Abstract
Phenotypic analysis of hematopoietic stem and progenitor cells (HSCs) has been an invaluable tool in defining the biology of stem cell populations. We have recently described the production of AC133, a monoclonal antibody (MoAb) that binds to a novel cell surface antigen present on a CD34bright subset of human HSCs. This antigen is a glycosylated protein with a molecular weight of 120 kD. Here, we report the molecular cloning of a cDNA encoding this antigen and show that it does not share homology with any previously described hematopoietic or other cell surface antigen(s). The AC133 polypeptide has a predicted size of 97 kD and contains five-transmembrane (5-TM) domains with an extracellular N-terminus and a cytoplasmic C-terminus. Whereas the expression of tetraspan (4-TM) and 7-TM molecules is well documented on mature and immature hematopoietic cells and leukocytes, this 5-TM type of structure containing two large (255–amino acid [aa] and 290-aa) extracellular loops is unique and does not share sequence homology with any known multi-TM family members. Expression of this protein appears limited to bone marrow in normal tissue by immunohistochemical staining; however, Northern analysis suggests that the mRNA transcript is present in a variety of tissues such as the kidney, pancreas, placenta, and fetal liver. The AC133 antigen is also expressed on subsets of CD34+ leukemias, suggesting that it may be an important early marker for HSCs, as well as the first described member of a new class of TM receptors.
THE ISOLATION OF hematopoietic stem and progenitor cells (HSCs) from bone marrow and peripheral blood using immunoselection with CD34 antibodies has been described1,2 and is considered an important advance for transplantation therapy in a number of disease states. Antibodies to CD34 recognize primitive, non–lineage-committed cells, as well as subsets of lineage-committed cells.3,4 We have described a monoclonal antibody (MoAb), AC133, that recognizes only CD34bright cells in bone marrow, fetal liver, cord, and peripheral blood that is efficient for immunoselection of HSCs using a magnetic bead system or fluorescence-activated cell sorting (FACS).5 The antigen recognized by MoAb AC133 is novel, and it is of significant interest because its expression on primitive stem and progenitor cell populations indicates that it may have a potentially important role in hematopoiesis and/or cell differentiation.
Four-transmembrane (4-TM) and 7-TM molecules form two major families of cell surface receptors that span the cell membrane multiple times and have important roles in the function of hematopoietic cells. 7-TM molecules such as the interleukin-8 receptors are well described, and are believed to interact with G-proteins as a result of binding chemotactic agonists.6-8 A 7-TM family member highly expressed in leukocytes9 has recently been identified as fusin, a molecule that renders CD4+ cells susceptible to HIV-1 ENV-mediated membrane fusion.10 The tetraspan or 4-TM superfamily includes molecules such as CD9 and CD37,11 and while specific roles have not been elucidated, it is speculated that they may function in the regulation of cell proliferation and activation.11 Members of both 4- and 7-TM families share significant degrees of homology with other family members, particularly in the TM domains, which has been the basis for successful cloning of additional family members.11,12 The size of the extracellular loops is relatively conserved within families, although the amino acid (aa) sequences and degree of glycosylation of these loops can be divergent.11
Here, we report the isolation, cDNA cloning, and tissue distribution of the antigen recognized by the AC133 antibody. The aa sequence of this protein, present on CD34bright HSCs, suggests a unique, 5-TM motif. Additionally, this protein does not share sequence homology with the TM domains of either the 4- or 7-TM families, and contains two extracellular loops that are twice as large as those proposed for 4- or 7-TM proteins. These data indicate that the AC133 antigen represents a new class of 5-TM receptors.
MATERIALS AND METHODS
Reagents. MoAb AC133 was prepared and purified in this laboratory. MoAb AC133-phycoerythrin (PE) conjugate was also prepared in this laboratory.13 Cyanogen bromide–activated Sepharose was purchased from Pharmacia (Alameda, CA), and MoAb AC133 affinity resin was prepared as directed with a 25-minute ligand-coupling reaction. The COS-7, Y79, and WERI-Rb-1 retinoblastoma cell lines were obtained from the American Type Culture Collection (Rockville, MD) and maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 2 mmol/L L-glutamine, and 15 mmol/L HEPES in a 9% CO2 atmosphere. COS-7 cells were removed from the flasks by Trypsin-EDTA digestion (GIBCO-BRL, Gaithersburg, MD). Phorbol myristate acetate (PMA) 1 ng/mL (GIBCO-BRL) was used to activate Y79 cells for 96 hours before harvesting for AC133 antigen isolation. Lysyl endopeptidase was supplied by Wako Chemicals (Chicago, IL). Custom primers were synthesized by Operon Technologies (Alameda, CA). AC133 magnetic conjugate was obtained from Miltenyi Biotec (Bergisch Gladbach, Germany).
Immunoprecipitation of AC133 antigen. PMA-activated Y79 cells or WERI-Rb-1 cells were washed with phosphate-buffered saline (PBS), biotinylated with NHS-Biotin (Pierce, Rockford, IL), and lysed in 0.125 mol/L NaCl, 25 mmol/L Tris, 0.005% NaN3 , 2.5 mmol/L EDTA, and 2.5% Brij 99/96 (2:1) detergent, pH 8, containing 1 mmol/L PMSF and 0.005 trypsin inhibitor U/μL aprotinin (extraction buffer). The lysates were incubated with MoAb AC133 after preclearing. Immune complexes were collected on Pansorbin cells (Calbiochem, La Jolla, CA) and run on 8% to 16% sodium dodecyl sulfate (SDS) gels (Novex, San Diego, CA).14 The protein was transferred to nitrocellulose filters and visualized with Streptavidin–horseradish peroxidase (HRP) (Amersham, Arlington Heights, IL) and the Super-Signal detection system (Pierce).
Purification of AC133 antigen. The AC133 antigen was isolated from 96-hour PMA-activated Y79 cells. The cells (2 × 109) were washed with PBS and lysed in extraction buffer. Cells were vortexed intermittently for 5 minutes at room temperature and then left on ice for 20 minutes. Cell nuclei and debris were removed by centrifugation at 10,000g for 10 minutes at 4°C. The lysate was filtered through a 0.2-μm filter before loading onto 0.5 mL MoAb AC133 affinity column equilibrated in wash buffer (0.125 mol/L NaCl, 25 mmol/L Tris, pH 8.0, 0.01% NaN3 , 2.5 mmol/L EDTA, and 0.1% Brij). The column was then washed extensively with wash buffer, and the antigen was eluted in 50 mmol/L ethanolamine, pH 11.5, 0.1% Brij and 0.01% NaN3 . The pH was immediately adjusted to neutral with HCl. Passage of the antigen eluate over a 300-μL bed volume DEAE column equilibrated in wash buffer removed many of the contaminating proteins, and a second affinity-chromatography step resulted in greater than 95% pure AC133 antigen amenable to proteolysis and protein sequence analysis. The purity and identity of AC133 antigen was confirmed by SDS-PAGE and Western analysis.15
Endoglyconase treatment of purified AC133 antigen. AC133 antigen (1 μg) was resuspended in 50 μL water, 125 μL 0.1-mol/L 2-mercaptoethanol, and 0.5% SDS. The protein was denatured at 100°C for 5 minutes. The denatured mixture (35 μL) was added to each of five tubes, and 25 μL 0.5-Mol/L Tris, pH 8, 10 μL water, and 10 μL 10% Nonidet P-40 were added to each tube. Zero to 0.1 U PNGase F (Sigma, St Louis, MO) was added to each tube, and the tubes were incubated at 30°C overnight. Deglycosylated antigen was visualized on a silver-stained16 SDS polyacrylamide gel.
Lysyl endopeptidase digestion of AC133 antigen and isolation of peptides. AC133 antigen was precipitated from 1.4 mL 2-μg/mL affinity column eluate by addition of trichloroacetic acid to 10%. The precipitated dry protein was suspended in 25 μL digest buffer (8 mol/L urea and 400 mmol/L Tris, pH 7.8). Five microliters of 45-mmol/L DTT was added, and the mixture was incubated at 50°C for 15 minutes. After cooling to room temperature, 5 μL 100-mmol/L iodoacetamide was added and the mixture was incubated for an additional 15 minutes. dH2O (70 μL) was added, diluting the urea to 2 mol/L, and 2 pmol LysC was added. The digestion was performed at 37°C for 24 hours. Peptides were isolated by high-performance liquid chromatography (HPLC) separation on a Vydac (Hesperia, CA) narrow-bore C18 reverse-phase column with a 4% to 32% acetonitrile gradient in 0.1% trifluoroacetic acid.
Protein sequence analysis of AC133 antigen peptides.N-terminal sequence analysis was determined using Edman chemistry17 18 on an Applied Biosystems (Foster City, CA) 477A or 473A liquid pulse protein sequenator. PTH-aa were separated on a Brownlee (Foster City, CA) C-18 reverse-phase column (2.1 mm × 22 cm) at 55°C in buffer A (3.5% tetrahydrofuran with addition of 2% to 4% ABI premix buffer concentrate) to buffer B (acetonitrile), with a 12% to 36% buffer B linear gradient over 18 minutes, followed by a 13-minute isocratic period at 36% buffer B.
cDNA cloning. Total RNA was isolated from WERI-Rb-1 retinoblastoma cells using TRI Reagent (MRC Inc, Cincinnati, OH),19 and poly A+ RNA was prepared using the Poly A+ Tract System (Promega Corp, Madison, WI). cDNA was synthesized20 using superscript reverse transcriptase (GIBCO BRL) and an oligo dT primer. The blunted cDNA was ligated to non–self-complementary Bst XI adaptors and gel-purified to remove unligated adaptors and small fragments. The linker cDNA was then ligated into the pcDNA-I expression vector (Invitrogen, San Diego, CA) and electroporated into Escherichia coli strain MC1061/P3.21,22 One hundred nanograms of the WERI-Rb-1 cDNA library was used as a polymerase chain reaction (PCR) template (per reaction) with 100 pmol each of degenerate sense and antisense primers designed from the protein sequence of four AC133 antigen peptides. PCRs were performed in 50 mmol/L KCl, 10 mmol/L Tris-HCI, pH 9.0, 0.1% Triton X-100, and 1.5 mmol/L MgCl2 with 5 U Taq DNA polymerase per reaction (Promega Corp, Madison, WI).23 Amplification was performed in an MJ Research Instrument (Watertown, MA) as follows: 92°C for 1 minute, 55° to 37°C for 1 minute, and 72°C for 3 minutes, for 35 cycles. After amplification, the reaction mixtures were run on 1% agarose gels, and unique bands not appearing in the individual primer controls were gel-purified and cloned into pCR 2.1 using the TA Cloning Kit (Invitrogen, San Diego, CA). The 5′ and 3′ ends of the gene were isolated by hemispecific PCR with nested sets of AC133 antigen gene-specific primers and library-specific primers. Twenty cycles of single-stranded PCR were performed with each gene-specific primer in a 50-μL reaction with 100 ng of the cDNA library and 10 pmol of each primer in PCR buffer with 5 U Taq polymerase. Ten microliters of this reaction mixture was removed and used as template for a second 35-cycle PCR using both the gene-specific primer and the library-specific primer. Five microliters of this PCR mixture was then used for another 35-cycle reaction using nested library- and gene-specific primers. Bands corresponding to the 5′ and 3′ ends of the gene were gel-purified and cloned into pCR 2.1. The overlapping cDNA clones were sequenced by the dideoxy chain reaction using fluorescent dye terminators and an ABI sequencer (Applied Biosystems, Foster City, CA).24
Northern analysis of AC133 antigen transcript. Total RNA was isolated from WERI-Rb-1 cells and Y79- and PMA-activated Y79 cells, and MACS separated HSCs from fetal liver as previously described.5 Northern blot analysis was performed using Clontech (Palo Alto, CA) multiple tissue Northern blots, and by resolving RNA samples on a 1% agarose–2-mol/L formaldehyde gel and capillary-blotting overnight onto nylon membrane. Total RNA was isolated with Tri Reagent, and 15 μg was loaded per lane. Staining of the blot with methylene blue was used to monitor RNA concentrations. An 800-bp EcoRI fragment of the cDNA was labeled with 32P-dCTP by random priming and used as a probe.
Expression of AC133 antigen in transfected COS-7 cells. Total RNA was isolated from WERI-Rb-1 and AC133+ fetal liver cells as already described. RT-PCR was performed using the Promega Access RT-PCR system (Promega Corp) with 1.0 μg total RNA template and primers directed before the start methionine and after the stop codon. The 2.8-kb band corresponding to the coding region of the gene was cloned into the Invitrogen directional eukaryotic TA cloning vector (pCR 3.1) containing the CMV promoter. Subconfluent COS-7 cells were removed from flasks with trypsin/EDTA, transfected with 5 μg cloned DNA by electroporation, and incubated for 48 hours before flow cytometric analysis. Transfected COS-7 cells were stained with 50 ng/106 cells MoAb AC133-PE, and analyzed on a FACScan (Becton Dickinson, San Jose, CA).
Immunohistochemical staining of paraffin-embedded tissue. Paraffin section immunohistochemistry was performed using a biotin-streptavidin method.25 In brief, incubation with antibody AC133 was followed by incubation with a biotinylated goat antimouse second-stage antibody (Jackson Immunoresearch Laboratories Inc, West Grove, PA) followed by HRP-conjugated streptavidin (Jackson Immunoresearch Laboratories Inc). Diaminobenzidene was used as the chromogenic substrate for HRP.
Expression of AC133 antigen on leukemia. Bone marrow aspirates were obtained from 12 adult and pediatric patients with acute leukemia after informed consent. Immunophenotyping was performed using multiparameter flow cytometry and various combinations of directly conjugated MoAbs, among which were CD7, CD19, CD33, CD34, CD38, CD45, HLA-DR (Becton Dickinson), CD90 (5E10; Pharmigen), CD117 (AMAC Inc, Westbrook, ME), and AC133, as well as the appropriate isotype-matched MoAb controls (Becton Dickinson). All samples were prepared using a stain-and-lyse method (R&D Systems, Minneapolis, MN) followed by fixation in 1% paraformaldehyde. List mode files of 50,000 events were acquired on a FACS caliber flow cytometer capable of four-color analysis. The leukemia cell population in each sample was identified by an aberrant pattern of hematopoietic marker expression on a blast cell population, and the leukemia was then classified as AML or ALL based on the pattern of lymphoid and myeloid markers. Data files were analyzed using Paint-a-Gate and Cell Quest software (Becton Dickinson Immunocytometry Systems), and the pattern of CD34, AC133, CD38, and CD90 expression on the leukemia cell population was determined. The staining pattern for leukemia cells using isotype-matched control antibodies was typically in the first decade of fluorescence intensity; these levels were then used to set staining thresholds on which to determine negative, dim, and positive patterns of marker expression.
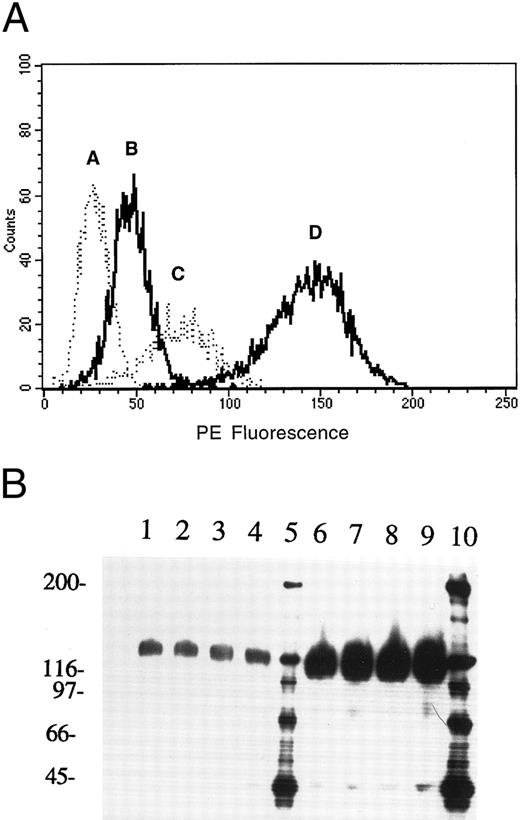
Expression of AC133 on retinoblastoma cell lines. (A) FACS analysis of Y79 and WERI-Rb-1 cell lines stained with AC133-PE: A, isotype control; B, Y79 cells; C, PMA-activated Y79 cells; D, WERI-Rb-1 cells. (B) Immunoprecipitation of AC133 antigen from PMA-activated Y79 and WERI-Rb-1 cells. Lanes 1 to 4, PMA-activated Y79 cells immunoprecipitated with 2.5, 5, 10, and 20 μg AC133, respectively. Lanes 6 to 9, WERI-Rb-1 cells immunoprecipitated with 2.5, 5, 10, and 20 μg AC133. Lanes 5 and 10, molecular weight markers.
Expression of AC133 on retinoblastoma cell lines. (A) FACS analysis of Y79 and WERI-Rb-1 cell lines stained with AC133-PE: A, isotype control; B, Y79 cells; C, PMA-activated Y79 cells; D, WERI-Rb-1 cells. (B) Immunoprecipitation of AC133 antigen from PMA-activated Y79 and WERI-Rb-1 cells. Lanes 1 to 4, PMA-activated Y79 cells immunoprecipitated with 2.5, 5, 10, and 20 μg AC133, respectively. Lanes 6 to 9, WERI-Rb-1 cells immunoprecipitated with 2.5, 5, 10, and 20 μg AC133. Lanes 5 and 10, molecular weight markers.
RESULTS
FACS analysis of retinoblastoma cell lines. The expression pattern of AC133 on a wide variety of cell lines and hematopoietic tissues is described in detail elsewhere.5 Expression of the AC133 antigen was limited to a subset of HSCs, two retinoblastoma cell lines (WERI-Rb-1 and Y79), and a teratocarcinoma cell line, NT-2. Both retinoblastoma cell lines were used in cloning the AC133 antigen gene. Expression of the AC133 antigen is upregulated by PMA activation on Y79 cells, but not on WERI-Rb-1 cells (Fig 1A). WERI-Rb-1 cells express approximately 10-fold more antigen than PMA-activated Y79 cells, as determined by FACS analysis and immunoprecipitation analysis (Fig 1A and B).
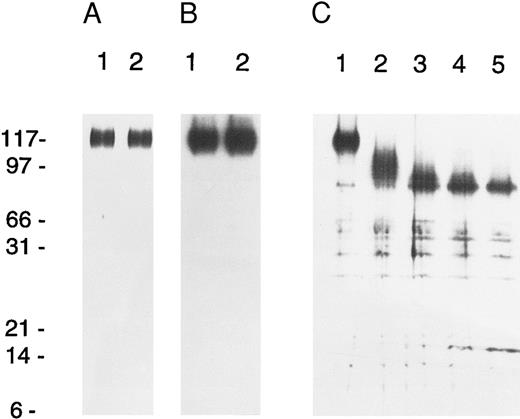
Isolation and protein sequencing of AC133 antigen. The 120-kd AC133 antigen was isolated by immunoaffinity chromatography from the Y79 retinoblastoma cell line, which had been PMA-activated for 96 hours before harvest. Sequential affinity chromatography and DEAE chromatography were used to generate more than 95% pure AC133 antigen by SDS-PAGE and silver staining (Fig 2A), and the identity of the purified molecule as AC133 antigen was confirmed by Western blotting (Fig 2B). Deglycosylation of the antigen with PGNase F to remove N-linked sugar shows that approximately 20 kd of the molecular weight is due to glycosylation (Fig 2C). Digestion of the purified antigen with lysyl endopeptidase followed by reverse-phase HPLC yielded four peptide sequences with lengths of 12 to 16 aa (Table 1). Searches of the major protein and nucleic acid databases with the peptide and resulting degenerate oligonucleotide sequences indicated that AC133 antigen did not share identity with any described molecules.
Isolation and analysis of AC133 protein. (A) Lanes 1 and 2, silver-stained gel of purified AC133 antigen prepared for protein sequence analysis. (B) Lanes 1 and 2, Western blot of partially purified AC133 antigen using AC133 as the primary antibody with an anti-mouse IgG-HRP conjugate as secondary antibody. Secondary antibody was detected by chemiluminescence with the Super-Signal system (Pierce). (C) Endoglyconase treatment of partially purified AC133 antigen. Lane 1, 0 U PNGase F; lane 2, 0.002 U; lane 3, 0.01; lane 4, 0.02 U; lane 5, 0.1 U. AC133 antigen loses approximately 20 to 25 kD upon N-linked deglycosylation.
Isolation and analysis of AC133 protein. (A) Lanes 1 and 2, silver-stained gel of purified AC133 antigen prepared for protein sequence analysis. (B) Lanes 1 and 2, Western blot of partially purified AC133 antigen using AC133 as the primary antibody with an anti-mouse IgG-HRP conjugate as secondary antibody. Secondary antibody was detected by chemiluminescence with the Super-Signal system (Pierce). (C) Endoglyconase treatment of partially purified AC133 antigen. Lane 1, 0 U PNGase F; lane 2, 0.002 U; lane 3, 0.01; lane 4, 0.02 U; lane 5, 0.1 U. AC133 antigen loses approximately 20 to 25 kD upon N-linked deglycosylation.
Peptide Sequences of AC133 Antigen
| Peptide . | Peptide Length . | aa Sequence . |
|---|---|---|
| -mer | ||
| 1 | 12 | (A, G, S)XNYELPAXNYE |
| 2 | 12 | LHQQDTQLRXXL |
| 3 | 12 | (P, A)LRTLLNETPEQ |
| 4 | 16 | XPAGVNLLXFAYDLEA |
| Peptide . | Peptide Length . | aa Sequence . |
|---|---|---|
| -mer | ||
| 1 | 12 | (A, G, S)XNYELPAXNYE |
| 2 | 12 | LHQQDTQLRXXL |
| 3 | 12 | (P, A)LRTLLNETPEQ |
| 4 | 16 | XPAGVNLLXFAYDLEA |
Peptides were obtained by Endo lys-C digestion of purified AC133 antigen. The protein sequence was obtained by standard Edman degradation in an ABI sequenator. Cycles in parentheses indicate the presence of multiple aa, and X indicates that no clear aa was determined for a cycle.
Isolation of a cDNA clone of AC133 antigen. To isolate the cDNA for this protein, a cDNA library was prepared from the WERI-Rb-1 retinoblastoma cell line. Degenerate primers were used in low-stringency PCR with the library to yield a reproducible 1.7-kb fragment. DNA sequence analysis of this fragment revealed that it contained a continuous open reading frame with the correct sequence of peptide 3 at the 5′ end and the correct sequence of peptide 4 at the 3′ end. Additionally, the sequence of peptide 2 was found within the fragment in the correct reading frame.
The 5′ and 3′ ends of AC133 cDNA were isolated using a hemispecific PCR approach. Nested pairs of AC133 gene-specific primers and vector primers were used to amplify 1.2- and 2-kb fragments overlapping with the initial 1.7-kb clone and corresponding to the 5′ and 3′ ends of the gene. Sequencing of the three partial overlapping clones predicted a 3.9-kb cDNA containing a long open reading frame of 2.7 kb with an ATG initiation codon at nucleotide 38, a UGA stop codon at nucleotide 2763, and a splice/poly (A) addition signal at nucleotide 3906 with a subsequent 20-base poly (A) tail. To isolate a full-length clone, primers were designed that flanked the start and stop codons and the 2.7-kb fragment cloned into the pCR3 vector.
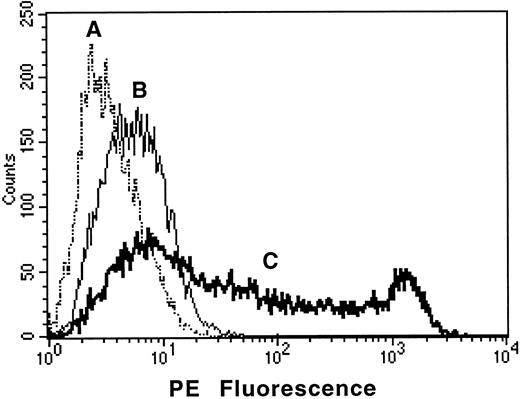
Surprisingly, when this clone was transfected into COS-7 cells, no expression of AC133 antigen was detected. Further sequence analysis revealed an Alu-like element at nucleotide 1178, which included several stop codons and thus would generate a truncated protein. Using primers flanking this region, PCR analysis of the WERI-Rb-1 cDNA library revealed only fragments that contained this element. However, using the same primers in RT-PCR analysis of total RNA derived from the WERI-Rb-1 cell line revealed that most of the amplified fragments had a consistent deletion of this element. Using RT-PCR with primers flanking the start and stop codons, a full-length clone was isolated from WERI-Rb-1 total RNA. This clone lacked the Alu-like element, and when transfected into COS-7 cells, it conferred expression of the AC133 antigen as assessed by flow cytometric and immunoprecipitation analysis (Fig 3). A similar clone was also isolated from the total RNA derived from human AC133+ HSCs isolated from fetal liver.
Transfection of COS-7 cells with the AC133 gene confers expression of AC133 antigen. COS-7 cells were transfected with AC133 antigen cDNA or with irrelevant CD8 cDNA and stained with AC133-PE after 48 hours of culture. (A), AC133 antigen–transfected COS stained with irrelevant isotype control; (B), CD8-transfected COS stained with AC133-PE; (C), AC133 antigen–transfected COS stained with AC133-PE. COS cells transfected with the irrelevant CD8 cDNA were nonreactive with AC133-PE, and AC133 antigen–transfected cells were nonreactive with a PE-conjugated isotype control.
Transfection of COS-7 cells with the AC133 gene confers expression of AC133 antigen. COS-7 cells were transfected with AC133 antigen cDNA or with irrelevant CD8 cDNA and stained with AC133-PE after 48 hours of culture. (A), AC133 antigen–transfected COS stained with irrelevant isotype control; (B), CD8-transfected COS stained with AC133-PE; (C), AC133 antigen–transfected COS stained with AC133-PE. COS cells transfected with the irrelevant CD8 cDNA were nonreactive with AC133-PE, and AC133 antigen–transfected cells were nonreactive with a PE-conjugated isotype control.
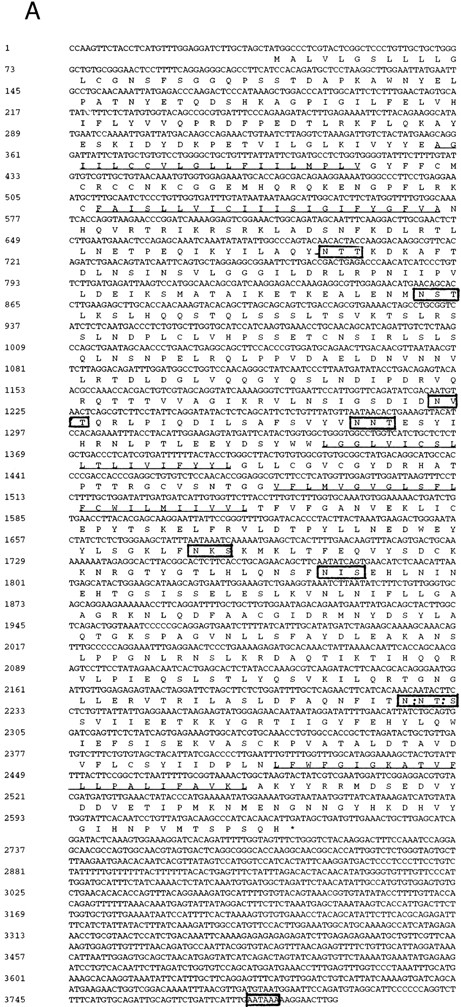
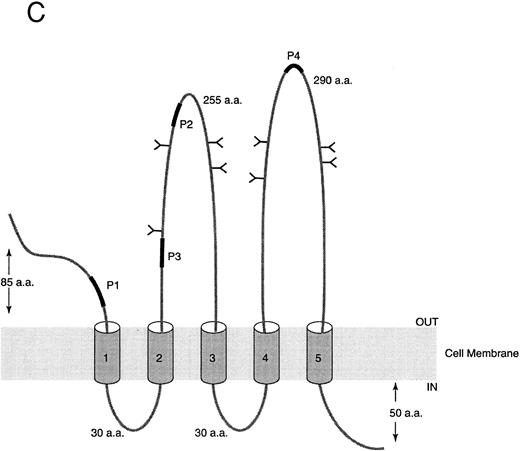
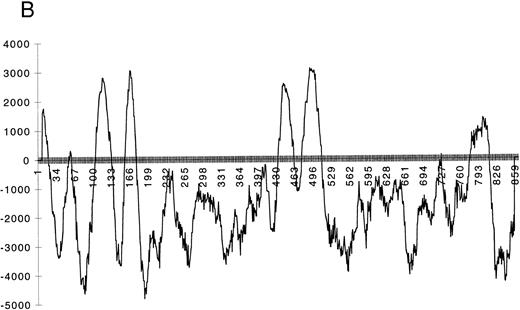
Sequence analysis of AC133 antigen. (A) Sequence of AC133 antigen cDNA. The 5-TM domains are underlined, the 8 N-linked glycosylation sites are in boxes (the seventh and eighth glycosylation sites are in 1 larger box; dashed lines indicate where they overlap), and the polyadenylation signal is in a small box. (B) Hydrophobicity analysis of the AC133 antigen protein sequence. The hydrophobic signal peptide is notable along with 5 very hydrophobic TM domains. The last TM domain contains a single lysine in the middle of the TM sequence. (C) Graphic of the proposed structural model of AC133 antigen. This protein is modeled as having an extracellular N-terminus, a cytoplasmic C-terminus (containing 5 tyrosine residues), 2 small cysteine-rich cytoplasmic loops, and 2 very large extracellular loops each containing 4 consensus sequences for N-linked glycosylation. The position of the 4 original peptides is indicated in bold.
Sequence analysis of AC133 antigen. (A) Sequence of AC133 antigen cDNA. The 5-TM domains are underlined, the 8 N-linked glycosylation sites are in boxes (the seventh and eighth glycosylation sites are in 1 larger box; dashed lines indicate where they overlap), and the polyadenylation signal is in a small box. (B) Hydrophobicity analysis of the AC133 antigen protein sequence. The hydrophobic signal peptide is notable along with 5 very hydrophobic TM domains. The last TM domain contains a single lysine in the middle of the TM sequence. (C) Graphic of the proposed structural model of AC133 antigen. This protein is modeled as having an extracellular N-terminus, a cytoplasmic C-terminus (containing 5 tyrosine residues), 2 small cysteine-rich cytoplasmic loops, and 2 very large extracellular loops each containing 4 consensus sequences for N-linked glycosylation. The position of the 4 original peptides is indicated in bold.
Characterization of AC133 cDNA and deduced protein. The cDNA encoding AC133 antigen is 3,794 nucleotides in length, with a 37-nucleotide 5′ untranslated region (UTR) and an 1,159-nucleotide 3′ UTR. There is a single long open reading frame of 2,598 nucleotides that predicts a protein of 865 aa with a molecular weight of 96.8 kd (Fig 4A). There is an ATG codon at nucleotide 38 flanked by a Kozak consensus sequence, with a guanine at position −3 and +4. This ATG is also followed by a consensus 19-aa signal peptide. There is also one upstream ATG at nucleotide 15 not flanked by a Kozak consensus sequence, and there is a stop codon in the same frame at nucleotide 33 just prior to the presumed functional ATG.
Hydrophobicity analysis of the sequence suggests that the protein is likely to have 5-TM domains in addition to the signal peptide (Fig 4B and C). The predicted protein contains a short 85-aa extracellular domain followed by two closely spaced TM domains, a large 255-aa extracellular domain, another pair of closely spaced TM domains, another large 290-aa extracellular domain, a final TM domain, and then a 50-aa C-terminal cytoplasmic tail. Prosite analysis of the protein reveals eight N-linked glycosylation sites, all within the putative extracellular domains, and a leucine zipper motif in the first extracellular domain. In addition, there are clusters of cysteine residues associated with the paired TM domains, including a cysteine residue within the TM segment of each of the four. The last TM domain contains a lysine residue within the middle of the TM segment. The cytoplasmic tail includes five tyrosine residues.
Comparison of the AC133 protein and nucleotide sequence with known sequences in the Genebank/EMBL databases reveals it to be a novel protein. However, there were nucleotide matches with several unpublished EST fragments in the dbEST. Matching clones were derived from retina (5), placenta (2), and infant brain, pancreatic islets, and colon (1).
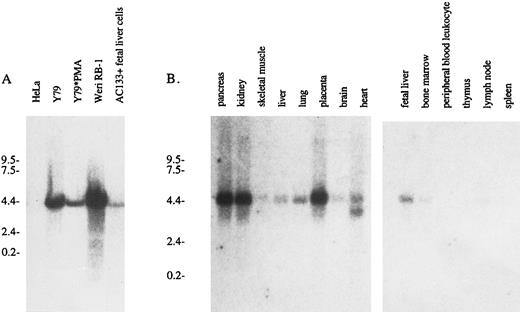
AC133 expression in various lymphoid and nonlymphoid cell lines. The presence of AC133 antigen transcript in a variety of cell lines was assessed by Northern analysis (Fig 5). A 4.4-kb mRNA transcript was detectable in WERI-Rb-1 cells and in Y79 cells and MACS-isolated AC133+ fetal liver cells. Surprisingly, while expression of AC133 antigen is enhanced in Y79 cells upon PMA activation, the corresponding mRNA appears to be downregulated. In normal hematopoietic tissue, the AC133 antigen message is detectable in fetal liver and weakly in adult bone marrow, as expected, due to the fact that AC133+ cells in these tissues are in a minority.5 The AC133 antigen transcript was also noted in nonlymphoid tissues, particularly the pancreas, kidney, and placenta. Weaker signals were observed for the liver, lung, brain, and heart. This is in contrast to immunohistochemical staining of paraffin tissue sections, where AC133 antigen expression was detectable only in bone marrow (Table 2).
Northern analysis of AC133 antigen mRNA expression. (A) Total RNA was extracted from HeLa cells, Y79 cells, PMA-activated Y79 cells, WERI-Rb-1 cells, and MACS-isolated AC133+ fetal liver cells. Each lane was loaded with 15 μg total RNA, and a 32P-labeled 800-bp cDNA fragment was used as a probe. (B) Multiple tissue poly A+ Northern blots were purchased from Clontech and probed with 32P-AC133 antigen cDNA.
Northern analysis of AC133 antigen mRNA expression. (A) Total RNA was extracted from HeLa cells, Y79 cells, PMA-activated Y79 cells, WERI-Rb-1 cells, and MACS-isolated AC133+ fetal liver cells. Each lane was loaded with 15 μg total RNA, and a 32P-labeled 800-bp cDNA fragment was used as a probe. (B) Multiple tissue poly A+ Northern blots were purchased from Clontech and probed with 32P-AC133 antigen cDNA.
Expression of AC133 and CD34 Antigens on Human Tissue
| Tissue . | AC133 . | CD34 . |
|---|---|---|
| Stomach | − | − (+ vessels) |
| Colon | − | − (+ vessels) |
| Liver | − | − (+ vessels) |
| Pancreas | − | − (+ vessels) |
| Uterus (endometrium) | − | − (+ vessels) |
| Uterus (myometrium) | − | − (+ CT) |
| Ovary | − | − (+ vessels) |
| Prostate | − | − (+ CT, vessels) |
| Testis | − | − (+ CT, vessels) |
| Brain | − | − (+ vessels) |
| Lung | − | − (+ vessels) |
| Heart | − | − (+ vessels) |
| Thyroid | − | − (+ vessels) |
| Parathyroid | − | − (+ vessels) |
| Skeletal muscle | − | − (+ vessels) |
| Tonsil | − | − (+ vessels) |
| Bone marrow | + (subset) | + (subset) |
| Kidney | − | − (+ vessels) |
| Placenta | − | − (+ vessels) |
| Fetal heart | − | − (+ vessels) |
| Fetal lung | − | − (+ vessels) |
| Fetal brain | − | − (+ vessels) |
| Seminoma* | − | − (+ vessels) |
| Embryonal carcinoma* | − | − (+ vessels) |
| Choriocarcinoma* | − | − (+ vessels) |
| Tissue . | AC133 . | CD34 . |
|---|---|---|
| Stomach | − | − (+ vessels) |
| Colon | − | − (+ vessels) |
| Liver | − | − (+ vessels) |
| Pancreas | − | − (+ vessels) |
| Uterus (endometrium) | − | − (+ vessels) |
| Uterus (myometrium) | − | − (+ CT) |
| Ovary | − | − (+ vessels) |
| Prostate | − | − (+ CT, vessels) |
| Testis | − | − (+ CT, vessels) |
| Brain | − | − (+ vessels) |
| Lung | − | − (+ vessels) |
| Heart | − | − (+ vessels) |
| Thyroid | − | − (+ vessels) |
| Parathyroid | − | − (+ vessels) |
| Skeletal muscle | − | − (+ vessels) |
| Tonsil | − | − (+ vessels) |
| Bone marrow | + (subset) | + (subset) |
| Kidney | − | − (+ vessels) |
| Placenta | − | − (+ vessels) |
| Fetal heart | − | − (+ vessels) |
| Fetal lung | − | − (+ vessels) |
| Fetal brain | − | − (+ vessels) |
| Seminoma* | − | − (+ vessels) |
| Embryonal carcinoma* | − | − (+ vessels) |
| Choriocarcinoma* | − | − (+ vessels) |
Deparaffinized Formalin-fixed tissue sections were stained for AC133 and CD34 with a biotin-streptavidin method.
Abbreviation: CT, connective tissue.
Germ cell tumors.
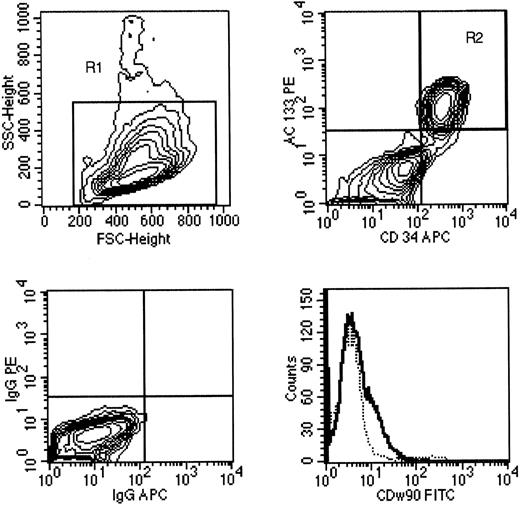
AC133 expression on leukemia. Eleven of 12 samples of acute leukemia expressed CD34 antigen by four-color FACS analysis; one sample was CD34−. Among the 11 CD34+ leukemias, three expressed high levels of AC133, four expressed low levels of AC133, and four did not express AC133 (Table 3). The majority of leukemia cells expressed CD38, a hematopoietic marker associated with lineage differentiation, with a small fraction of CD34+ cells (usually 10% to 30%) lacking CD38 expression (Table 4). The CD34+CD38− cells were presumed to be leukemic based on light-scatter characteristics and the pattern of expression for other informative hematopoietic markers; the pattern of AC133 and CD90 expression on CD38+ and CD38− subsets of leukemic cells was similar in each case. Figure 6 is a FACS analysis of AC133 expression on a bone marrow aspirate from case no. 3, a patient with AML. The light-scatter properties of leukemia cells showed a blast population (upper left panel) and expression of AC133, CD34, and CD90 (upper and lower right panels). AC133 was coexpressed with CD34 on the leukemic blast cell population that comprised 46% of the cells in the sample (upper right panel). A subset of leukemia cells (which were mainly the CD34+CD38− fraction) expressed CD90 at low levels (lower right panel) compared with an isotype control antibody.
Expression of AC133 on Acute ALL and AML
| . | CD34+ . | CD34− . |
|---|---|---|
| AC133++ (third decade) | Case 1 AML, M4 | None |
| Case 2 AML, M5 | ||
| Case 3 AML, M4 | ||
| AC133+ (second decade) | Case 4 ALL | None |
| Case 5 AML | ||
| Case 6 ALL | ||
| Case 7 ALL | ||
| AC133− | Case 8 ALL | Case 12 AML |
| Case 9 AML | ||
| Case 10 ALL | ||
| Case 11 ALL |
| . | CD34+ . | CD34− . |
|---|---|---|
| AC133++ (third decade) | Case 1 AML, M4 | None |
| Case 2 AML, M5 | ||
| Case 3 AML, M4 | ||
| AC133+ (second decade) | Case 4 ALL | None |
| Case 5 AML | ||
| Case 6 ALL | ||
| Case 7 ALL | ||
| AC133− | Case 8 ALL | Case 12 AML |
| Case 9 AML | ||
| Case 10 ALL | ||
| Case 11 ALL |
Twelve samples of bone marrow from patients with acute leukemia were obtained by bone marrow aspiration. Expression of AC133 and CD34 was determined by multiparameter flow cytometry using directly conjugated MoAbs. Eleven of 12 acute leukemias expressed CD34 and were subclassified according to whether they expressed bright levels of AC133 (between the second and third decade of fluorescence intensity); dim levels of AC133 (between the first and second decade of fluorescence), or no AC133 (between the second and third decade of fluorescence), or no AC133 (fluorescence was limited to the first decade of fluorescence intensity). The single CD34− leukemia (case no. 12) lacked AC133 expression. Only myelomonocytic acute leukemia (FAB M4 or M5) expressed the highest level of AC133 (cases no. 1, 2, and 3).
Expression of CD38, CD90, and AC133 on CD34+Acute Leukemia
| . | % CD38+ . | % CD90+ . | AC133 Staining . |
|---|---|---|---|
| Case 1 AML | 93.9 | 22.7 | ++ |
| Case 2 AML | 93.4 | 16.9 | ++ |
| Case 3 AML | 98.6 | <1 | ++ |
| Case 4 ALL | 91.0 | <1 | + |
| Case 5 AML | 56.2 | <1 | + |
| Case 6 ALL | 86.5 | 48.5 | + |
| Case 7 ALL | 80.1 | <1 | + |
| Case 8 ALL | 87.9 | <1 | − |
| Case 9 AML | 99.5 | <1 | − |
| Case 10 ALL | 77.3 | <1 | − |
| Case 11 ALL | 69.8 | <1 | − |
| . | % CD38+ . | % CD90+ . | AC133 Staining . |
|---|---|---|---|
| Case 1 AML | 93.9 | 22.7 | ++ |
| Case 2 AML | 93.4 | 16.9 | ++ |
| Case 3 AML | 98.6 | <1 | ++ |
| Case 4 ALL | 91.0 | <1 | + |
| Case 5 AML | 56.2 | <1 | + |
| Case 6 ALL | 86.5 | 48.5 | + |
| Case 7 ALL | 80.1 | <1 | + |
| Case 8 ALL | 87.9 | <1 | − |
| Case 9 AML | 99.5 | <1 | − |
| Case 10 ALL | 77.3 | <1 | − |
| Case 11 ALL | 69.8 | <1 | − |
Patterns of CD38, CD90, and AC133 expression on 11 cases of CD34+ acute leukemia were determined. The cases are numbered as in Table 3. The percentage of leukemia cells expressing CD38 or CD90 is shown and compared with the level of AC133 expression on the leukemia blast population. The pattern of CD90 and AC133 expression on CD38− and CD38+ fractions of the leukemia cells was similar in each case (data not shown).
A sample of AML present in a bone marrow aspirate (case no. 3) was stained with CD34 (APC). AC133-PE and CD90-FITC MoAbs, as well as isotype control antibodies conjugated with FITC, PE, or APC. Upper left, the gate on forward scatter v side scatter (R1) that contained the leukemia blast cell population is shown on a 10% probability contour plot. Upper right, the pattern of CD34 and AC133 expression on cells gated for R1 is shown; 46% of the cells were contained within the upper right quadrant (R2). Lower left, the pattern of IgG-APC and IgG-PE isotype MoAb expression on the R1 population. Lower right, the pattern of CD90 expression on the leukemia cell population (gated on R1 and R2) compared with the pattern of staining using an isotype control antibody (- - - -).
A sample of AML present in a bone marrow aspirate (case no. 3) was stained with CD34 (APC). AC133-PE and CD90-FITC MoAbs, as well as isotype control antibodies conjugated with FITC, PE, or APC. Upper left, the gate on forward scatter v side scatter (R1) that contained the leukemia blast cell population is shown on a 10% probability contour plot. Upper right, the pattern of CD34 and AC133 expression on cells gated for R1 is shown; 46% of the cells were contained within the upper right quadrant (R2). Lower left, the pattern of IgG-APC and IgG-PE isotype MoAb expression on the R1 population. Lower right, the pattern of CD90 expression on the leukemia cell population (gated on R1 and R2) compared with the pattern of staining using an isotype control antibody (- - - -).
DISCUSSION
MoAb AC133 is an antibody with specificity for a novel cell surface antigen that is expressed on CD34bright subpopulations of HSCs found in adult bone marrow, fetal bone marrow and liver, cord blood, and adult peripheral blood.5 MoAb AC133 can be used for magnetic bead immunoselection of HSC populations for transplantation, as well as for phenotypic analysis of stem and progenitor cell populations using flow cytometric techniques. To further characterize the nature of this novel molecule, AC133 antigen was purified by immunoaffinity chromatography. The AC133 antigen consists of a single polypeptide chain with a reduced molecular weight of 120 kD, and is a glycoprotein with 20-kD N-glycosidic–linked polysaccharides.
Degenerate oligonucleotides, designed from four peptide sequences of the proteolytically digested AC133 antigen, yielded a 1.7-kb fragment that was then used to isolate cDNA for the entire gene. Analysis of this cDNA suggests the presence of 5-TM domains, resulting in two large extracellular loops with a total of eight consensus sites for N-linked glycosylation. We have modeled the AC133 antigen as a 5-TM protein with an extracellular N-terminus and a cytoplasmic C-terminus. Immunoprecipitation of AC133 antigen from tunicamycin-treated cells and FACS analysis of these cells indicate that MoAb AC133 recognizes a glycosylated structure (data not shown). These data are consistent with our model suggesting that the two large loops are extracellular.
A 5-TM cell surface antigen has not been described previously, and the structure of this molecule differs markedly from known multi-TM proteins with respect to molecular weight and size of the extracellular loops. Additionally, AC133 antigen does not share sequence homology with other 4- or 7-TM proteins, while family members do share significant homology with each other, particularly within the TM domains. Therefore, we believe the AC133 antigen is representative of a new family of TM receptors.
Short fragments of the AC133 gene are present in Genebank as ESTs (expressed sequence tags), and have been cloned from adult retina, pancreatic islets, colon, placenta, and fetal brain. While histologic and phenotyping data suggest that expression of AC133 antigen is limited to CD34bright HSCs, the AC133 antigen transcript is detectable by Northern analysis in a variety of normal, nonlymphoid tissues such as the pancreas, kidney, and placenta. PMA-activated Y79 cells upregulate expression of the antigen as detected by FACS analysis; however, Northern blots clearly demonstrate a reduction in the corresponding AC133 transcript. These data indicate that AC133 antigen expression is likely to be regulated posttranscriptionally. CD34 is also likely to be regulated, in part, posttranscriptionally. Nuclear run-on assays demonstrate that aborted transcription of CD34 occurs in some CD34− cell lines.26 The CD34 transcript is detectable in HuVEC cells without concordant expression of CD34 after culture in vitro,27 and may be due to secondary structure in the 5′ UTR of the gene for CD34.28-30 AC133 antigen is also expressed on NT-2 teratocarcinoma cells, but expression is lost as these cells terminally differentiate into neurons after treatment with retinoic acid.5,31 The presence of the AC133 antigen transcript in retina is intriguing and could be explained by the presence of more primitive retinal ganglion cells (RGCs) in retinal tissue. Embryonic RGCs retain the regenerative capacity to grow axons, although this ability is lost at some point in development.32 AC133 antigen is present on subsets of CD34+ leukemias, and the presence of AC133 antigen on leukemias is consistent with our data5 suggesting that this is an early hematopoietic marker. It is important to note that 30% to 40% of CD34+ leukemias are AC133−. These patients would not ordinarily be able to benefit from CD34-based selection and autologous transplantation; however, isolation of HSCs via MoAb AC133 may allow stem cell transplantation as a viable alternative.
Currently, there is no known function for AC133 antigen. There is no significant homology to 7-TM G-protein–coupled receptors or other cell surface antigens that might indicate a potential function. Its presence on early, undifferentiated cells is suggestive of a growth factor receptor, and the presence of five tyrosine residues on the 50-aa cytoplasmic tail may indicate that the protein is phosphorylated in response to ligand binding and initiates a signal transduction cascade. Future studies will have to address the physiologic ligand of this receptor, as well as any intracellular signaling pathway associated with it.
ACKNOWLEDGMENT
We thank Kerry Magee and Michael Brown for excellent technical assistance, Jeff Fairman for advice on molecular biology protocols and primer design, Aaron Kantor for helpful discussions, and Sarah Blanch for graphic design expertise.
Genebank submission no. is AF027208.
Address reprint requests to David W. Buck, AmCell Corp, 1190 Bordeaux Dr, Sunnyvale, CA 94089.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal