Abstract
The short-chain fatty acid butyrate has been shown to elevate fetal hemoglobin (HbF ) by inducing expression of the γ-globin gene. Regulation of gene expression by butyrate is thought to proceed via inhibition of the enzyme histone deacetylase, leading to elevated levels of core histone acetylation which affect chromatin structure and transcription rates. To determine whether changes in histone acetylation are critical for the regulation of the γ-globin gene, we tested three potent and specific inhibitors of histone deacetylase, the cyclic tetrapeptides trapoxin and Helminthsporium carbonum toxin (HC toxin), and the antifungal antibiotic trichostatin A for their ability to induce fetal hemoglobin expression in erythroid cells. These compounds induced fetal hemoglobin in both primary erythroid cell cultures and human erythroleukemia (K562) cells. A butyrate-responsive element spanning the duplicated CCAAT box region of the γ-globin promoter has been identified in transient transfection assays using a reporter construct in K562 cells, and we show that the same promoter region is required for response to trapoxin and trichostatin. Mutational analysis of the γ-globin promoter indicates that the distal CCAAT box and 3′ flanking sequence (CCAATAGCC) is critical for activation by butyrate, trapoxin, and trichostatin, whereas the proximal element (CCAATAGTC) plays a less important role. These results show that inhibition of histone deacetylase can lead to transcriptional activation of γ-globin promoter reporter gene constructs through proximal promoter elements, and suggest that butyrate induces γ-globin expression via such changes in histone acetylation.
THE EMBRYONIC, fetal, and adult globin genes in the human β-globin cluster are regulated in a stage-specific manner with the fetal-to-adult switch occurring soon after birth.1 The pharmacologic elevation of fetal γ-globin gene expression is being explored as a treatment for patients suffering from β-globin deficiencies. One inducer of HbF, hydroxyurea, has shown clinical efficacy for treatment of sickle cell anemia,2 making it the first real treatment option for some patients. A separate group of compounds currently under investigation in the clinic are short-chain fatty acids, including butyrate. Butyrate treatment increases levels of fetal hemoglobin in experimental models and in humans,3 4 but the mechanism of butyrate action is not understood.
In addition to regulating γ-globin expression, butyrate inhibits growth of a variety of cell lines and induces expression of a number of different genes. The pleiotropic effects of butyrate appear to be due, at least in part, to the inhibition of histone deacetylating enzymes in cells. Core histones are subject to reversible acetylation of the ε-amino group of specific lysine residues located in conserved amino terminus regions.5 Changes in histone acetylation are thought to cause changes in chromatin structure by altering the way in which histones bind to DNA or to other nuclear proteins.5 Such changes appear to be important for transcriptional regulation of genes in general, but the precise molecular mechanisms by which butyrate activates specific genes in individual cell types are largely unknown. Butyrate is a fairly weak inhibitor of histone deacetylase activity in crude cell extracts (Ki = 60 μmol/L, ref 6), and in intact cells millimolar concentrations of butyrate are required to cause accumulation of hyperacetylated histones.7 The requirement for treatment with high levels of butyrate to obtain optimal effects raises the question of whether butyrate has other, perhaps nonspecific, effects on cells.
The effort to understand the action of butyate and the role of histone acetylation in control of cell growth and gene expression has been facilitated by the discovery of potent, specific inhibitors of histone deacetylase.8 The cyclic tetrapeptide trapoxin is an irreversible inhibitor of histone deacetylase at nanomolar concentrations.9 Trapoxin contains an unusual amino acid that may act as a lysine substrate mimic and whose reactive epoxy ketone side chain forms a covalent complex with histone deacetylase.9 Trapoxin belongs to a family of cyclic tetrapeptides that all contain a similar reactive residue and at least two other members of this family, Helminthsporium carbonum (HC toxin) and chlamydocin, are also histone deacetylase inhibitors.10 The high affinity and specificity of trapoxin for histone deacetylase was exploited by using trapoxin as an affinity probe to isolate a histone deacetylase catalytic subunit for the first time.11 Trichostatin A is another potent histone deacetylase inhibitor that is structurally distinct from trapoxin. Trichostatin is a reversible inhibitor of partially purified histone deacetylase (Ki = 3.4 nmol/L, ref 12). Histone deacetylase appears to be the specific target for trichostatin in cells because selection of cells for resistance to the cytostatic effects of trichostatin results in the appearance of cells containing histone deacetylase activity that is less sensitive to trichostatin inhibition.12
To determine whether inhibition of histone deacetylase is involved in the induction of the γ-globin gene by butyrate, we studied the effects of butyrate, trapoxin, HC toxin, and trichostatin on primary erythroid progenitor cells and K562 cells. Primary erythroid progenitor cells in culture provide an in vitro model closely approximating hemoglobin (Hb) production that occurs under physiological conditions.13 These cultures respond to agents known to affect HbF levels, including butyrate, and have been used to assess the ability of compounds to elevate γ-globin gene expression.14,15 The K562 cell line constitutively expresses embryonic (ε-) and fetal (γ-) globin from the β-globin locus, and K562 cells can be induced to express higher levels of ε- and γ-globin by differentiation-inducing agents such as hydroxyurea and butyrate.16 K562 cells have an advantage over primary cells of being easily cultured and transfected, facilitating studies of transcriptional regulation. Previous studies of butyrate regulation of γ-globin expression in K562 cells show that the regulation occurs at least in part at the transcriptional level, because butyrate can increase expression of a γ-globin promoter construct fused to a reporter gene in a transient transfection assay.17 Activation by butyrate and other short-chain fatty acids is dependent on a duplicated CCAAT box sequence located just upstream of the transcription start site.17 18
We report here that the specific histone deacetylase inhibitors trapoxin, trichostatin, and HC toxin can induce fetal Hb expression in primary erythroid cell cultures and increase Hb expression in K562 cells. In addition, the inhibitors increase activity of the γ-globin promoter in K562 cells by acting on CCAAT box sequences identical to those required for butyrate induction. These results show that discrete γ-globin promoter elements can mediate a transcriptional response to changes in histone acetylation. Furthermore, inhibition of histone deacetylase activity appears sufficient to activate γ-globin gene expression in both primary cells and in K562 cells, suggesting that histone deacetylase is the relevant target for butyrate action in elevating HbF.
MATERIALS AND METHODS
Cell culture and transfections.Cultures of erythroid progenitor cells were established as previously described.13 HbF in mature cultures was analyzed by high-performance liquid chromatography (HPLC).14 The human erythroleukemia cell line K562 was obtained from the American Type Culture Collection (ATCC; Rockville, MD) and grown in RPMI (GIBCO, Grand Island, NY) supplemented with 10% fetal bovine serum (Biowhittaker, Walkersville, MD), 50 U/mL penicillin, and 50 μg/mL streptomycin (GIBCO). Trapoxin and trichostatin were provided by Shionogi and Co (Osaka, Japan). HC toxin was purchased from Sigma (St Louis, MO). For Hb induction assays, K562 cells were plated at a dilution of 2 × 105 per mL and inducing agents were added. Three days later the cells were harvested by centrifugation, washed twice with phosphate-buffered saline (PBS), and lysed by resuspending in water. Cell lysates were assayed for Hb using the plasma Hb assay kit (Sigma) in a microtiter plate format. Protein concentration was determined in cell extracts using the Coomassie Plus assay reagent (Pierce, Rockford, IL), and Hb content was expressed as nanograms of Hb per micrograms of protein.
For transfection experiments, K562 cells were plated at a dilution of 2 × 105 per mL. Cells were collected the next day and washed in sterile 1× PBS. Fifty micrograms of supercoiled CAT reporter plasmid (prepared by CsCl gradient or LiCl/PEG precipitation) and 7 μg supercoiled pCMV-βgal transfection control plasmid (Clontech, Palo Alto, CA) were added to 1 × 107 cells in 800 μL RPMI. Each sample of cells was electroporated in a 0.4-cm gap cuvette (Biorad, Hercules, CA) at 260 V/960 μFD using a Biorad Gene Pulser. After electroporation, cells were cultured in serum-supplemented RPMI for 6 hours, after which the culture was split into aliquots and drugs were added. After 36 hours, the cells were harvested by centrifugation and lysed by freeze-thawing in 250 mmol/L Tris, pH 7.5. Butyrate has been shown to activate several viral promoters, and preliminary experiments revealed that expression of the internal control plasmid, CMV-β-gal, was increased by butyrate. Therefore, for determination of transfection efficiencies, β-galactosidase activity was assayed19 in extracts from untreated cultures only. CAT assays were performed as described20 after normalizing for the amount of β-galactosidase activity in the untreated cultures.
γ-Globin promoter constructs.Deletions were generated by polymerase chain reaction (PCR) using six different oligonucleotides designed to give the endpoints shown in Fig 3, paired with an oligonucleotide extending to +45 bp relative to the transcription intitation site. Human genomic DNA was used as a template, and the resulting PCR products contained sequences derived from both the Aγ and Gγ globin genes. These genes share extensive sequence identity in their promoter regions and appear to be regulated identically.1 PCR products were cloned into the pCAT Basic vector (Promega, Madison, WI), and sequenced. Promoter sequences derived from both γ-globin genes could be identified for some fragments, and Aγ promoter sequences were selected for transfection experiments. Mutations were introduced into the −198 γ-globin promoter-CAT plasmids by PCR methods.21
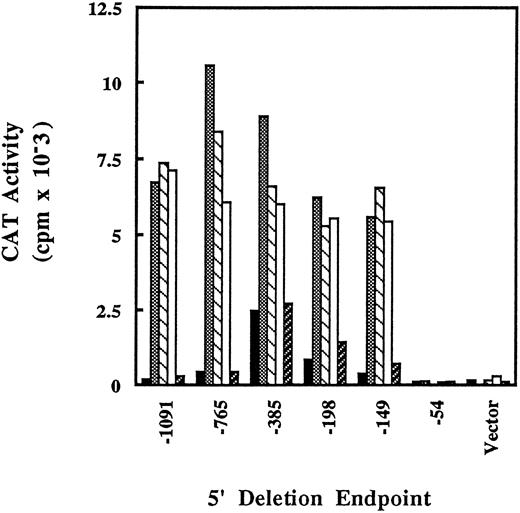
Deletion analysis of γ-globin promoter-CAT fusion constructs in K562 after induction by butyrate, histone deacetylase inhibitors, or hydroxyurea. K562 cells were transfected (see Materials and Methods) with γ-globin promoter-CAT plasmids containing sequences from −1091 to +45 bp relative to the transcription start site with the indicated 5′ ends. The transfected cells were treated with arginine butyrate (, 1 mmol/L), trapoxin (▧, 2.5 nmol/L), trichostatin A (□, 100 nmol/L), or hydroxyurea (▨, 1 mmol/L) for 36 hours, then harvested and assayed for CAT activity. (▪), Untreated. The experiment shown is representative of three transfections done with the deletion series.
Deletion analysis of γ-globin promoter-CAT fusion constructs in K562 after induction by butyrate, histone deacetylase inhibitors, or hydroxyurea. K562 cells were transfected (see Materials and Methods) with γ-globin promoter-CAT plasmids containing sequences from −1091 to +45 bp relative to the transcription start site with the indicated 5′ ends. The transfected cells were treated with arginine butyrate (, 1 mmol/L), trapoxin (▧, 2.5 nmol/L), trichostatin A (□, 100 nmol/L), or hydroxyurea (▨, 1 mmol/L) for 36 hours, then harvested and assayed for CAT activity. (▪), Untreated. The experiment shown is representative of three transfections done with the deletion series.
Histone preparation and gel fractionation.Histones were prepared by a modification of a previously described method.6 K562 cells were seeded at a density of 2 × 105 per mL and drugs were added immediately. Cells (5 × 106) were procured 24 hours later and washed twice in PBS. Cells were lysed by Dounce homogenization in 1 mL ice-cold lysis buffer (10 mmol/L Tris pH 8, 50 mmol/L NaCl, 1% Triton X-100, 10 mmol/L MgCl2 , 8.6% sucrose, and 0.5 mmol/L dithiothreitol [DTT]). Nuclei were collected by centrifugation for 5 minutes at 6,000 rpm in a microcentrifuge. The pellet was washed twice with 500 μL lysis buffer, once with 500 μL 10 mmol/L Tris pH 8.0/13 mmol/L EDTA, and resuspended in 125 μL water. Sulfuric acid was added to a concentration of 0.4 N and the tubes were incubated on ice for 1 hour. Debris was pelleted by centrifugation, and the supernatant was collected. Histones were precipitated by addition of 10 vol of acetone and incubation at −20°C overnight. Pellets were collected by centrifugation, briefly dried under vacuum, and resuspended in gel loading buffer (6 mol/L urea, 0.9 mol/L acetic acid, 5% 2-mercaptoethanol, 0.2% methylgreen).
Histone acetylation was evaluated by fractionating histones on acid/urea/acrylamide gels. Gels were poured using 20 ×20-cm glass plates with 1.4-mm spacers. The lower gel was 15% acrylamide, 2.5 mol/L urea, 5% acetic acid with 0.5% TEMED, and 0.12% ammonium persulfate. The stacking gel was 7.5% acrylamide, 2.5 mol/L urea, 5% acetic acid, 0.75% TEMED, and 0.15% ammonium persulfate. After polymerization, gels were prerun overnight at 80 V in 5% acetic acid running buffer. Before loading samples the running buffer was replaced, and the gel loaded and run overnight at 90 V and stopped 30 minutes after the slow-moving dye component had run out of the gel. Gels were fixed and stained for 1 hour in 0.25% Coomassie blue/10% acetic acid/40% methanol, then destained with repeated changes of acetic acid/methanol.
RESULTS
When trapoxin, HC toxin, and trichostatin A were added to primary erythroid progenitor cell cultures at optimal concentrations, the compounds induced a 1.5- to 1.8-fold increase in fetal Hb (Table 1). The concentrations required for maximal induction of HbF by these inhibitors were in the nanomolar range, compared to millimolar for butyrate, consistent with their higher potencies as histone deacetylase inhibitors. These results show the ability of these compounds to specifically induce γ-globin in a normal erythroid progenitor cell assay.
Fetal Hb Induction in Primary Erythroid Cell Cultures
| . | % HbF Fold Increase . | % HbF Fold Increase . | ||
|---|---|---|---|---|
| . | . | . | . | . |
| Treatment . | (Experiment 1) . | (Experiment 2) . | ||
| . | . | . | . | . |
| No addition | 4.18 | 1.35 | ||
| 1 mmol/L butyrate | 6.56 | 1.6 | ||
| 100 μmol/L hydroxyurea | 6.85 | 1.6 | ||
| 250 nmol/L trichostatin | 6.46 | 1.5 | 2.42 | 1.8 |
| 10 nmol/L trapoxin | 2.30 | 1.7 | ||
| 10 nmol/L HC toxin | 2.39 | 1.8 | ||
| . | % HbF Fold Increase . | % HbF Fold Increase . | ||
|---|---|---|---|---|
| . | . | . | . | . |
| Treatment . | (Experiment 1) . | (Experiment 2) . | ||
| . | . | . | . | . |
| No addition | 4.18 | 1.35 | ||
| 1 mmol/L butyrate | 6.56 | 1.6 | ||
| 100 μmol/L hydroxyurea | 6.85 | 1.6 | ||
| 250 nmol/L trichostatin | 6.46 | 1.5 | 2.42 | 1.8 |
| 10 nmol/L trapoxin | 2.30 | 1.7 | ||
| 10 nmol/L HC toxin | 2.39 | 1.8 | ||
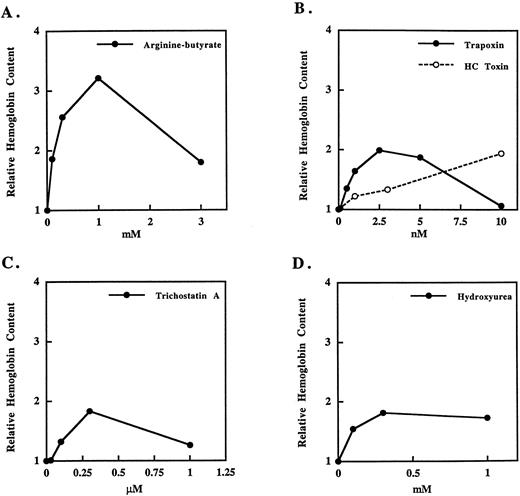
We used K562 cells to further study the mechanism of induction of γ-globin by histone deacetylase inhibitors. Treatment with butyrate has been shown to cause the differentiation of K562 cells as indicated by inhibition of cell growth and increased expression of Hb consisting of predominantly ε- and γ-globin chains.16 22 We observe a dose-dependent increase in total Hb in K562 cultures after a 3-day treatment with the arginine salt of butyrate with a maximal effect at 1 mmol/L butyrate (Fig 1A). At higher butyrate concentrations, cell number and Hb content of cultures is decreased because of the cytotoxic effects of butyrate. Treatment with the cyclic tetrapeptide deacetylase inhibitors trapoxin or HC toxin (Fig 1B) or trichostatin A (Fig 1C) also causes an increase in the total Hb content of the cultures. Like butyrate, these compounds all cause cell death at concentrations only slightly higher than those required for Hb induction. Cells treated with histone deacetylase inhibitors show consistently lower levels of Hb than butyrate-treated cells, but show Hb induction levels similar to those seen with hydroxyurea treatment (Fig 1D).
Induction of Hb in K562 cells by butyrate, histone deacetylase inhibitors, or hydroxyurea. K562 cells were treated for 3 days with the indicated compounds and assayed for total Hb as described in Materials and Methods. The curves shown are representative of at least three experiments with each compound.
Induction of Hb in K562 cells by butyrate, histone deacetylase inhibitors, or hydroxyurea. K562 cells were treated for 3 days with the indicated compounds and assayed for total Hb as described in Materials and Methods. The curves shown are representative of at least three experiments with each compound.
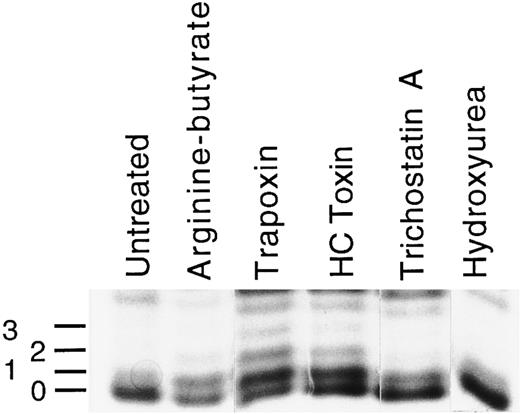
Histone acetylation in K562 cells after treatment with butyrate, histone deacetylase inhibitors, or hydroxyurea. Cells were treated for 24 hours with the arginine-butyrate (1 mmol/L), trapoxin (2.5 nmol/L), HC toxin (10 nmol/L), trichostatin A (0.5 μmol/L), or hydroxurea (1 mmol/L), after which histones were isolated (see Materials and Methods) and fractionated on acid/urea gels. A gel stained with Coomassie blue is shown. The identities of the H4 bands were verified by protein microsequencing. The lowest band in each lane is unacetylated H4, and the numbers to the left indicate the number of acetylated lysines in each of the higher bands.
Histone acetylation in K562 cells after treatment with butyrate, histone deacetylase inhibitors, or hydroxyurea. Cells were treated for 24 hours with the arginine-butyrate (1 mmol/L), trapoxin (2.5 nmol/L), HC toxin (10 nmol/L), trichostatin A (0.5 μmol/L), or hydroxurea (1 mmol/L), after which histones were isolated (see Materials and Methods) and fractionated on acid/urea gels. A gel stained with Coomassie blue is shown. The identities of the H4 bands were verified by protein microsequencing. The lowest band in each lane is unacetylated H4, and the numbers to the left indicate the number of acetylated lysines in each of the higher bands.
To confirm that these compounds can act as histone deacetylase inhibitors in K562 cells, we assessed the level of histone acetylation in cells after treatment with these agents. Histones were isolated from cells and fractionated on acid/urea/polyacrylamide gels, and histone bands were visualized by staining with Coomassie blue. Changes in histone H4 acetylation in treated cells could be detected readily (Fig 2). The H4 band at the bottom of each lane (marked 0) represents unacetylated H4, with the mono-, di-, and tri-acetylated forms (marked 1, 2, and 3) migrating with successively lower mobility. This particular gel ran unevenly, so that the mobility of unacetylated H4 varies from lane to lane; the lowest band visible in each lane represents unacetylated H4. Unacetylated H4 is the predominant species in untreated cells, with a smaller amount of the monoacetylated form present. In cells treated with butyrate (1 mmol/L), there is a modest increase in H4 acetylation such that the mono-acetylated form is more abundant, the relative amount of unacetylated H4 decreases, and a di-acetylated form becomes visible. Treatment with either trapoxin or HC toxin at nanomolar levels results in the clear accumulation of di- and tri-acetylated H4. In some gels, a tetra-acetylated H4 band was clearly resolved; in this gel it is obscured by a comigrating band (uppermost band in each lane). Trichostatin (0.5 μmol/L) has a similar effect to butyrate, increasing slightly the amount of mono- and di-acetylated H4. The effects of trapoxin and HC toxin on H4 acetylation are more dramatic than those of arginine butyrate or trichostatin A, perhaps because of their action as irreversible inhibitors. In contrast to the effects of these agents, treatment of cells with hydroxyurea at 1 mmol/L has no effect on histone acetylation.
Sequences in the proximal promoter of the γ-globin gene are important for developmental and butyrate-induced gene expression in vivo.1,23 Butyrate treatment of K562 cells has been shown to activate transcription from reporter gene constructs bearing these proximal promoter elements.17 To study the regulation of specific proximal promoter elements by specific histone deacetylase inhibitors, we constructed reporter gene fusion plasmids with up to 1091 bp relative to the transcription start site fused to a CAT reporter gene. These plasmids were transfected into K562 cells by electroporation. Cells were treated with butyrate or other histone deacetylase inhibitors, obtained 36 hours later, and cell extracts were assayed for CAT activity. A fusion plasmid containing −1091 bp of the γ-globin promoter was highly inducible by butyrate, trapoxin, and trichostatin (Fig 3). Deletion of bases to −765 did not significantly affect activity, whereas a further deletion to −385 caused an increase in basal (unstimulated) transcription levels but did not affect the level of activity in stimulated cells. Further deletions to −198 and −149 did not decrease activity of the promoter significantly compared with the −1091 construct. However, deletion of about 100 bp to −54 caused a total loss of both basal and stimulated promoter activity. Levels of activity with the −54 construct were similar to that seen with the CAT plasmid containing no promoter sequences (Vector). These results indicate that important transcription regulatory elements are located between −149 and −54 in the γ-globin promoter, and that this region is required for the response not only to butyrate but also for the response to other inhibitors of histone deacetylase. Hydroxyurea does not increase activity of any of the promoter constructs tested. Thus, the ability of butyrate, trapoxin, and trichostatin to activate the −54 to −149 region of the γ-globin promoter appears to be specific, presumably resulting from their ability to inhibit histone deacetylase. Further, these results are consistent with hydroxyurea increasing Hb in K562 cells by a mechanism that is distinct from that of butyrate and that may not involve activation of transcription from proximal promoter elements.
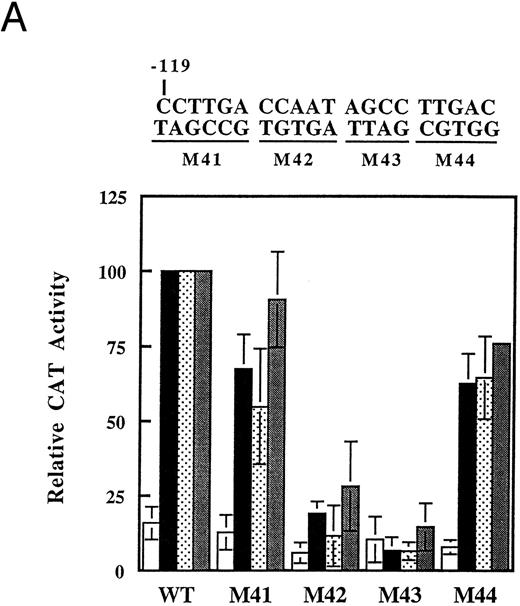
To further define the regions of the γ-globin promoter that are responsive to inhibitors of histone deacetylase, we introduced a series of 10-bp replacement mutations in the promoter between −149 and −55 (Fig 4A). Each construct was transfected into K562 cells and tested for induction by butyrate, trapoxin, and trichostatin. Mutation of the 20-bp region between −149 and −130 (M1 and M2) completely abolished the activity of the promoter, lowering both basal and inducible activity (Fig 4B). In addition, mutation of the 20-bp region between −119 and −100 (M4 and M5) also had a significant effect on promoter inducibility, whereas mutation of the region between −89 and −80 (M7) had a smaller effect. Induction of this series of mutants by trapoxin or trichostatin followed a similar pattern. These results show that two major and one minor elements (defined by M1/M2, M4/M5, and M7, respectively) are required for full activity in response to butyrate. Furthermore, the same promoter elements are essential for activation by trapoxin and trichostatin, consistent with the idea that these three agents use a common mechanism for γ-globin promoter induction.
Mutational analysis of the γ-globin promoter. (A) Sequence of the γ-globin promoter between −149 and −55. The 10-bp substitution mutations are indicated below. In addition to the 10-bp mutation, M1 has one base changed upstream of −149 in a region that does not appear to be important for butyrate induction (see Fig 3). (B) Activity of mutated γ-globin promoter constructs in K562 cells treated with butyrate, trapoxin, or trichostatin. After transfection, cells were cultured for 36 hours with no addition (Untreated) or with 1 mmol/L arginine-butyrate, 2.5 nmol/L trapoxin, or 100 nmol/L trichostatin. After harvesting, CAT activity was determined in the cell extracts. The CAT activity for each mutant is expressed relative to the activity of the unmutated −198 plasmid (WT) stimulated with butyrate, whose activity is set to 100%. Each construct was tested a minimum of three times with butyrate and the height of the bars represents the mean of all experiments, with the error bars indicating the standard deviation. Each construct was tested two or three times with trapoxin or trichostatin, and a representative experiment is shown.
Mutational analysis of the γ-globin promoter. (A) Sequence of the γ-globin promoter between −149 and −55. The 10-bp substitution mutations are indicated below. In addition to the 10-bp mutation, M1 has one base changed upstream of −149 in a region that does not appear to be important for butyrate induction (see Fig 3). (B) Activity of mutated γ-globin promoter constructs in K562 cells treated with butyrate, trapoxin, or trichostatin. After transfection, cells were cultured for 36 hours with no addition (Untreated) or with 1 mmol/L arginine-butyrate, 2.5 nmol/L trapoxin, or 100 nmol/L trichostatin. After harvesting, CAT activity was determined in the cell extracts. The CAT activity for each mutant is expressed relative to the activity of the unmutated −198 plasmid (WT) stimulated with butyrate, whose activity is set to 100%. Each construct was tested a minimum of three times with butyrate and the height of the bars represents the mean of all experiments, with the error bars indicating the standard deviation. Each construct was tested two or three times with trapoxin or trichostatin, and a representative experiment is shown.
The upstream region (M1 and M2) contains a CCACCC box that has previously been shown to function as a basal transcriptional element for the γ-globin promoter in K562 cells24,25 and in transgenic mice.26 The downstream site (M4 and M5) spans the distal of two CCAAT boxes, whereas the M7 mutation occurs over the proximal of these two CCAAT boxes. Our results confirm previous findings that the distal CCAAT box is the major butyrate-responsive element in the γ-globin promoter,27 and further show that the response to specific histone deacetylase inhibitors is also mediated by these sequences. To test the ability of histone deacetylase inhibitors to activate the CCACCC box and the duplicated CCAAT boxes separately, we made multimers of each site that were inserted upstream of the −54 γ-globin promoter-CAT plasmid. The activity of these multimers was compared to the activity of the −198 promoter fragment (Fig 5). In these experiments, a CCAAT box dimer([−126/−66]X2) is inducible by butyrate, trichostatin, and trapoxin (Fig 5 and Table 2). Although the basal and induced activity of the CCAAT box dimer is lower than that of the −198 construct (Fig 5), the fold induction is identical (Table 1, n = 3 experiments). In contrast, the multimer of the CCACCC box ([−148/−135]X3) was far less inducible. These data are consistent with the idea that the CCACCC box is required for basal promoter activity, while the duplicated CCAAT boxes are required for induction by butyrate and other inhibitors of histone deacetylase.
Activity of multimers of the CCACCC and CCAAT box regions of the γ-globin promoter. A trimer of the CCACCC box region (−148/−135) or a dimer of the CCAAT box region (−126/−66) was cloned into the −54 γ-globin promoter-CAT plasmid. These constructs were then transfected into K562 cells that were left untreated (▪) or treated with 1 mmol/L arginine butyrate (). A representative experiment of three trials for each multimer is shown.
Activity of multimers of the CCACCC and CCAAT box regions of the γ-globin promoter. A trimer of the CCACCC box region (−148/−135) or a dimer of the CCAAT box region (−126/−66) was cloned into the −54 γ-globin promoter-CAT plasmid. These constructs were then transfected into K562 cells that were left untreated (▪) or treated with 1 mmol/L arginine butyrate (). A representative experiment of three trials for each multimer is shown.
Induction of Multimeric γ-Globin Promoter Fragments
| . | Promoter construct . | ||
|---|---|---|---|
| . | −198 . | CCACCC . | CCAAT . |
| . | . | (−148/−135)X3 . | (−126/−66)X2 . |
| . | . | ||
| . | (fold induction) . | . | . |
| +Arginine-butyrate | 10.2 ± 1.1 | 2.3 ± 0.1 | 11.8 ± 1.3 |
| +Trapoxin | 1.9 | 9.9 ± 1.6 | |
| +Trichostatin | 2.9 | 13.8 | |
| . | Promoter construct . | ||
|---|---|---|---|
| . | −198 . | CCACCC . | CCAAT . |
| . | . | (−148/−135)X3 . | (−126/−66)X2 . |
| . | . | ||
| . | (fold induction) . | . | . |
| +Arginine-butyrate | 10.2 ± 1.1 | 2.3 ± 0.1 | 11.8 ± 1.3 |
| +Trapoxin | 1.9 | 9.9 ± 1.6 | |
| +Trichostatin | 2.9 | 13.8 | |
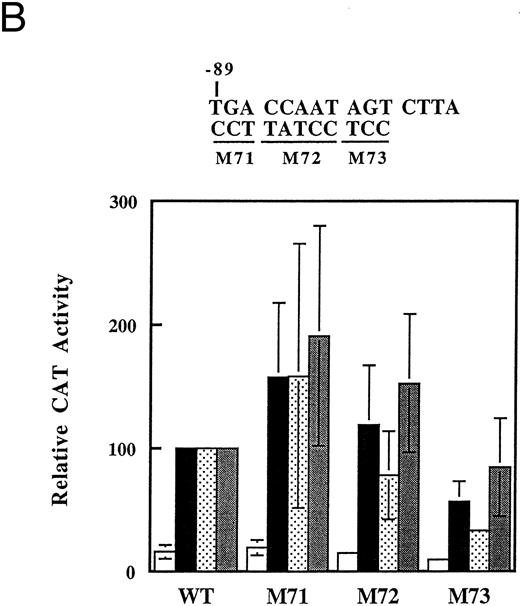
A more detailed mutational mapping of the CCAAT boxes indicates that it is the CCAAT sequence plus additional bases just 3′ to this sequence that are necessary for histone deacetylase inhibitor induction of the promoter. Base-substitution mutations were made in the distal and proximal CCAAT boxes in the context of the −198 promoter fragment, and tested for butyrate responsiveness in transient transfections (Fig 6). In the case of the distal CCAAT box sequence, mutation of nucleotides just 3′ to the CCAAT box (M43) had the most severe deleterious effect on promoter activity (Fig 6A). Mutation of the CCAAT sequence itself (M42) also had a profound effect and mutations of flanking sequences (M41, M44) had little or no effect on promoter activity. In all cases, induction by trapoxin or trichostatin was not significantly different for the mutants (Fig 6A). Experiments with mutations in the proximal CCAAT box gave much more variable results (Fig 6B). None of the three mutations tested dramatically decreased promoter activity, and in some experiments promoters with mutations in the CCAAT and 5′ residues (M71 and M72) displayed increased activity. Promoters with mutations in the bases just 3′ to the CCAAT sequence (M73) consistently gave lower activity than the unmutated promoter, but the effect was not as large as that seen with distal CCAAT box. These results indicate that response of the γ-globin promoter to butyrate and to specific inhibitors of histone deacetylase is for the most part dependent on a small area of the promoter within the distal CCAAT box region having the sequence CCAATAGCC. Definition of a discrete region in the promoter that is required for induction of the γ-globin promoter should allow identification and characterization of the relevant protein factors that bind to this region.
Mutational analysis of the CCAAT box region of the γ-globin promoter. The indicated base substitution mutations were introduced into the −198 γ-globin-CAT plasmid in the distal (A) or the proximal (B) CCAAT box. Transfections were performed as usual, and cells were left untreated (□) or treated with 1 mmol/L arginine-butyrate (▪), 2.5 nmol/L trapoxin (▧), or 100 nmol/L trichostatin () as indicated. CAT activity is expressed relative to the activity of the −198 construct (WT). The data shown are the average of two or three transfections done with each construct; error bars show the standard deviation where three trials were done.
Mutational analysis of the CCAAT box region of the γ-globin promoter. The indicated base substitution mutations were introduced into the −198 γ-globin-CAT plasmid in the distal (A) or the proximal (B) CCAAT box. Transfections were performed as usual, and cells were left untreated (□) or treated with 1 mmol/L arginine-butyrate (▪), 2.5 nmol/L trapoxin (▧), or 100 nmol/L trichostatin () as indicated. CAT activity is expressed relative to the activity of the −198 construct (WT). The data shown are the average of two or three transfections done with each construct; error bars show the standard deviation where three trials were done.
DISCUSSION
This work shows that specific inhibitors of histone deacetylase can mimic the effects of butyrate on γ-globin induction in primary erythroid cell cultures and in K562 cells. We have shown that the histone deacetylase inhibitors trapoxin and trichostatin induce accumulation of Hb and activate the γ-globin promoter in K562 cells. Furthermore, the promoter elements that are necessary for induction of γ-globin by butyrate are also required for induction by trapoxin and trichostatin. These results show that the γ-globin promoter can respond to changes in histone acetylation, and suggest that changes in levels of histone acetylation may play an important role in the regulation of γ-globin gene expression by butyrate.
Like butyrate, hydroxyurea induces fetal Hb expression in K562 cells16 as well as in primary erythroid cell cultures and in humans.14,28 However, hydroxyurea does not affect bulk histone acetylation nor does it activate any of the γ-globin promoter constructs tested. These results suggest that butyrate and hydroxyurea have different mechanisms for fetal Hb induction. This is consistent with the observed ability of butyrate to act additively or synergistically with drugs such as hydroxyurea or 5-azacytidine to induce fetal Hb in vivo.29 30
Our functional analysis of the activation of the γ-globin promoter by butyrate agrees with previous studies that have implicated two distinct promoter elements in expression of the γ-globin promoter in K562 cells.17,25 Both the CCACCC and CCAAT box regions appear to be bound by protein factors based on in vivo footprinting experiments.31 The upstream CCACCC box appears to function as a basal promoter element.24,25,32 The CCACCC box binds to and can be transactivated by Sp1 proteins, consistent with the activity of this element in several cell types.25,32 The region of the promoter containing the duplicated CCAAT boxes appears to be responsible for the ability of butyrate17 and other histone deacetylase inhibitors to activate the promoter, and furthermore the distal CCAAT box is functionally more important than the proximal element.27 Our results confirm the importance of the distal CCAAT box, and further define a minimal butyrate-responsive sequence as the CCAAT bases plus the four adjacent 3′ residues.
The mechanism by which changes in histone acetylation cause specific transcriptional activation through the CCAATAGCC sequence are not known. Acetylation of histone H4 in nucleosome cores has been shown to allow increased binding of transcription factors to their target DNA sequences.33 Thus, γ-globin promoter activation by histone deacetylase inhibitors could be due simply to increased binding of available transcription factors to exposed promoter sequences. Another possibility is that changes in histone acetylation could cause changes in the expression of factors that bind to this region. Butyrate has been reported to decrease expression of CCAAT displacement protein (CDP),34 a putative repressor of γ-globin transcription.17 However, it has not been shown that CDP is in fact a repressor of globin gene expression in K562 cells. In addition, Superti-Furga et al35 showed that mutation of 4 bp just upstream of the distal CCAAT sequence abolished binding of CDP to this sequence, whereas in our experiments a similar mutation (M41, see Fig 6) had only a small effect on promoter activity. These results suggest that proteins other than CDP may be relevant for the activation of the γ-globin promoter in response to butyrate, and at least two other proteins have been identified that bind to this region in vitro.27,35 36 Our fine mutational analysis of the duplicated CCAAT box region should allow the identification of proteins that regulate γ-globin gene transcription by binding to this region, and elucidation of the mechanism by which histone acetylation affects their activity.
The activity of the γ-globin promoter in K562 cells that have not been treated with butyrate resembles the activity of the γ-globin promoter as studied in transgenic mice, with gradually increasing activity with the −149, −189, and −385 fragments followed by a decrease in activity when sequences out to −765 are included. These results are consistent with the presence of a negative regulatory element between −382 and −730 that has been observed in transgenic mice.37 However, induction of transgenes by butyrate in these mice places butyrate responsive elements between −382 and −1030,23 in contrast to our results that implicate sequences proximal to −149 in the response to butyrate. The discrepancies in these results could reflect differences in regulation of the γ-globin promoter when it is introduced as a transfected plasmid in an erythroid cell line versus when it is integrated chromosomally in a normal cell. It is also possible that the mechanism of action of butyrate in cell lines and in a whole organism might differ. Our results in primary erythroid cell cultures and K562 cells strongly support the idea that the relevant biochemical target for butyrate action is inhibition of histone deacetylase. Therefore, it would be interesting to assess the effects of histone deacetylase inhibitors other than butyrate on γ-globin expression in the transgenic mouse model.
Although all of the histone deacetylase inhibitors elevated levels of H4 acetylation in K562 cells (Fig 2), the absolute levels of histone acetylation observed in the gel assay do not correlate with the levels of Hb expression. Thus, the irreversible inhibitors trapoxin and HC toxin induce the highest levels of histone acetylation, yet cause less Hb induction than butyrate. Histone acetylation is regulated in a complex manner, as suggested by the identification to date of four different histone acetylases and five deacetylases.38 Different deacetylase activities appear differentially inhibited by butyrate39 and could potentially show distinctive responses to trapoxin and trichostatin. The recent demonstration that some transcription factors contain intrinsic histone acetylase activity or recruit histone acetylases to transcription complexes38 raises the possibility of dynamic and specific regulation of histone acetylation at sites of active transcription. Thus, the lack of correlation between levels of histone acetylation in the gel assay and levels of gene expression could result from regulation of the γ-globin gene by small, specific changes in histone acetylation. In fact, our demonstration that four different histone deacetylase inhibitors from three structural classes (short-chain fatty acid, cyclic tetrapeptide, and trapoxin) all induce γ-globin in K562 cells and primary erythroid cells is strong evidence for an important role of histone acetylation in γ-globin gene regulation.
Although butyrate requires high concentrations (millimolar) to elicit optimal Hb induction in cells, trapoxin and trichostatin induce γ-globin at very low (nanomolar) concentrations. Despite their potency, all of these agents may be limited by their cytotoxicity, which occurs at concentrations only slightly higher than those required to detect Hb induction. Hyperacetylation of histones is associated with cell-cycle arrest,8 and butyrate has been shown to induce apoptosis in several cell lines.40 41 It is possible that limited and specific changes in histone acetylation are required for transcriptional regulation of the γ-globin gene, while excessive or inappropriate histone acetylation leads to aberrant changes in chromatin structure that can cause cells to arrest and undergo apoptosis. A greater understanding of the regulation of histone acetylation and its function in γ-globin gene expression will be instrumental for the development of more potent and specific agents for the pharmacological modulation of fetal Hb.
ACKNOWLEDGMENT
We thank Mark Fleming for sequencing histone samples and Guy Bemis for helpful discussions.
Address reprint requests to Patricia G. McCaffrey, PhD, Vertex Pharmaceuticals Incorporated, 130 Waverly St, Cambridge, MA 02139.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal