To the Editor:
The Rhesus (RH) blood group system, one of the most complex polymorphic systems in humans, encompasses at least 45 antigens. These antigens are carried by at least two red blood cell membrane proteins that are encoded by two homologous genes, RHD and RHCE.1 The RHD antigen is a mosaic structure of at least 37 epitopes. Rearrangements of the RHD gene with the RHCE gene or point mutations in the RHD gene result in the loss of one or more D epitopes. Individuals with partial D phenotypes can produce antibodies to the missing epitopes in response to transfusion with D-positive blood or by pregnancy with a D-positive fetus. Until now, the following partial D phenotypes have been described: DII, DIIIa, DIIIb, DIIIc, DIVa, DIVb, DVa, DVI, DVII, DDFR, DDBT, DDNU, DHMi, DHMii, and RoHar.2
Of the partial DVI phenotype, the partial D category that is most frequently leading to alloimmunization, two genotypes have been described3: the conversion type and the deletion type. In the conversion type, exons 4, 5, and 6 of the RHD gene are replaced by RHCE equivalents occurring in individuals of the DVI Ccee phenotype. Individuals of the DVIccEe phenotype were originally described as belonging to the deletion type in which exons 4, 5, and 6 of the RHD gene are lost. Recent evidence suggests that these two types can also be distinguished at the serologic level using anti-BARC serum4 or several monoclonal antibodies (IgG MoAbs NOI, SAL17-4E8, BRAD-3 and IgM MoAb CA27-4C5, all distributed via the Third International Workshop on Monoclonal Antibodies5 ).
In contrast to our previous results,3 we show now that exon replacement is involved in both DVI genotypes. In the present study, 12 individuals of the DVI phenotype were analyzed: 9 individuals of the DVICcee phenotype and 3 individuals of the DVIccEe phenotype, including the 2 individuals (307 and DEL) who were described as having the deletion type of DVI.3
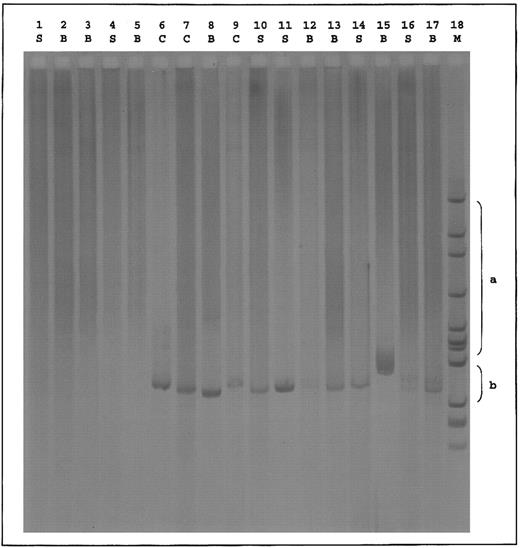
Standard RHD genotyping6 suggested the absence of RHD exon 4 and RHD intron 4 and the presence of RHD exon 7 and RHD 3′-noncoding region in all DVI individuals. In addition, six exon-specific primer sets amplifying RHD exons 3, 4, 5, 6, 7, and 9 were developed. From DNA of 3 individuals of the DVICe haplotype, RHD exons 3, 7, and 9 could be amplified, whereas exons 4, 5, and 6 could not. This is in agreement with the DVI exon 4/5/6 conversion genotype. From the DNA of 2 individuals of the DVIcE haplotype, RHD exons 3, 6, 7, and 9 could be amplified, whereas exons 4 and 5 could not. Furthermore, in polymerase chain reaction (PCR) experiments that were performed on the reticulocyte transcripts from the DVI variants previously described,3 we amplified hybrid D(exon 3)-CE(exon 4) fragments from both DVIccEe and DVICcee samples and a hybrid CE(exon 5)-D(exon 6) fragment only from the DVIccEe sample (Fig 1). From genomic DNA, a hybrid CE(exon 6)-D(exon 7) fragment could only be amplified from DVICcee samples.7 As expected, no amplification product was obtained with control dccee and DccEE samples.
Hybrid D-CE and CE-D PCR of reticulocyte transcripts from DVI phenotype individuals. cDNAs derived from reticulocytes of donors with the indicated phenotypes were amplified between two sets of primers in which one oligonucleotide is specific of the RHD gene and the other one of the RHCE gene. Set 1: 5′TTTGTCGGTGCTGATCTCAGTGGA3′ (RHD, exon 3); 5′GAACACGTAGAAGTGCCTCAG3′ (RHCE, exon 4). Set 2: 5′GGATGTTCTGGCCAAGTG3′ (RHCE, exon 5); 5′AGGTACTTGGCTCCCCCGGAC3′ (RHD, exon 6). PCR products were resolved by electrophoresis on 2.5% agarose gel and characterized by hybridization with the Rh cDNA probe.
Hybrid D-CE and CE-D PCR of reticulocyte transcripts from DVI phenotype individuals. cDNAs derived from reticulocytes of donors with the indicated phenotypes were amplified between two sets of primers in which one oligonucleotide is specific of the RHD gene and the other one of the RHCE gene. Set 1: 5′TTTGTCGGTGCTGATCTCAGTGGA3′ (RHD, exon 3); 5′GAACACGTAGAAGTGCCTCAG3′ (RHCE, exon 4). Set 2: 5′GGATGTTCTGGCCAAGTG3′ (RHCE, exon 5); 5′AGGTACTTGGCTCCCCCGGAC3′ (RHD, exon 6). PCR products were resolved by electrophoresis on 2.5% agarose gel and characterized by hybridization with the Rh cDNA probe.
Together with the sequence analysis of full-length RHD cDNA of individual 307, previously described as having the DVI deletion genotype,3 these results showed replacement of RHD exons 4 and 5 by their equivalent exons from the RHCE gene and not the deletion of exons 4, 5, and 6 in variants with the DVIcE haplotype.
The different conclusions from our previous published results could be explained by (1) the comigration of the rearranged CE(exon 4-5)-D(exon 6) BamHI genomic fragment with the restriction fragment of 5.3 kb carrying exons 4, 5, and 6 of the RHCE gene, which resulted in a higher intensity of this band in the DVIccEe (DEL and 307) as compared with the other DVICcee samples and D controls; and (2) the fact that the original sequence analysis of the DVI DEL transcripts was most likely performed on a splice variant of the Rh transcripts lacking exon 4-5-6.
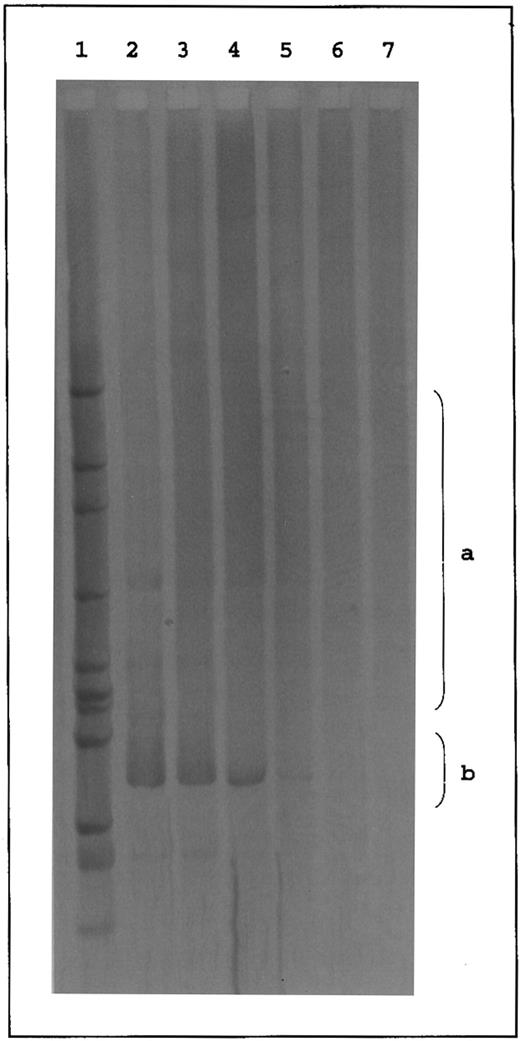
Western blot analysis performed with a monoclonal antibody recognizing a nonconformation epitope of the RhD antigen8 showed a 30- to 34-kD RhD polypeptide in the red blood cell membrane of DVIccEe (DEL and 307) and DVICcee (861 and BOU) variants, as in control DccEE samples (Fig 2). These results provided the definitive proof that the variant phenotypes of the DVIccEe samples previously investigated3 did not result from the expression of a deleted isoform of the RhD polypeptide.
Western blot analysis of RhD proteins from red blood cells of different DVI and common Rh phenotypes. Total membrane proteins from red blood cells of different DVI (DVI ccEe and DVI Ccee) and common (dccee and DccEE) Rh phenotypes were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12.5%) in nonreducing conditions, transferred to a nitrocellulose membrane (0.45 mm; Schleicher & Schuell, Dassel, Germany), and immunostained with LOR-15C9 monoclonal antibody.8 Immunoblots were finally stained with antihuman IgG peroxidase-tagged antibodies (Biosys, Compiegne, France) and the peroxidase activity was shown by the ECL chemoluminescence system from Amersham (Bucks, UK).
Western blot analysis of RhD proteins from red blood cells of different DVI and common Rh phenotypes. Total membrane proteins from red blood cells of different DVI (DVI ccEe and DVI Ccee) and common (dccee and DccEE) Rh phenotypes were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12.5%) in nonreducing conditions, transferred to a nitrocellulose membrane (0.45 mm; Schleicher & Schuell, Dassel, Germany), and immunostained with LOR-15C9 monoclonal antibody.8 Immunoblots were finally stained with antihuman IgG peroxidase-tagged antibodies (Biosys, Compiegne, France) and the peroxidase activity was shown by the ECL chemoluminescence system from Amersham (Bucks, UK).
In conclusion, our results show that the DVI deletion genotype does not exist. In the conversion type described before, exons 4, 5, and 6 of the RHD gene are replaced by RHCE equivalents occurring in individuals of the DVICcee phenotype. Individuals of the DVIccEe phenotype have the newly described conversion type in which RHD exons 4 and 5 are replaced by RHCE equivalents. Our results confirm the recent analysis of unrelated DVIccEe samples performed by Avent et al9 and Huang.10 Based on the serologic heterogeneity among DVI variants, it may be possible that, in the future, more rare DVI genotypes will be described.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal