and KCL22 also expressed detectable amounts of C/EBPε mRNA (Fig 1, Table 1). The T-cell leukemic cell lines HSB-2 and CEM also had detectable levels of C/EBPε transcripts (Table 2). No PCR products were observed in either the erythroblast cell lines HEL and K562, the megakaryoblast cell line Mega 1, or the basophilic blast cell line KU812 (Fig 1, Table 1) indicating these lines do not express C/EBPε mRNA. Also, except for the CEM and HSB-2 T-cell lines, 22 nonmyeloid cell lines did not express C/EBPε (Table 2). The β-actin PCR products (internal control) were easily detected in all of these samples for each PCR analysis (Fig 1 and data not shown). The PCR-reactions were retested two to three times per sample to ensure accuracy.
mRNA expression of various genes including C/EBPε in hematopoietic cell lines. The mRNA was amplified using RT-PCR. Each PCR reaction included primers specific for the analyzed gene and primers for β-actin as an internal control. PCR conditions yielded amplification in the linear range. PCR products were blotted, hybridized using either cDNA probes or internal oligonucleotides specific for the amplified genes, and autoradiography was performed. All samples were examined at least twice for all genes.
mRNA expression of various genes including C/EBPε in hematopoietic cell lines. The mRNA was amplified using RT-PCR. Each PCR reaction included primers specific for the analyzed gene and primers for β-actin as an internal control. PCR conditions yielded amplification in the linear range. PCR products were blotted, hybridized using either cDNA probes or internal oligonucleotides specific for the amplified genes, and autoradiography was performed. All samples were examined at least twice for all genes.
The synthesis of enzymes that are stored in azurophilic or primary granules occurs only during the late myeloblast and promyelocyte stage of development; myeloperoxidase (MPO) is an abundant enzyme of these granules, which is important in defense against microbes. Most of the cell lines that expressed C/EBPε expressed MPO, with two exceptions (U937 and KCL-22, Fig 1). However, the ML-1 and AML-1 cells expressed low levels of C/EBPε and abundant levels of MPO, C/EBP-α, and -δ (Fig 1).
Each member of the C/EBP family of transcription factors has a similar basic region followed by a leucine zipper. These proteins potentially can homo- and hetero-dimerize. The cell line, NB4, which is most differentiated down the granulocytic pathway, expressed C/EBPε, but not C/EBP-α, -β, and -δ (Fig 1). Therefore, indirect evidence suggests that at least at the stage of the promyelocyte, expression of C/EBP-α,-β, or -δ and their heterodimerization with C/EBPε may not be required. Furthermore, NB4, HL-60, and GFD8 expressed easily detectable levels of C/EBPε, but not C/EBP-β, suggesting that the late myeloblastic and promyelocytic stages of differentiation may require C/EBPε, but not C/EBP-β. The expression pattern of C/EBP-δ in general paralleled that of C/EBP-β, except for the AML-1 and GPD8 cell lines. None of the C/EBPs (C/EBP-α,-β,-δ,-ε) were expressed in the red blood cell precursors (HEL, K562).
We also examined the expression pattern of other transcription factors important in hematopoietic cell differentiation (Fig 1). The PU.1 (spi-1) mRNA was readily detected in the myelomonocytic blast cells (ML-1, U937, THP-1), as well as in macrophages. TF-1 (myeloblasts) and Mega 1 (megakaryocyte precursors) also expressed PU.1. In cell lines with prominent expression of C/EBPε (NB4, HL-60, GFD8), PU.1 was not readily detected suggesting that its interaction with C/EBPε at the late myeloblast and promyelocyte stages of development is unlikely. The AML-1 mRNA was expressed in most of the cell lines (Fig 1). As expected, the GATA-1 mRNA was expressed in erythroblasts (K562, HEL), as well as in the basophil and megakaryocyte precursors (KU812, Mega, respectively). The GM150 growth factor–dependent myeloblasts expressed GATA-1, but none of the other early or late myeloid (granulocytic/monocytic) precursors expressed GATA-1.
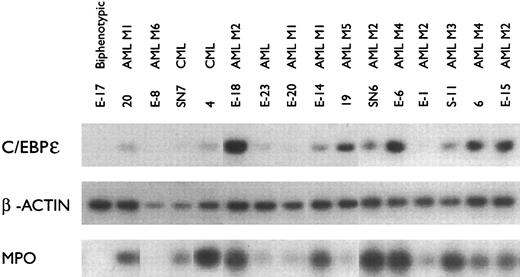
C/EBPε mRNA expression in fresh leukemia samples.Of the fresh leukemia samples, most of the AML samples were positive for C/EBPε (10/12) and MPO (11/12) (Fig 2). In contrast, the samples E-17 (biphenotypic [early blasts with myeloid and lymphoid characteristics]), E-8 (M6, erythroblastic leukemia), and E-1 (M2, myeloblastic leukemia) were negative for C/EBPε mRNA. In addition, E-17 and E-8 had undetectable levels of MPO mRNA. All samples were positive for β-actin mRNA (Fig 2). These results indicate that C/EBPε mRNA is expressed in acute myelogenous leukemia samples from patients.
C/EBPε and MPO mRNA expression in fresh leukemia samples. The mRNA was amplified using RT-PCR and examined as described in Fig 1. β-actin expression was used as an internal control.
C/EBPε and MPO mRNA expression in fresh leukemia samples. The mRNA was amplified using RT-PCR and examined as described in Fig 1. β-actin expression was used as an internal control.
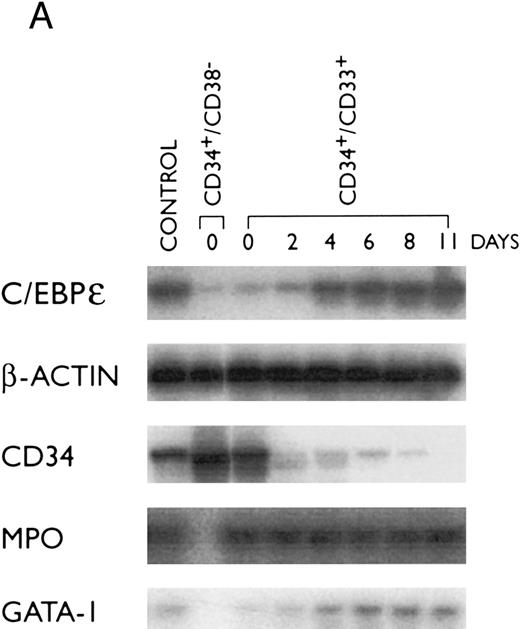
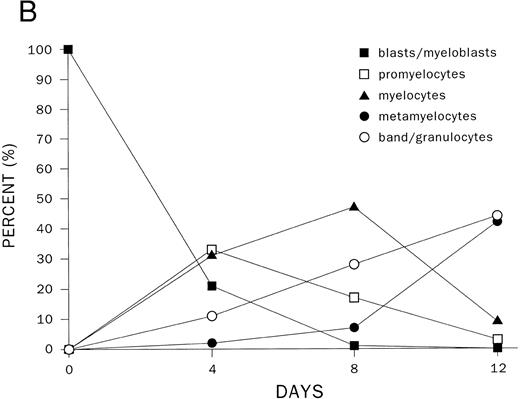
Modulation of C/EBPε mRNA expression during differentiation of human hematopoietic stem cells.One of the earliest normal human hematopoietic stem cells is CD34+/CD38−; these cells can proceed down all lineages of hematopoietic development. A purified CD34+/CD38− population was found to express low levels of C/EBPε, MPO, and GATA-1 mRNAs (Fig 3A and B); but as expected, these cells expressed high levels of CD34 mRNA. The CD34+/CD33+ stem cells are more mature and can no longer differentiate down the lymphoid pathway. These cells also expressed low levels of C/EBPε mRNA (Fig 3). As the cells differentiated to promyelocytes (day 4, 30%), myelocytes (day 8, 40%) and metamyelocytes, and granulocytes (day 11, 80%), the C/EBPε mRNA reached an elevated plateau level and expression of CD34 mRNA markedly decreased (Fig 3). This suggests an important role for C/EBPε at the myeloblast/promyelocyte and later stages of myeloid differentiation.
Expression of transcription factors C/EBPε, PU.1, GATA-1, and differentiation markers (CD34 and MPO) during myeloid differentiation of CD34+ cells. Total RNA was extracted from CD34+CD38+ and CD34+/CD33+ cells before and after culture with stem cell factor and PIXY321. Cells were harvested at the days indicated. (A) The mRNA expression was studied by RT-PCR and expression of β-actin mRNA was used as an internal control. (B) Differentiation of CD34+/CD33+ cells as determined by morphology. For each slide, 200 cells were counted, and the number of cells at different stages of differentiation were plotted on a graph.
Expression of transcription factors C/EBPε, PU.1, GATA-1, and differentiation markers (CD34 and MPO) during myeloid differentiation of CD34+ cells. Total RNA was extracted from CD34+CD38+ and CD34+/CD33+ cells before and after culture with stem cell factor and PIXY321. Cells were harvested at the days indicated. (A) The mRNA expression was studied by RT-PCR and expression of β-actin mRNA was used as an internal control. (B) Differentiation of CD34+/CD33+ cells as determined by morphology. For each slide, 200 cells were counted, and the number of cells at different stages of differentiation were plotted on a graph.
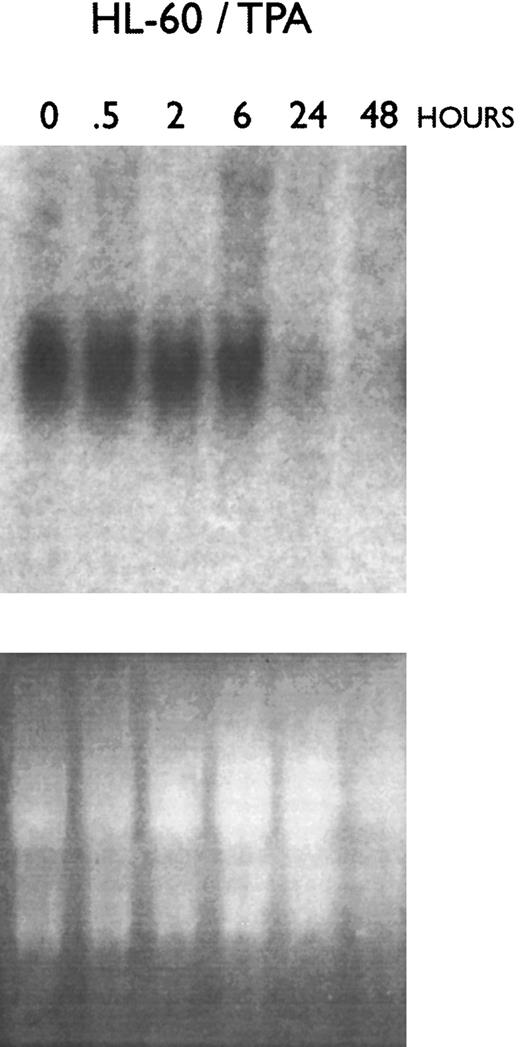
Increased C/EBPε mRNA and protein expression during granulocytic differentiation of myeloid leukemia cell lines.Several of the myeloid leukemia cell lines can undergo differentiation towards macrophages and/or granulocytes. This experimental system eliminates the risk of contamination with other cell types. Northern blot and RT-PCR analyses showed a time-dependent decrease in C/EBPε mRNA expression after exposure to TPA in both HL60 (Fig 4) and KG1 cells (data not shown). Nearly 100% of these TPA-exposed cells differentiated toward macrophage-like cells with greater than 95% becoming adherent to plastic, having the morphology of macrophages and staining for nonspecific esterase, which is specific for this cell type. Furthermore, macrophages from normal human bone marrow expressed very low levels of C/EBPε (Fig 1).
Effect of macrophage-like differentiation of HL-60 cells on their expression of C/EBPε mRNA. HL-60 cells were cultured with TPA (2 × 10−8 mol/L) and cells were harvested for RNA extraction after 0.5, 2, 6, 24, and 48 hours. Northern blot analysis was performed using a 32P-labeled cDNA probe for C/EBPε.
Effect of macrophage-like differentiation of HL-60 cells on their expression of C/EBPε mRNA. HL-60 cells were cultured with TPA (2 × 10−8 mol/L) and cells were harvested for RNA extraction after 0.5, 2, 6, 24, and 48 hours. Northern blot analysis was performed using a 32P-labeled cDNA probe for C/EBPε.
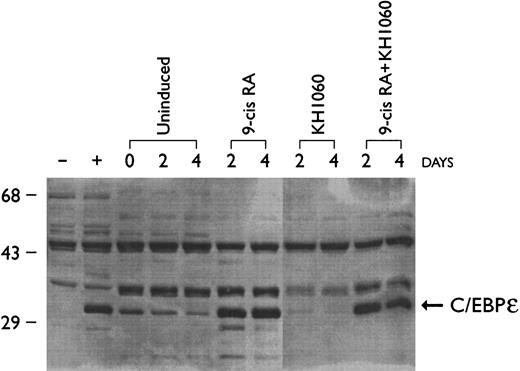
Protein expression of C/EBPε in hematopoietic cell lines correlated with the RT-PCR data with highest levels of C/EBPε protein expression in the promyelocytic NB4 cell line (Fig 5) and myeloblastic HL60 cells (data not shown) and undetectable levels in KG-1 (early myeloblasts) and K562 (erythroblasts) (data not shown). Upregulation of C/EBPε protein expression was observed as the NB4 cells were induced for 2 and 4 days with 9-cis–RA to differentiate down the granulocytic pathway (Fig 5). Differentiation was confirmed by measuring expression of CD11b (<5% at day 0 and >90% by day 2) and by morphologic examination of Giemsa-stained cytoprepartions (>50% at myelocyte or more mature stage after exposure to 9-cis–RA [10−7 mol/L, 4 days]). NB4 cells can differentiate towards monocytes/macrophages when cultured with a potent vitamin D3 analog known as KH1060 (>70% adherent cells and ≈20% CD14-positive cells after exposure to KH1060 [10−7 mol/L, 4 days] compared with <1% adherent cells and <10% expression of CD14 on wild-type NB4 cells)29 (and data not shown). Expression of C/EBPε protein did not increase as NB-4 cells differentiated down the monocytic pathway; in fact, levels appeared to diminish (Fig 5). Exposure of NB-4 cells to the combination of 9-cis–RA (10−7 mol/L) plus KH1060 (10−7 mol/L) for 2 days was slightly more potent in inducing myeloid differentiation than was either agent alone (CD11b > 95%, CD14 ≈35%, and ≈70% morphologically at the myelocyte or a more mature stage); levels of C/EBPε in these NB4 cells were comparable to those present in the cells cultured with 9-cis–RA alone.
Effect of 9-cis retinoic acid (9-cis–RA) and vitamin D analog (KH1060) on C/EBPε protein expression in NB4 cells. NB4 cells were cultured with 9-cis-RA (10−7 mol/L), KH1060 (10−7 mol/L) and 9-cis–RA + KH1060 (10−7 mol/L). Rat-1 cell transfected with either C/EBPε expression vector or empty vector were used as positive (+) and negative (−) controls, respectively. Cells were harvested for protein extraction on days 0, 2, and 4 of culture and analyzed by Western blotting.
Effect of 9-cis retinoic acid (9-cis–RA) and vitamin D analog (KH1060) on C/EBPε protein expression in NB4 cells. NB4 cells were cultured with 9-cis-RA (10−7 mol/L), KH1060 (10−7 mol/L) and 9-cis–RA + KH1060 (10−7 mol/L). Rat-1 cell transfected with either C/EBPε expression vector or empty vector were used as positive (+) and negative (−) controls, respectively. Cells were harvested for protein extraction on days 0, 2, and 4 of culture and analyzed by Western blotting.
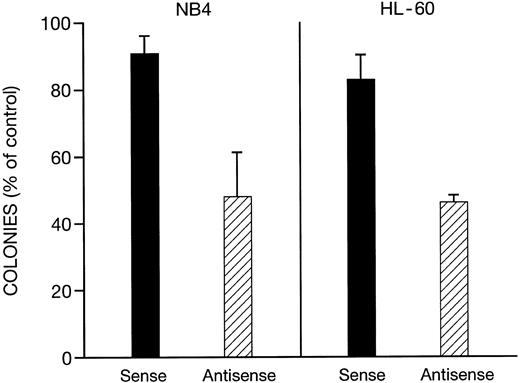
Effects of antisense C/EBPε phosphothiolated oligonucleotides on clonal proliferation of NB-4 and HL-60 cells.We examined the effect of C/EBPε phosphothiolated antisense oligonucleotides on soft-agar clonal growth of NB4 and HL-60 cells. Initial experiments showed that the antisense oligonucleotides were able to suppress clonal growth in a dose-dependent manner (0.1 to 20 μmol/L, data not shown). At 0.25 μmol/L of C/EBPε antisense oligonucleotides, the clonal growth of both NB-4 and HL-60 cells was inhibited approximately 55%; sense oligonucleotides at the same concentration caused less than 17% clonal growth inhibition (Fig 6). At the same concentration (0.25 μmol/L), the antisense oligonucleotide decreased expression of C/EBPε by 85% and 80% in NB-4 and HL-60 cells, respectively, while sense oligonucleotides had no effect on levels of C/EBPε, as determined by Western blot (data not shown).
Effect of sense and antisense C/EBPε oligonucleotides on clonal growth of NB4 and HL-60 cells. The cells (103 dish) were placed in soft agar that contained either sense or antisense C/EBPε phosphothiolated oligonucleotides (0.25 μmol/L) or diluant control; and dishes were incubated at 37°C in 5% CO2 and colonies were enumerated on day 12 of culture. The results represent mean ± standard deviation (SD) of three independent experiments with each experimental point having three plates. Results are normalized to control plates containing diluant alone (100%).
Effect of sense and antisense C/EBPε oligonucleotides on clonal growth of NB4 and HL-60 cells. The cells (103 dish) were placed in soft agar that contained either sense or antisense C/EBPε phosphothiolated oligonucleotides (0.25 μmol/L) or diluant control; and dishes were incubated at 37°C in 5% CO2 and colonies were enumerated on day 12 of culture. The results represent mean ± standard deviation (SD) of three independent experiments with each experimental point having three plates. Results are normalized to control plates containing diluant alone (100%).
DISCUSSION
The hypothesis that transcriptional factors are central to the development of differentiated cells from multipotential progenitors has prompted the identification and characterization of several transcriptional regulators of myelopoiesis.31 Members of the major transcription factor families have been implicated in regulation of various stages of myelopoiesis and/or functional activation of mature myeloid cells. Examples include the AML1 transcriptional factor complex,32-34 several members of the Myb,35-38and Ets family of transcriptional regulators, in particular PU.1,39-43 as well as zinc-finger44-46 and helix-loop-helix proteins.47 Identification of a number of transcriptional regulators that have roles in activation and repression of transcription in myeloid cells suggests the importance of an interactive network of differentially expressed transcriptional regulators and signal transduction pathways that result in cooperative activation and inactivation of target genes. Genes that are selectively expressed in immature myeloid cells (eg, neutrophil elastase, myeloperoxidase, granulocyte colony-stimulating factor receptor) are regulated by at least four families of transcriptional activators: C/EBPs, PU.1, c-MYB, and PEBP2/CBFs, including AML-1. None of the four groups of factors have been identified as purely myeloid specific. C/EBPε does appear to be specifically expressed in myeloid cells and a selected group of T cells. It is not expressed in erythroid, megakaryocyte, or basophil precursor cells, B lymphocytes, or nonhematopoietic cells of various tissues.
Examination of the C/EBP family showed that the avian NF-M is the structural homolog of the mammalian C/EBP-β (NF-IL-6) gene, but this homology does not extend to their expression patterns: avian NF-M is exclusively expressed in promyelocytes; mammalian C/EBP-β is expressed in many tissues in response to inflammatory stimuli such as lipopolysaccharides, IL-1, tumor necrosis factor, and IL-6. Therefore, C/EBP-β appears to be more important for functional activation of cells of different lineages rather than involved in the determination of the differentiation pathway. For example, mice with targeted disruption of the C/EBP-β gene appear to have normal myeloid differentiation, but have defects in activation of their macrophages.48,49 The C/EBP-α does appear to be critical for neutrophil development because C/EBP-α “knock-out” mice do not have neutrophils.50 Furthermore, C/EBP-α is able to upregulate the promoter activities of the receptor for granulocyte, granulocyte-monocyte, and monocyte colony-stimulating factor, as well as the neutrophil elastase promoter.7,51-53 Additional investigations have observed a pattern of coordinated regulation of the three C/EBP-like proteins, C/EBP-α, C/EBP-β, and C/EBP-δ in adipocytes, as well as differentiating myeloid cells.4,23,54 Expression of C/EBP-α was higher in early myeloid cells and it decreased with granulocytic differentiation; while in adipocytes, levels of expression of C/EBP-α paralleled their differentiation.55,56 In contrast, levels of C/EBP-β (NF-IL-6) increased as myeloid cells differentiated towards macrophages, while expression of C/EBP-β decreased during adipocyte development. Levels of C/EBP-δ expression often paralleled levels of C/EBP-β. Another C/EBP-like factor, C/EBP-γ (Ig-C/EBP) was expressed ubiquitously.9
Since each of the C/EBP-like factors can cooperate in the avian system in the induction of myeloid-specific genes,57 by binding to the CCAAT consensus sequence, we hypothesized that the amino acid sequence homology of the C/EBP proteins may play a secondary role in determining functional homology among family members. Instead, regulation of expression of a particular C/EBP isoform may be of primary importance. Therefore, in this study, we determined the expression pattern of C/EBPε, as well as other members of the C/EBP family by examining 28 hematopoietic and 14 nonhematopoietic cell lines, 18 fresh leukemia samples, and normal human peripheral blood granulocytes and macrophages using RT-PCR. Expression of C/EBPε mRNA was detected in the late-myeloblast, promyelocyte cell lines HL-60, NB4, and GFD8 by RT-PCR (Fig 1) and Northern blot (Fig 4).1 C/EBPε mRNA expression was detected by RT-PCR in myelomonoblastic THP-1 and U937 cells; but was nearly undetectable in the early myeloblastic lines such as KG-1. No expression was found in the erythroid, megakaryocytic, and basophil precursor cell lines. Two T-cell lines (CEM, HSB-2) had weak expression and eight other lymphoid lines had no detectable levels of C/EBPε. No expression was found in any of the nonhematopoietic samples. This expression pattern was distinctly different from the other members of the C/EBP family. In contrast to C/EBPε, no expression of C/EBP-α, -β, -δ was detected in the NB4 cell line, which is blocked at the promyelocyte stage of development.
To elucidate expression patterns of C/EBPε in cells that are not transformed or immortalized, we purified human hematopoietic stem cells. The very early hematopoietic stem cells (CD34+/CD38−) and the myeloid stem cells (CD34+/CD33+) had weak expression of C/EBPε similar to the very early myeloblast cell line (eg, KG-1). Levels of C/EBPε mRNA increased as these cells differentiated towards granulocytes. Likewise, purified granulocytes also expressed C/EBPε (Fig 1); and this expression appeared to be greater than C/EBPε levels present in macrophages. Furthermore, levels of C/EBPε protein increased as NB4 cells differentiated toward granulocytes. Levels of C/EBPε mRNA decreased as HL-60 and KG-1 cells differentiated toward macrophages. Levels of C/EBPε protein expression slightly declined as NB4 cells were treated with the potent vitamin D3 analog (KH1060) as they underwent partial macrophage-differentiation (Fig 5). In summary, C/EBPε expression was most robust in myeloid committed stem cells as they differentiated down the granulocytic pathway of development.
At the late myeloblastic and promyelocytic stages of development, one of the key events is the development of primary, azurophilic granules, which contain newly synthesized antimicrobial enzymes, including MPO. Although the correlation was not perfect, most cell lines that expressed C/EBPε also expressed MPO, and vice versa. Furthermore, we have shown that ectopic expression of C/EBPε in CEM cells can result in a fivefold to sixfold upregulation of a reporter gene containing the MPO promoter.1 Expression of myeloid differentiation products probably involves the combinatorial interaction of a variety of transcriptional factors. Further studies to define the myeloid-specific target genes that are modulated by C/EBPε will require a series of mice having single and combined disruption of transcriptional factors including C/EBPε.
The C/EBPε protein has a leucine zipper; this motif is the site for homo- and heterodimerization to other proteins with leucine zippers. The known leucine zipper-containing proteins that are expressed during myelopoiesis include Jun, Fos, and C/EBP-α, -β, -δ. We previously showed that Jun and Fos transcripts were induced as HL-60 myeloblasts differentiated towards macrophages, but not towards granulocytes.58 Mice with disruption of the Fos gene have abnormalities of bone formation, but not abnormalities in development of granulocytes.59 60 Therefore, heterodimerization of C/EBPε to these two proteins is unlikely to be critical for granulocyte differentiation. The present studies showed that the promyelocytic myeloid cell line, NB4, expressed prominent levels of C/EBPε, but not C/EBP-α, -β, -δ; therefore, C/EBPε probably does not require heterodimerization to these leucine-zipper proteins for activity at least at the promylocyte stage of differentiation. We have recently begun to identify protein partners of C/EBPε using the yeast two-hybrid system. Preliminary studies suggest that C/EBPε homodimerizes poorly (data not shown).
Forced expression of C/EBPε in the NB4 cells increased their cloning efficiency in soft-agar1 and decreased expression of C/EBPε in HL-60 and NB4 cells using antisense olignucleotides to C/EBPε decreased their clonal growth in soft agar. These experiments suggest that C/EBPε may play a role in growth of myeloid cells at the myeloblast and promyelocyte stage of development. Earlier myeloid cells such as 34+/38− stem cells and KG-1 early myeloblasts express nearly undetectable levels of this transcription factor suggesting it is probably less important at this earlier stage of development. Development of C/EBPε “knock-out” mice will help define the role of C/EBPε in hematopoiesis in vivo.
Taken together, our results show that expression of C/EBPε is restricted to hematopoietic tissues; higher levels of expression were detected in hematopoietic cells of myeloid lineage; and the highest levels were observed as late myeloblasts and promyelocytes differentiated towards granulocyte-like cells. Thus, this transcriptional activator may play an important role in the regulation of granulocytic differentiation. Further studies of C/EBPε and its partners may yield important new information about the regulatory mechanism that coordinates the simultaneous processes of normal myeloid proliferation and differentiation.
Supported by National Institutes of Health (NIH) grants and also in part by the Concern Foundation (Los Angeles, CA) and the Parker Hughes Trust (Los Angeles, CA). H.P.K. is a member of the UCLA Jonsson Comprehensive Cancer Center and holds an endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center/UCLA School of Medicine.
Address reprint requests to H. Phillip Koeffler, MD, Division of Hematology/Oncology, Cedars-Sinai Medical Center/UCLA School of Medicine, 8700 Beverly Blvd, B-213, Los Angeles, CA 90048.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal