Abstract
The mRNA encoding full-length erythropoietin (EPO) receptor (EPOR-F ) comprises exons I through VIII. Another membrane-bound EPOR (EPOR-T) isoform has a truncated cytoplasmic region and is encoded by the mRNA containing unspliced intron VII (EPOR-T mRNA). EPOR-T is believed to have a dominantly negative function against EPOR-F. We show that EPOR-T mRNA is markedly decreased in the blood cells of patients with polycythemia vera (PV). We also show that EPOR-T mRNA is not detected in erythroid/megakaryocytic leukemia cell lines, but is expressed in nonerythroid/nonmegakaryocytic lines, suggesting the presence of a cell type–specific system by which intron VII of the EPOR transcript is spliced. Deregulation of this splicing system in early hematopoietic progenitors possibly explains the profound decrease in EPOR-T mRNA and consequent pathophysiology of PV.
INTERACTION between erythropoietin (EPO) and the cell surface receptor for EPO (EPOR) plays a fundamental role in erythropoiesis.1 EPOR is known to transduce signals via ligand-mediated homo-oligomerization of EPOR protein2 and activation of JAK2 protein,3 a cytoplasmic protein tyrosine kinase.
The major EPOR protein, designated here as EPOR-F, is encoded by an mRNA comprising exons I through VIII with all the introns spliced out.4,5 However, in addition to this mRNA, expression of another mRNA species has been reported. The latter mRNA contains a small intervening sequence between exons VII and VIII, derived from an incomplete intron, and encodes a protein with a truncated cytoplasmic domain.6 Nakamura et al6 7 have claimed that this mRNA species containing intron VII (EPOR-T mRNA) is expressed only in early hematopoietic progenitors (ie, CD34+/CD71−) in the bone marrow and that the EPOR protein (EPOR-T) encoded by this mRNA has a dominant-negative function against EPOR-F. They showed that transfection with a cDNA for EPOR-T decreased the responsiveness to EPO in EPOR-F cDNA–transfected Ba/F3, a murine pro-B cell line. Since such interference was observed only at low (ie, physiologic) EPO concentration, they proposed an attractive hypothesis that EPOR-T is a key regulator in maintaining homeostasis of the red blood cell mass. They speculated that if the concentration of EPO is low, most of the early hematopoietic progenitors cannot be mobilized to undergo erythropoiesis because of the coexpression of EPOR-T (physiologic erythropoiesis). However, these immature populations could be mobilized despite expression of EPOR-T if the concentration of EPO becomes elevated in pathologic states such as anemia and hypoxia (stress erythropoiesis). Since EPOR-T is downregulated in the maturing erythroid precursor cells, these cells can respond to the physiologic concentration of EPO, thus maintaining normal erythropoiesis.
Given these considerations, one may expect that the decreased expression of EPOR-T in early hematopoietic progenitors may cause deregulated red blood cell production. Polycythemia vera (PV) is a disease characterized by an absolute increase in red blood cell mass in the circulation while EPO level in the plasma is usually within or less than the normal range. Therefore, the deregulated erythropoiesis in PV may result from decreased EPOR-T. Here, we show that EPOR-T is in fact markedly decreased in patients with PV.
We also propose that erythroid cells may specifically have a trans-activation system that facilitates splicing of intron VII. Consequently, there may be a cis-acting element recognized by such a cell type–specific trans-activation system. We discuss the localization of the presumed cis-element, which might be a cell type–specific splice enhancer for intron VII.
SUBJECTS AND METHODS
Patients and clinical materials.Five patients treated at the University of Tokyo Hospital and three at the Yale-New Haven Hospital were evaluated in this study, all of whom were diagnosed with PV according to the Polycythemia Vera Study Group (PVSG) criteria.8 Repeated phlebotomy was necessary for all eight patients, among whom three were managed in combination with hydroxyurea. None had been treated with busulfan or 32P. In two patients, bone marrow aspiration was performed for diagnostic purposes while this study was in progress. A portion of peripheral blood and bone marrow aspirates was used for these studies after obtaining informed consent. Peripheral blood and bone marrow cells were also obtained from seven normal volunteers after obtaining consent. Additional peripheral blood cells were obtained from two patients with essential thrombocythemia (ET) and three patients with chronic myeloid leukemia (CML) in chronic phase (CML-CP). Diagnosis of ET and CML was made by the PVSG criteria for diagnosis of ET8 and the presence of a Ph1 chromosome and a major bcr-abl transcript, respectively. Frozen leukemia cells obtained from a patient with CML in blast crisis (this patient's profile is described in Mitani et al9 ) are also used.
Cell lines.Seventy-one human leukemia cell lines were used in this study. Most were from a cell line collection maintained at the Fujisaki Cell Center.10 Additional cell lines used were Meg-01,11 TF-1,12 and F-36E.13 We studied 24 myeloid/monocytic, five erythroid, two megakaryocytic, 15 B-lymphoid, and 22 T-lymphoid cell lines.10 Mouse NIH3T3 cells and monkey COS-7 cells were purchased from the American Type Culture Collection (Rockville, MD). KBM-5 is a cell line established from another patient with CML in blast crisis.
Plasmid construction and transfection.Human EPOR cDNA (encoding EPOR-F ) in the expression vector BCMGSneo was a kind gift from H. Nakauchi (Tsukuba, Japan).6 A HindIII-Pst I fragment encompassing the entire coding sequence for EPOR-F was placed behind the CMV promoter (Invitrogen, San Diego, CA; the resulting plasmid, pCMV-F ). To construct the cDNA encoding EPOR-T, a 453-bp fragment comprising a 3′ part of intron VI, exon VII, intron VII, and a 5′-part of exon VIII was generated by polymerase chain reaction (PCR) using human genomic DNA as the template. Primers used were as follows: sense-1, AGTGGAGGGGGAATTGCTGA, and antisense-1, TGTCCAGCACCAGATAGGTA. The annealing temperature was 60°C. The 343-bp BglII-Nsp I fragment therein was then used to replace a 248-bp BglII-Nsp I fragment in the pCMV-F. Integrity of the replaced region in the resulting plasmid, pCMV-T, was verified by sequencing.
For transient transfection of these plasmids, Lipofectamine (GIBCO-BRL, Gaithersburg, MD) was used in accordance with the manufacturer's instructions. In brief, adherent cells grown to subconfluence in a six-well dish were washed with serum-free medium, and 0.8 mL Opti-MEM (GIBCO-BRL) was overlaid onto the cells. Cells grown in suspension were also washed in the same manner and resuspended in Opti-MEM at a concentration of 3 × 106/mL, and 0.8 mL cell suspension was seeded in a six-well dish. One microgram of plasmid and 5 to 10 μL Lipofectamine were diluted into 0.1 mL Opti-MEM, respectively. These solutions were combined, gently mixed, and left standing at room temperature for 15 to 45 minutes, and the lipid-DNA complex of 0.2 mL was added to the wells containing the cells. The cells in 1 mL were then incubated for 6 to 24 hours at 37°C in a 5% CO2 incubator. Transfection was terminated by adding 2 mL normal growth medium containing 20% fetal calf serum, and the cells were incubated further for 24 hours before harvest.
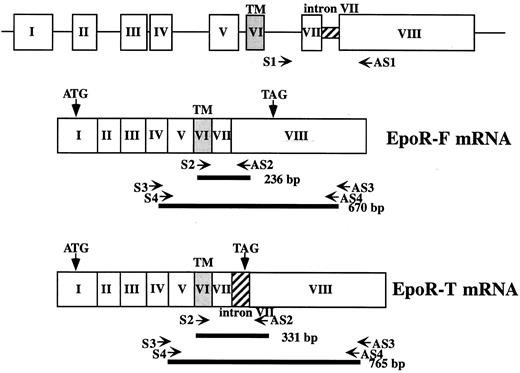
RT-PCR.Total cellular RNA was isolated from bone marrow and peripheral blood mononuclear cells and the cell lines described earlier by a single-step method using acid guanidinium thiocyanate.14 Where indicated, cytoplasmic RNA was prepared using Nonidet P-40 to exclude the possibility that we were detecting immature RNA present in the nucleus. PolyA(+) RNA was purified using an oligo dT system (Oligotex-dT30 Super; Takara Shuzo, Otsu, Shiga, Japan) according to the manufacturer's instructions. One microgram of total RNA or 0.5 μg polyA(+) RNA was reverse-transcribed using oligo dT and Moloney murine leukemia virus reverse transcriptase (GIBCO-BRL) under the recommended conditions in a total volume of 25 μL. One to 2 μL cDNA was amplified by PCR using primers as follows: sense-2, 5′-TGGTCATCCTGGTGCTGCTGA-3′, and antisense-2, 5′-AGCGCTCTGAGAGGACTTCCA-3′. The expected sizes of the PCR product with and without intron VII were 331 bp and 236 bp, respectively. Cycling conditions were 1 minute at 94°C, 1 minute at 65°C, and 1 minute at 72°C. The number of cycles was 20 in transfection experiments, 30 in experiments using samples from patients and volunteers, and 40 in experiments to study endogenous expression in the cell lines. In some experiments, we used a nested PCR strategy. The primer sets used were as follows: sense-3, 5′-TGAGACACCCATGACGTCCA-3′, and antisense-3, 5′-TATTGGATCCCTGATCATCT-3′, for the first PCR; and sense-4, 5′-CAGAAGATCTGGCCTGGCAT-3′, and antisense-4, 5′-AGCCTGGTGTCCTAAGAGCAA-3′, for the second PCR. The expected size in this case was 765 bp and 670 bp with and without intron VII, respectively. The scheme of the PCR is illustrated in Fig 1.
Diagram of EPOR gene and positions of PCR primers. Top, structure of the human EPOR gene; middle and bottom, structure of the 2 mRNA species. Positions of PCR primers are shown by arrows, and the product of each PCR is shown by bold lines along with expected length.
Diagram of EPOR gene and positions of PCR primers. Top, structure of the human EPOR gene; middle and bottom, structure of the 2 mRNA species. Positions of PCR primers are shown by arrows, and the product of each PCR is shown by bold lines along with expected length.
Ribonuclease protection assay.A cDNA fragment of EPOR comprising a part of exon VII, intron VII, and a part of exon VIII was generated by PCR and subcloned into pBluescript vector (Stratagene, La Jolla, CA). The sequence of the insert was verified. This plasmid was digested with BglII, the site of which is in the exon VII segment of EPOR cDNA, and 1 μg thereof was used as a template to generate a 420-bp antisense-strand riboprobe labeled with [α-32P]UTP (ICN Biomedicals, Costa Mesa, CA) using T7 RNA polymerase. Ten micrograms of polyA(+) RNA was coprecipitated with 1 to 2 × 105 cpm of the gel-purified probe. The pellet was resuspended in 20 μL hybridization buffer (80% formamide, 100 mmol/L sodium citrate, pH 6.4, 300 mmol/L sodium acetate, pH 6.4, and 1 mmol/L EDTA), heated at 90°C for 3 minutes, and incubated at 45°C overnight. After incubation, 200 μL RNase A (2.5 U/mL) and RNase T1 (10 U/mL) (Ambion, Austin, TX) was added to the hybridization mixture and incubated at 37°C for 30 minutes. The protected RNA was precipitated, resuspended in 80% formamide, and loaded onto 5% polyacrylamide/8 mol/L urea gel. The gel was dried and autoradiographed.
RESULTS
Expression of intron VII–spliced and –unspliced mRNAs in blood cells from patients with PV versus normal volunteers.Figure 2A and B shows the results of RNA-based RT-PCR using peripheral blood mononuclear cells obtained from PV patients and normal volunteers. Figure 2C shows the result of RT-PCR for bone marrow mononuclear cells obtained from PV patients no. 1 and 2, and a normal volunteer. In these experiments, we confirmed that two species of EPOR mRNA are expressed in peripheral blood and bone marrow mononuclear cells prepared from normal volunteers. The size of each amplified band matched that of species with and without intron VII. In contrast, only a single species of EPOR mRNA was detected in all the samples prepared from peripheral blood and bone marrow mononuclear cells in six PV patients. Both mRNA species were demonstrated in peripheral blood from patients with ET and CML-CP, myeloproliferative disorders distinct from PV (Fig 2D).
Expression of intron VII–spliced and –unspliced mRNAs for EPOR in peripheral blood (A, B, D, and E) and bone marrow (C) mononuclear cells from PV patients, normal volunteers, ET and CML-CP patients, and a cell line. Total cellular RNA was reverse-transcribed and subjected to PCR using the primer sets of sense-3 and antisense-3, and then sense-4 and antisense-4 (nested PCR, [A]) or sense-2 and antisense-2 (B, C and D), respectively. In (E), cytoplasmic RNA was reverse-transcribed and subjected to PCR using the primers sense-2 and antisense-2. (A) Lane a, PV patient no. 1; b, PV patient no. 3; c, normal no. 1 (RNA lot 1); d, normal no. 1 (RNA lot 2); e, normal no. 2. The size of the intron VII–spliced and –unspliced mRNAs is 670 bp and 765 bp, respectively. (B) Lane a, PV patient no. 4; b, PV patient no. 5; c, PV patient no. 6; d, normal no. 4; e, cell line KBM-5. (C) Lane a, normal volunteer no. 3; b, PV patient no. 2; and c, PV patient no. 1. (D) Lane a, ET no. 1; b, ET no. 2; c, CML-CP no. 1; d, CML-CP no. 2; e, CML-CP no. 3. (E) Lane a, PV patient no. 7; b, PV patient no. 8; c, normal no. 5; d, normal no. 6; and e, negative control without template. In (B) to (E), the sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively.
Expression of intron VII–spliced and –unspliced mRNAs for EPOR in peripheral blood (A, B, D, and E) and bone marrow (C) mononuclear cells from PV patients, normal volunteers, ET and CML-CP patients, and a cell line. Total cellular RNA was reverse-transcribed and subjected to PCR using the primer sets of sense-3 and antisense-3, and then sense-4 and antisense-4 (nested PCR, [A]) or sense-2 and antisense-2 (B, C and D), respectively. In (E), cytoplasmic RNA was reverse-transcribed and subjected to PCR using the primers sense-2 and antisense-2. (A) Lane a, PV patient no. 1; b, PV patient no. 3; c, normal no. 1 (RNA lot 1); d, normal no. 1 (RNA lot 2); e, normal no. 2. The size of the intron VII–spliced and –unspliced mRNAs is 670 bp and 765 bp, respectively. (B) Lane a, PV patient no. 4; b, PV patient no. 5; c, PV patient no. 6; d, normal no. 4; e, cell line KBM-5. (C) Lane a, normal volunteer no. 3; b, PV patient no. 2; and c, PV patient no. 1. (D) Lane a, ET no. 1; b, ET no. 2; c, CML-CP no. 1; d, CML-CP no. 2; e, CML-CP no. 3. (E) Lane a, PV patient no. 7; b, PV patient no. 8; c, normal no. 5; d, normal no. 6; and e, negative control without template. In (B) to (E), the sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively.
PCR products obtained from PV patients no. 1 and 2 were excised from the gel, subcloned into a plasmid, and sequenced. No mutation was found in these cDNAs compared with the reported EPOR cDNA sequence (data not shown), indicating that sequences in both splice-acceptor sites for intron VII were intact in PV patients. We extracted genomic DNA from each material obtained from PV patients, and the DNA samples were subjected to PCR using the primer set of sense-2 and antisense-2. The amplified DNA migrated at 331 bp, suggesting that intron VII of 95 bp was present in the genomic DNA. By sequencing the DNA excised from the gel, it was found that intron VII was present without any mutations (data not shown).
Cytoplasmic RNA was prepared from two additional PV patients and two normal volunteers to exclude the possibility that the longer PCR product resulted from immature RNA present in the nucleus. The result of the RT-PCR experiment using these RNA samples showed again that the two RNA species were expressed at a readily detectable level in normal volunteers, but only the intron VII–spliced mRNA (EPOR-F mRNA) was detected in PV patients (Fig 2E).
Expression of intron VII–spliced and –unspliced mRNAs in leukemia cells.We next determined the spectrum of expression for each EPOR mRNA species using various human leukemia cell lines. The same RT-PCR strategy was applied using the primer set of sense-2 and antisense-2. Unexpectedly, we found that when the number of PCR cycles was increased to 40, all 71 cell lines studied expressed EPOR mRNAs regardless of lineage. Expression of the two EPOR mRNA species was evident in 64 lines. In the remaining seven lines, only EPOR-F mRNA was detected; these seven cell lines were all of erythroid or megakaryocytic lineage. The classification of the 71 cell lines and the pattern of EPOR mRNA expression are summarized in Table 1. Representative ethidium bromide staining of the RT-PCR is shown in Fig 3A and B. For some cell lines, cytoplasmic RNA was prepared as described in the methods. In RT-PCR experiments using these samples, the results obtained were the same as when using total cellular RNA (data not shown).
Expression of EPOR-F and EPOR-T mRNA Species in Various Human Leukemia Cell Lines
| Cell Line . | EPOR-F mRNA . | EPOR-T mRNA . |
|---|---|---|
| Myelocytic/monocytic | 24/24 | 24/24 |
| Erythroid | 5/5 | 0/5 |
| Megakaryocytic | 2/2 | 0/2 |
| B-lymphocytic | 15/15 | 15/15 |
| T-lymphocytic | 22/22 | 22/22 |
| Non–T, non–B-lymphocytic | 3/3 | 3/3 |
| Cell Line . | EPOR-F mRNA . | EPOR-T mRNA . |
|---|---|---|
| Myelocytic/monocytic | 24/24 | 24/24 |
| Erythroid | 5/5 | 0/5 |
| Megakaryocytic | 2/2 | 0/2 |
| B-lymphocytic | 15/15 | 15/15 |
| T-lymphocytic | 22/22 | 22/22 |
| Non–T, non–B-lymphocytic | 3/3 | 3/3 |
Expression of the intron VII–spliced and –unspliced mRNAs for EPOR in various leukemia cell lines and leukemia cells. Total cellular RNA (A, B, and C) or polyA(+) RNA (C) was reverse-transcribed and subjected to PCR using the primer sets of sense-2 and antisense-2. (A) Lane a, BV-173 (lymphoid); b, NALM-16 (lymphoid); c, NALM-24 (lymphoid); d, K562 (erythroid); e, MEG-01 (megakaryocytic); f, HEL (erythroid); g, JOSK-1 (monocytic); h, SU-DHL-4 (lymphoid); and i, CCRF-CEM (lymphoid). (B) Lane a, KOPT-K1 (lymphoid); b, PEER (lymphoid); c, F-36E (erythroid); d, F-36P (erythroid); e, UT-7 (megakaryocytic); and f, TF-1 (erythroid). (C) One microgram total RNA prepared from U-937 cells, 0.5 μg polyA(+) RNA therefrom, and 0.5 μg polyA(+) RNA prepared from peripheral blood leukemia cells from a patient with CML in blast crisis9 was reverse-transcribed and subjected to the PCR protocol with the indicated number of cycles. Total RNA was used in lanes a to c and polyA(+) RNA in d to i. The number of cycles was 30 (lanes a, d, and g), 35 (b, e, and h), and 40 (c, f, and i). The sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively. (D) RNase protection assay shows expression of EPOR-F and EPOR-T mRNAs in U-937 and leukemia cells obtained from the same patient as described in C. A 420-bp [32P]-labeled riboprobe was hybridized with 10 μg polyA(+) RNA samples, digested with the mixture of RNase A and RNase T1, and analyzed in 5% polyacrylamide/8 mol/L urea gel. Lane a, undigested probe; b, yeast RNA; c, U-937; d, CML blast crisis.
Expression of the intron VII–spliced and –unspliced mRNAs for EPOR in various leukemia cell lines and leukemia cells. Total cellular RNA (A, B, and C) or polyA(+) RNA (C) was reverse-transcribed and subjected to PCR using the primer sets of sense-2 and antisense-2. (A) Lane a, BV-173 (lymphoid); b, NALM-16 (lymphoid); c, NALM-24 (lymphoid); d, K562 (erythroid); e, MEG-01 (megakaryocytic); f, HEL (erythroid); g, JOSK-1 (monocytic); h, SU-DHL-4 (lymphoid); and i, CCRF-CEM (lymphoid). (B) Lane a, KOPT-K1 (lymphoid); b, PEER (lymphoid); c, F-36E (erythroid); d, F-36P (erythroid); e, UT-7 (megakaryocytic); and f, TF-1 (erythroid). (C) One microgram total RNA prepared from U-937 cells, 0.5 μg polyA(+) RNA therefrom, and 0.5 μg polyA(+) RNA prepared from peripheral blood leukemia cells from a patient with CML in blast crisis9 was reverse-transcribed and subjected to the PCR protocol with the indicated number of cycles. Total RNA was used in lanes a to c and polyA(+) RNA in d to i. The number of cycles was 30 (lanes a, d, and g), 35 (b, e, and h), and 40 (c, f, and i). The sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively. (D) RNase protection assay shows expression of EPOR-F and EPOR-T mRNAs in U-937 and leukemia cells obtained from the same patient as described in C. A 420-bp [32P]-labeled riboprobe was hybridized with 10 μg polyA(+) RNA samples, digested with the mixture of RNase A and RNase T1, and analyzed in 5% polyacrylamide/8 mol/L urea gel. Lane a, undigested probe; b, yeast RNA; c, U-937; d, CML blast crisis.
To further exclude the possibility that detection of the longer RT-PCR product was generated from immature RNA-based cDNA, we compared total and polyA(+) RNAs prepared from U-937 (monocytic) cells in a RT-PCR experiment. The amount of 0.5 μg polyA(+) RNA–based cDNA produced distinctly more abundant PCR product than 1 μg total RNA–based cDNA in each number of PCR cycles, suggesting that the longer PCR product is derived from mature polyA(+) RNA coding for EPOR-T (Fig 3C).
To see if the ratio of each RT-PCR product reflects the actual abundance of each mRNA species, we performed a RNase protection assay. A 365-bp RNase-protected band representing EPOR-T mRNA was seen in addition to protected bands of 198 bp and 72 bp representing EPOR-F mRNA (Fig 3D) in U-937 cells and leukemia cells from CML crisis. However, the ratio of the bands representing the two different RNA species was different in these samples, with the ratio of EPOR-T and EPOR-F mRNA being much lower in the latter (lane d). This result corresponded well with the result of RT-PCR using the same RNA sample, ie, a very low ratio of EPOR-T to EPOR-F was also suggested by RT-PCR in the CML crisis sample (Fig 3C, lanes g to i).
Splicing of intron VII transfected into cell lines.The expression pattern in the leukemia cell lines led to an attractive speculation that splicing of intron VII of EPOR might be an event specific to the erythroid and megakaryocytic lineages. In this respect, Nakamura and Nakauchi7 found that when the human EPOR cDNA containing the unspliced intron VII in an expression vector was transfected into cell lines, both mRNA species derived from the cDNA were expressed in a pro-B cell line, Ba/F3, but little expression of the intron VII–spliced mRNA was seen in L cells, a fibroblast-like cell line. This result suggests that hematopoietic cells, which generally express EPOR (Figs 2 and 3), possess the cellular machinery needed to splice intron VII. Moreover, our results suggest that such machinery should be more effective in erythroid and megakaryocytic cells than in nonerythroid/nonmegakaryocytic cells. This speculation also suggests the presence of cis-acting element(s) recognized by the splicing machinery. The cis-element may be localized to the exons (and intron VII) of the EPOR gene, because cDNA was used in the transfection experiments reported by Nakamura and Nakauchi.7 Thus, identification of the cis-element responsible for the cell type–specific splicing of intron VII may be possible through the use of a cDNA construct containing intron VII in an appropriate erythroid/megakaryocytic cell line.
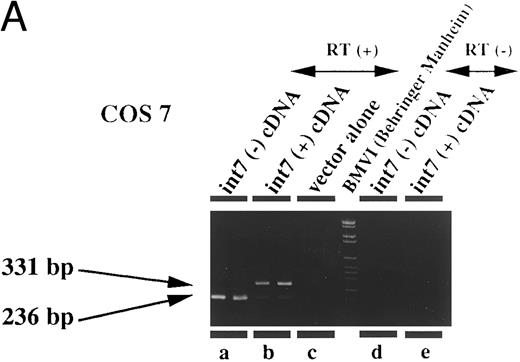
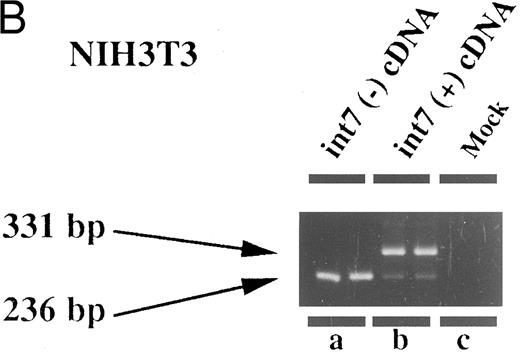
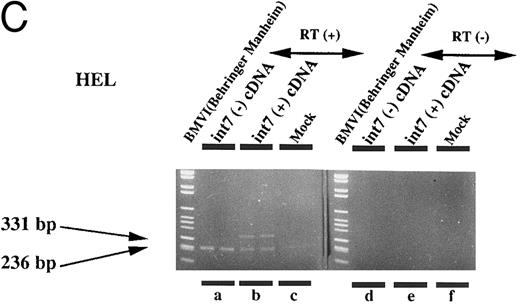
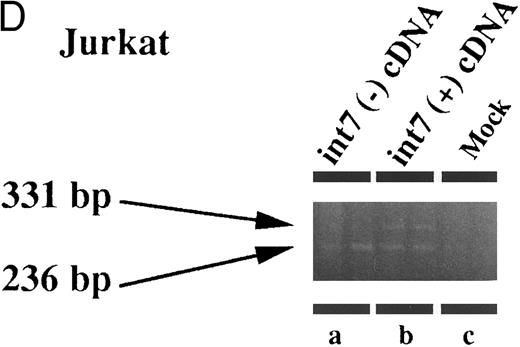
Therefore, we prepared an expression plasmid containing the EPOR cDNA including intron VII and transfected this plasmid (pCMV-T) into various cell lines including fibroblasts and lymphoid, erythroid, and megakaryocytic cells. In human leukemia cell lines, we found that both species of mRNA could be generated at comparable levels from the transfected pCMV-T (Fig 4C and D). In contrast, the same plasmid generated a much higher ratio of EPOR-T mRNA compared with EPOR-F mRNA in the nonhematopoietic cell lines (Fig 4A and B). These results suggest that the cell specificity to splice intron VII is present in the cDNA-transcribed RNA. However, the ratio of EPOR-F mRNA to EPOR-T mRNA was indistinguishable between Jurkat, a T-lymphoid cell line, and HEL, an erythroid cell line (Fig 4C and D), indicating that the splicing pattern of the endogenous EPOR pre-mRNA was not maintained in splicing of intron VII from pCMV-T–derived RNA.
Expression of intron VII–spliced and –unspliced mRNAs for EPOR in cell lines transfected with EPOR cDNA with (int7 cDNA) and without (-int cDNA) the intron VII sequence. Transfection was performed using the Lipofectamine reagent. Total RNA was prepared 24 hours after the transfection procedure was terminated. RT-PCR was performed using the primer set of sense-2 and antisense-2. Because the number of cycles was reduced, endogenous mRNA is not demonstrated even in the hematopoietic cells. Since the cDNA used for transfection could directly be a template for PCR to generate a product with the same size as that generated by mRNA, RT-negative reaction mixture was always used as a negative control (see lanes d and e in A and d to f in C). All steps were performed in duplicate. Figures shown are results obtained using COS 7 (A), NIH3T3 (B), HEL (C), and Jurkat (D) cells. The size marker used was BMVI (Boehringer-Mannheim). Among cells transfected with cDNA containing intron VII, EPOR-T mRNA is much more abundant than EPOR-F mRNA in COS 7 and NIH3T3 cells, whereas the ratio of EPOR-T mRNA to EPOR-F mRNA is about the same in hematopoietic cells.
Expression of intron VII–spliced and –unspliced mRNAs for EPOR in cell lines transfected with EPOR cDNA with (int7 cDNA) and without (-int cDNA) the intron VII sequence. Transfection was performed using the Lipofectamine reagent. Total RNA was prepared 24 hours after the transfection procedure was terminated. RT-PCR was performed using the primer set of sense-2 and antisense-2. Because the number of cycles was reduced, endogenous mRNA is not demonstrated even in the hematopoietic cells. Since the cDNA used for transfection could directly be a template for PCR to generate a product with the same size as that generated by mRNA, RT-negative reaction mixture was always used as a negative control (see lanes d and e in A and d to f in C). All steps were performed in duplicate. Figures shown are results obtained using COS 7 (A), NIH3T3 (B), HEL (C), and Jurkat (D) cells. The size marker used was BMVI (Boehringer-Mannheim). Among cells transfected with cDNA containing intron VII, EPOR-T mRNA is much more abundant than EPOR-F mRNA in COS 7 and NIH3T3 cells, whereas the ratio of EPOR-T mRNA to EPOR-F mRNA is about the same in hematopoietic cells.
DISCUSSION
Many investigators have characterized molecular mechanisms through which the EPO-EPOR system transduces various signals into the nucleus, such as the homo-oligomerization of the EPOR protein upon ligand binding2,15 and the activation of JAK2 protein tyrosine kinase.3 However, there are still some unresolved questions regarding events at the cell surface. One issue is whether another membrane protein(s) exists that can complex with EPO and EPOR.16-19 The second issue is the significance of the truncated form of EPOR, EPOR-T.6,7 The role of this protein, encoded by the mRNA containing intron VII, EPOR-T mRNA, might be to function in a dominantly negative fashion against EPOR-F,7,20 possibly by interfering with the homo-oligomerization of EPOR-F. A recent finding that transgenic mice overexpressing EPOR-T became anemic (H. Nakauchi, Y. Nakamura, and H. Nakauchi, personal communication, September 1996) supports this hypothesis; this result corresponds to the finding from knockout experiments that the absence of EPOR-mediated signals results in a defect in definitive erythropoiesis.21 22
PV is a disease characterized by an absolute increase in red blood cell mass in the circulation and an EPO level in the plasma that is within or less than the normal range.23 When assessed by the in vitro colony assay, the number of erythroid precursor cells that respond to erythropoietin is increased in the bone marrow of PV patients.24,25 Although mutations in the EPOR gene that result in C-terminal truncation of EPOR protein have been found in primary familial and congenital polycythemia,26,27 mutations in the coding sequence of the EPOR gene are rare in acquired PV.28 29
In the current study, we propose that the deregulated erythropoiesis in PV patients might result from a decrease in EPOR-T. We found that the expression of EPOR-T mRNA was extremely decreased or absent in all eight PV patients studied, whereas both the EPOR-F mRNA and EPOR-T mRNA species were expressed at a comparable level in normals, in agreement with our expectation. Decreased EPOR-T mRNA may reflect a shift in the maturation profile of PV erythroid progenitors. However, our finding is more likely to contribute to understanding the pathogenesis of PV, since CFU-GEMM, the most immature non-lymphoid progenitor that we believe normally expresses both mRNA-F and mRNA-T, is also known to be increased in PV patients.30
A deregulated EPOR-associated signal might be able to explain leukocytosis and thrombocytosis, which are also common characteristics of PV. There is increasing evidence suggesting that an EPOR-associated signal promotes megakaryopoiesis.31 EPOR mRNA is expressed in hematopoietic stem cells,32 and expression of a constitutively active EPOR in primary hematopoietic progenitors enhances granulocyte/macrophage progenitor growth in addition to erythroid progenitor growth.33 Thus, leukocytosis could also be explained by deregulation of the EPOR signal. On the other hand, blood samples from patients with other myeloproliferative disorders such as ET and CML showed a normal expression pattern of EPOR mRNA. This indicates that the decrease in EPOR-T mRNA is not a characteristic of myeloproliferative syndromes in general, but of PV specifically.
The significance of the expression of both mRNA species in nonerythroid/nonmegakaryocytic cell lines is unclear. Since these cells expressed EPOR mRNA at a much lower level and since the EPOR protein isoforms have not been extensively characterized, further analyses are required. There are several reports that EPO administered to patients undergoing dialysis modulated the immune system.34-36 This phenomenon may represent a direct effect of EPO on lymphocyte function, although we have not determined whether normal lymphocytes also express EPOR as the lymphoid leukemia cell lines do.
One may speculate that the splicing mechanism to process intron VII of the EPOR gene may be deregulated in PV patients. This speculation is supported by the two expression patterns of EPOR mRNAs in leukemia cell lines in accordance with the lineages. The expression patterns predict that the erythroid/megakaryocytic cells have a specific splicing system that facilitates processing of intron VII.
The presence of the splice trans-activation system further predicts a cis-acting element recognized by such a system in the genome. We transfected various cell lines with a plasmid containing the cDNA for EPOR containing intron VII (pCMV-T) to see if the expression patterns of EPOR-T mRNA and EPOR-F mRNA were the same as the endogenous expression patterns (Figs 3 and 4). The ratio of EPOR-F mRNA to EPOR-T mRNA was much lower in fibroblast lines than in hematopoietic lines. This suggests that hematopoietic cell lines possess a trans-activation system to splice intron VII in RNA derived from pCMV-T. Thus, a cis-element required for the hematopoietic cell-specific splicing may be present in the cDNA sequence used in transfection. Erythroid cell lines, including HEL, transfected with pCMV-T expressed EPOR-T mRNA in a pattern similar to a lymphoid cell line, Jurkat. Since the endogenous expression patterns are different between Jurkat (data not shown, but similar to other nonerythroid/nonmegakaryocytic cell lines shown in Fig 3A and B) and HEL (Fig 3A), splicing of intron VII from the pCMV-T–derived RNA did not perfectly reproduce the pattern from the endogenous EPOR pre-mRNA. Thus, the sequence contained in the cDNA used in transfection was not sufficient for recognition by the erythroid-specific trans-activation system. Sequences outside the coding region may be necessary for the correct splicing of intron VII.
Splicing of intron VII of the EPOR gene is likely to be biologically significant in both normal and pathologic erythropoiesis in humans. It may be of interest to identify cis-element(s) responsible for this splicing to investigate a trans-activation system functioning in a cell type–specific manner.
ACKNOWLEDGMENT
We thank H. Nakauchi for providing an EPOR cDNA and S. Ogawa for useful discussion.
Supported by scientific research grants from the Ministry of Education, Culture, and Science of Japan (H.H.), grants from the US National Institutes of Health (R01DK45118 to K.T. and R37GM09966 to F.H.R.), Special Coordination Funds from the Science and Technology Agency of Japan (J.M.), and the Leukemia Research Foundation (K.T.). S.C. was supported by the US-Japan cooperative program funded by the National Cancer Institute and Japan Foundation of Cancer Research, and the Donaghue Medical Research Foundation at Yale University.
Address reprint requests to Hisamaru Hirai, MD, Third Department of Internal Medicine, Faculty of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113, Japan.

![Fig. 2. Expression of intron VII–spliced and –unspliced mRNAs for EPOR in peripheral blood (A, B, D, and E) and bone marrow (C) mononuclear cells from PV patients, normal volunteers, ET and CML-CP patients, and a cell line. Total cellular RNA was reverse-transcribed and subjected to PCR using the primer sets of sense-3 and antisense-3, and then sense-4 and antisense-4 (nested PCR, [A]) or sense-2 and antisense-2 (B, C and D), respectively. In (E), cytoplasmic RNA was reverse-transcribed and subjected to PCR using the primers sense-2 and antisense-2. (A) Lane a, PV patient no. 1; b, PV patient no. 3; c, normal no. 1 (RNA lot 1); d, normal no. 1 (RNA lot 2); e, normal no. 2. The size of the intron VII–spliced and –unspliced mRNAs is 670 bp and 765 bp, respectively. (B) Lane a, PV patient no. 4; b, PV patient no. 5; c, PV patient no. 6; d, normal no. 4; e, cell line KBM-5. (C) Lane a, normal volunteer no. 3; b, PV patient no. 2; and c, PV patient no. 1. (D) Lane a, ET no. 1; b, ET no. 2; c, CML-CP no. 1; d, CML-CP no. 2; e, CML-CP no. 3. (E) Lane a, PV patient no. 7; b, PV patient no. 8; c, normal no. 5; d, normal no. 6; and e, negative control without template. In (B) to (E), the sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.97/5/m_bl_0022f2a.jpeg?Expires=1763477876&Signature=KXF9B4SHVlufor8336FgJSylNVi5W4qRD-cHBxfKdyr2J1GKZMx4ujhSIHlf3jX9Eneed-~DSN86B~Ygp-8vcVqJMYc7EBeWzFTM6eipnYtEQ-yKgOHHchdfMmaW4tiJjK3XWnWWfgSFLMkVi1xQL3YxXR6~VilunTBHLeHSv3hnjlaQmGq77uo6l3WfKiSycXUcUxOBWq-AiAaVZh0G9PDq6gzXtqVP37RPZJzibdJF4P91ZNPhTT6Fblc4jzSeDiFqQa-HACUTh-eRhbA~nEyN1uwOlfgqMpMy2cY361CD22WsqBFTJErxO8-Bqrkmof2pz1zQ~gjAYh5jJap1QA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Expression of intron VII–spliced and –unspliced mRNAs for EPOR in peripheral blood (A, B, D, and E) and bone marrow (C) mononuclear cells from PV patients, normal volunteers, ET and CML-CP patients, and a cell line. Total cellular RNA was reverse-transcribed and subjected to PCR using the primer sets of sense-3 and antisense-3, and then sense-4 and antisense-4 (nested PCR, [A]) or sense-2 and antisense-2 (B, C and D), respectively. In (E), cytoplasmic RNA was reverse-transcribed and subjected to PCR using the primers sense-2 and antisense-2. (A) Lane a, PV patient no. 1; b, PV patient no. 3; c, normal no. 1 (RNA lot 1); d, normal no. 1 (RNA lot 2); e, normal no. 2. The size of the intron VII–spliced and –unspliced mRNAs is 670 bp and 765 bp, respectively. (B) Lane a, PV patient no. 4; b, PV patient no. 5; c, PV patient no. 6; d, normal no. 4; e, cell line KBM-5. (C) Lane a, normal volunteer no. 3; b, PV patient no. 2; and c, PV patient no. 1. (D) Lane a, ET no. 1; b, ET no. 2; c, CML-CP no. 1; d, CML-CP no. 2; e, CML-CP no. 3. (E) Lane a, PV patient no. 7; b, PV patient no. 8; c, normal no. 5; d, normal no. 6; and e, negative control without template. In (B) to (E), the sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.97/5/m_bl_0022f2b.jpeg?Expires=1763477876&Signature=wcmTBUYu4km~HsuIF6bv86nt4~JXk4ZhUCU42AA0d-EoWKBEsfR9tUKRs5YqsvV-zJpJE2eOfNZ-d09z~nwO0QsIlCVRxChOcJbc4QFC8UCTAJVLnG-2Lz6jEN6ers~r1YJLxnMz0KXU1xHbuErqsVjJd~rInRIxL1~CLS-YLeszLa--h7Tqcisg2DCDwoTebegc5~NNRchIdoLVdexMVBzLcsXO8mHBLdDXI7ntwBRSByaFR-W14GDILWN6MROdf-RaOgGWDdLjhK7GQuJnKf2giP7DNHWPz4dj4POlX20duKpujUELMJ092HLvWejIlaiYkSfh3IsECeXYElWAlw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Expression of intron VII–spliced and –unspliced mRNAs for EPOR in peripheral blood (A, B, D, and E) and bone marrow (C) mononuclear cells from PV patients, normal volunteers, ET and CML-CP patients, and a cell line. Total cellular RNA was reverse-transcribed and subjected to PCR using the primer sets of sense-3 and antisense-3, and then sense-4 and antisense-4 (nested PCR, [A]) or sense-2 and antisense-2 (B, C and D), respectively. In (E), cytoplasmic RNA was reverse-transcribed and subjected to PCR using the primers sense-2 and antisense-2. (A) Lane a, PV patient no. 1; b, PV patient no. 3; c, normal no. 1 (RNA lot 1); d, normal no. 1 (RNA lot 2); e, normal no. 2. The size of the intron VII–spliced and –unspliced mRNAs is 670 bp and 765 bp, respectively. (B) Lane a, PV patient no. 4; b, PV patient no. 5; c, PV patient no. 6; d, normal no. 4; e, cell line KBM-5. (C) Lane a, normal volunteer no. 3; b, PV patient no. 2; and c, PV patient no. 1. (D) Lane a, ET no. 1; b, ET no. 2; c, CML-CP no. 1; d, CML-CP no. 2; e, CML-CP no. 3. (E) Lane a, PV patient no. 7; b, PV patient no. 8; c, normal no. 5; d, normal no. 6; and e, negative control without template. In (B) to (E), the sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.97/5/m_bl_0022f2c.jpeg?Expires=1763477876&Signature=XkmiJcs-~wl4jRJhA9RO6OSSplfqKivtMGYdD5t7ocXFqUufdaeUODBg9vLgiNXnqhGsDbvmEHBy95MqDM09OoaA08ULtxcLZP3sEEEmrH3D0lmEO9qawJ-61Dw14780Uz85JLxBScHiCBjfleEDgYeiRIlABknNC~4Xxx9GhJz6EsAnP3J540Zyu11xyCwIsjFHlJj~kci1tagArnDQi4OjmeYRSb3oSyAiiLbBuUI7XkQ-WWMYso4wCL7-zp4SqnDK8ZJPu2CBtAGMwDYK-IbUXfe9XvABoOVtph7qleQ14Yw43AVUK3AfI~HMufzX7Xxv8Lt~GB1Qt41zzrlIFA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Expression of intron VII–spliced and –unspliced mRNAs for EPOR in peripheral blood (A, B, D, and E) and bone marrow (C) mononuclear cells from PV patients, normal volunteers, ET and CML-CP patients, and a cell line. Total cellular RNA was reverse-transcribed and subjected to PCR using the primer sets of sense-3 and antisense-3, and then sense-4 and antisense-4 (nested PCR, [A]) or sense-2 and antisense-2 (B, C and D), respectively. In (E), cytoplasmic RNA was reverse-transcribed and subjected to PCR using the primers sense-2 and antisense-2. (A) Lane a, PV patient no. 1; b, PV patient no. 3; c, normal no. 1 (RNA lot 1); d, normal no. 1 (RNA lot 2); e, normal no. 2. The size of the intron VII–spliced and –unspliced mRNAs is 670 bp and 765 bp, respectively. (B) Lane a, PV patient no. 4; b, PV patient no. 5; c, PV patient no. 6; d, normal no. 4; e, cell line KBM-5. (C) Lane a, normal volunteer no. 3; b, PV patient no. 2; and c, PV patient no. 1. (D) Lane a, ET no. 1; b, ET no. 2; c, CML-CP no. 1; d, CML-CP no. 2; e, CML-CP no. 3. (E) Lane a, PV patient no. 7; b, PV patient no. 8; c, normal no. 5; d, normal no. 6; and e, negative control without template. In (B) to (E), the sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.97/5/m_bl_0022f2d.jpeg?Expires=1763477876&Signature=dMsedryMW1yBLgH65Qyl1HkvA2lAZEtDniv1sggkDfnjnk2deLkuGNFnpvHcv36AtLRr98HNBY~TOOI1-UGX~ceS0qos-FreG-ctaDe6mDHi6F45qzZtzHcc-4h9Eo-1i5M-CiT3OPe40M4uftKQgpuB-My2XuTWzYkD8D7r5x2v1lV8RZfR9p4XpYTCNY6ueURnvacxQWWLGm-tqdUHtdqTioDC8z~7ncFidIZ7OXTCCAYKR-vl0JqRgMTWeVKniDnum2kHjm3d8IN07pnvDNXPzsPDO-XvtqJI7PEA44X3ILK6GXqqxKaG6am6n4MHguB3CA~na-8bWkbTyyxIjw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Expression of intron VII–spliced and –unspliced mRNAs for EPOR in peripheral blood (A, B, D, and E) and bone marrow (C) mononuclear cells from PV patients, normal volunteers, ET and CML-CP patients, and a cell line. Total cellular RNA was reverse-transcribed and subjected to PCR using the primer sets of sense-3 and antisense-3, and then sense-4 and antisense-4 (nested PCR, [A]) or sense-2 and antisense-2 (B, C and D), respectively. In (E), cytoplasmic RNA was reverse-transcribed and subjected to PCR using the primers sense-2 and antisense-2. (A) Lane a, PV patient no. 1; b, PV patient no. 3; c, normal no. 1 (RNA lot 1); d, normal no. 1 (RNA lot 2); e, normal no. 2. The size of the intron VII–spliced and –unspliced mRNAs is 670 bp and 765 bp, respectively. (B) Lane a, PV patient no. 4; b, PV patient no. 5; c, PV patient no. 6; d, normal no. 4; e, cell line KBM-5. (C) Lane a, normal volunteer no. 3; b, PV patient no. 2; and c, PV patient no. 1. (D) Lane a, ET no. 1; b, ET no. 2; c, CML-CP no. 1; d, CML-CP no. 2; e, CML-CP no. 3. (E) Lane a, PV patient no. 7; b, PV patient no. 8; c, normal no. 5; d, normal no. 6; and e, negative control without template. In (B) to (E), the sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.97/5/m_bl_0022f2e.jpeg?Expires=1763477876&Signature=xRG6Kh6tW0~ikLD9rDP69Mk5myrobhtwnzeyRYjxQ10nKINugTZdwB3cAqhM8Q3Bw~2o~t5my-hHWwQQYrwUsAYN7IlupkWMbdXMhP-JGB~x28vpSxZkddlwPP~L2HfUZKu86ajur8jKS3Y~wy5X7D-qryv3QnQp5kCxflKPTxTo-L5c660C6wGWko5ErzjCJWQ170n9~W3~7cY9epRFSHOS0MaWIJVcy4ARGYFi32J1j5BzC8tFYjr1yboUTV84XYzrcPN4y9UhidK5vcnquIlFH4HRPREySHmHrGKj9O7MjGHl5KdhSeH7HscF9hTJVFZG7Gq7iPc0W09VYhwh0A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Expression of the intron VII–spliced and –unspliced mRNAs for EPOR in various leukemia cell lines and leukemia cells. Total cellular RNA (A, B, and C) or polyA(+) RNA (C) was reverse-transcribed and subjected to PCR using the primer sets of sense-2 and antisense-2. (A) Lane a, BV-173 (lymphoid); b, NALM-16 (lymphoid); c, NALM-24 (lymphoid); d, K562 (erythroid); e, MEG-01 (megakaryocytic); f, HEL (erythroid); g, JOSK-1 (monocytic); h, SU-DHL-4 (lymphoid); and i, CCRF-CEM (lymphoid). (B) Lane a, KOPT-K1 (lymphoid); b, PEER (lymphoid); c, F-36E (erythroid); d, F-36P (erythroid); e, UT-7 (megakaryocytic); and f, TF-1 (erythroid). (C) One microgram total RNA prepared from U-937 cells, 0.5 μg polyA(+) RNA therefrom, and 0.5 μg polyA(+) RNA prepared from peripheral blood leukemia cells from a patient with CML in blast crisis9 was reverse-transcribed and subjected to the PCR protocol with the indicated number of cycles. Total RNA was used in lanes a to c and polyA(+) RNA in d to i. The number of cycles was 30 (lanes a, d, and g), 35 (b, e, and h), and 40 (c, f, and i). The sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively. (D) RNase protection assay shows expression of EPOR-F and EPOR-T mRNAs in U-937 and leukemia cells obtained from the same patient as described in C. A 420-bp [32P]-labeled riboprobe was hybridized with 10 μg polyA(+) RNA samples, digested with the mixture of RNase A and RNase T1, and analyzed in 5% polyacrylamide/8 mol/L urea gel. Lane a, undigested probe; b, yeast RNA; c, U-937; d, CML blast crisis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.97/5/m_bl_0022f3a.jpeg?Expires=1763477876&Signature=4A7RVNrjcZKYAAFBi862P~a79EzE-jQf88VPPfro3kVVJ3VBD74mZd~3NbyfyQDMMTcYtWEYsFxCqw19Hdz-nKLc2Ruuh9ngkLo75oW7cTQ~oZ9JFF2-gwcJW6EflbVBlEIDaYOECNFtZ18xNoa0MypInmdj4tRrkfsXiyLY~Obxw1MJbpC4lAvoXFN-Wh5odYCoinRPkRSr9RegSY-NP8AJy7CN9lz8SaO33IPfsaQC3ehoaD6DCtEQ0jT61KJY2niF3D6vMd-fU0VdbJVNEU-7Q~DKKxUKSnZ3R26eAxVXosPIxyaOTXWOGtuQcYBkdkqSTfuEGVHwVCfls~6WpA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Expression of the intron VII–spliced and –unspliced mRNAs for EPOR in various leukemia cell lines and leukemia cells. Total cellular RNA (A, B, and C) or polyA(+) RNA (C) was reverse-transcribed and subjected to PCR using the primer sets of sense-2 and antisense-2. (A) Lane a, BV-173 (lymphoid); b, NALM-16 (lymphoid); c, NALM-24 (lymphoid); d, K562 (erythroid); e, MEG-01 (megakaryocytic); f, HEL (erythroid); g, JOSK-1 (monocytic); h, SU-DHL-4 (lymphoid); and i, CCRF-CEM (lymphoid). (B) Lane a, KOPT-K1 (lymphoid); b, PEER (lymphoid); c, F-36E (erythroid); d, F-36P (erythroid); e, UT-7 (megakaryocytic); and f, TF-1 (erythroid). (C) One microgram total RNA prepared from U-937 cells, 0.5 μg polyA(+) RNA therefrom, and 0.5 μg polyA(+) RNA prepared from peripheral blood leukemia cells from a patient with CML in blast crisis9 was reverse-transcribed and subjected to the PCR protocol with the indicated number of cycles. Total RNA was used in lanes a to c and polyA(+) RNA in d to i. The number of cycles was 30 (lanes a, d, and g), 35 (b, e, and h), and 40 (c, f, and i). The sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively. (D) RNase protection assay shows expression of EPOR-F and EPOR-T mRNAs in U-937 and leukemia cells obtained from the same patient as described in C. A 420-bp [32P]-labeled riboprobe was hybridized with 10 μg polyA(+) RNA samples, digested with the mixture of RNase A and RNase T1, and analyzed in 5% polyacrylamide/8 mol/L urea gel. Lane a, undigested probe; b, yeast RNA; c, U-937; d, CML blast crisis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.97/5/m_bl_0022f3b.jpeg?Expires=1763477876&Signature=tALmZY0EMiTcKcNAkx6C~aCnUBR2A177xr8jALiJ-fF-dnpdi5zSpk~dJE94SJmd7wHkzyGsvbYwfSZ9VpO~7vBRoQ8A9scKkGTwQhzXRwWyjg5LfNT0HnRYjjSNlIalPqBj9zYnG2ALrOUImhs6mUwO5lxD1fb88aWJVbttOsZr6hYgGIhKKHxFuz8jTrNKsoLaL75PFzRxwkgPae-~qFXIm96qN8-gFAL0-mgTy84ZG~woxi0V8FPndA8tL0f99hsi5rbqx42F54jMb1Q7la1c2QlWsBkbicO1wxMiZgyWWETE8tethT9AD9iuUMrzjx7PJM21BFjTst8f0HXrDw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Expression of the intron VII–spliced and –unspliced mRNAs for EPOR in various leukemia cell lines and leukemia cells. Total cellular RNA (A, B, and C) or polyA(+) RNA (C) was reverse-transcribed and subjected to PCR using the primer sets of sense-2 and antisense-2. (A) Lane a, BV-173 (lymphoid); b, NALM-16 (lymphoid); c, NALM-24 (lymphoid); d, K562 (erythroid); e, MEG-01 (megakaryocytic); f, HEL (erythroid); g, JOSK-1 (monocytic); h, SU-DHL-4 (lymphoid); and i, CCRF-CEM (lymphoid). (B) Lane a, KOPT-K1 (lymphoid); b, PEER (lymphoid); c, F-36E (erythroid); d, F-36P (erythroid); e, UT-7 (megakaryocytic); and f, TF-1 (erythroid). (C) One microgram total RNA prepared from U-937 cells, 0.5 μg polyA(+) RNA therefrom, and 0.5 μg polyA(+) RNA prepared from peripheral blood leukemia cells from a patient with CML in blast crisis9 was reverse-transcribed and subjected to the PCR protocol with the indicated number of cycles. Total RNA was used in lanes a to c and polyA(+) RNA in d to i. The number of cycles was 30 (lanes a, d, and g), 35 (b, e, and h), and 40 (c, f, and i). The sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively. (D) RNase protection assay shows expression of EPOR-F and EPOR-T mRNAs in U-937 and leukemia cells obtained from the same patient as described in C. A 420-bp [32P]-labeled riboprobe was hybridized with 10 μg polyA(+) RNA samples, digested with the mixture of RNase A and RNase T1, and analyzed in 5% polyacrylamide/8 mol/L urea gel. Lane a, undigested probe; b, yeast RNA; c, U-937; d, CML blast crisis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.97/5/m_bl_0022f3c.jpeg?Expires=1763477876&Signature=MZuZrK~mUjw0jnRkZZdizxs8VGCWZOesEN1ibZ3lN7rf-VIlIMT4DVeUUmEesYAg6U5H~3LPitcLRPg93olqsBzgMiyhILzMXdFF8a2A9GSiSTBDfYYZTKa9z6woEU9Y68jNF~qx~52LKeYxuhc7EHTVRkUTV20eDs1cz2ksR7WDcwhUzfqh0Uuj6Pz3jFfgW~Qwo7TzQWMz~S97~Km3Wp8LQQZsOMN9W6qt1eWeTxiHbu687F3Ha0jp3F1DGxB-lQ4rC9YxxzS0UVfYj25RqG-8E55y2IV5oCrsrawyc9F1cLGaVu~MDBmTOL6tvFiFF7xLSHFot41bZW9c~lD8fw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Expression of the intron VII–spliced and –unspliced mRNAs for EPOR in various leukemia cell lines and leukemia cells. Total cellular RNA (A, B, and C) or polyA(+) RNA (C) was reverse-transcribed and subjected to PCR using the primer sets of sense-2 and antisense-2. (A) Lane a, BV-173 (lymphoid); b, NALM-16 (lymphoid); c, NALM-24 (lymphoid); d, K562 (erythroid); e, MEG-01 (megakaryocytic); f, HEL (erythroid); g, JOSK-1 (monocytic); h, SU-DHL-4 (lymphoid); and i, CCRF-CEM (lymphoid). (B) Lane a, KOPT-K1 (lymphoid); b, PEER (lymphoid); c, F-36E (erythroid); d, F-36P (erythroid); e, UT-7 (megakaryocytic); and f, TF-1 (erythroid). (C) One microgram total RNA prepared from U-937 cells, 0.5 μg polyA(+) RNA therefrom, and 0.5 μg polyA(+) RNA prepared from peripheral blood leukemia cells from a patient with CML in blast crisis9 was reverse-transcribed and subjected to the PCR protocol with the indicated number of cycles. Total RNA was used in lanes a to c and polyA(+) RNA in d to i. The number of cycles was 30 (lanes a, d, and g), 35 (b, e, and h), and 40 (c, f, and i). The sizes of intron VII–spliced and –unspliced mRNAs are 236 bp and 331 bp, respectively. (D) RNase protection assay shows expression of EPOR-F and EPOR-T mRNAs in U-937 and leukemia cells obtained from the same patient as described in C. A 420-bp [32P]-labeled riboprobe was hybridized with 10 μg polyA(+) RNA samples, digested with the mixture of RNase A and RNase T1, and analyzed in 5% polyacrylamide/8 mol/L urea gel. Lane a, undigested probe; b, yeast RNA; c, U-937; d, CML blast crisis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.97/5/m_bl_0022f3d.jpeg?Expires=1763477876&Signature=ulwtJd557uVC7qB3WgF6hRiTO6z5KBdp2z44DquA7reHKx7gO6~Yc9NKBKiyUBOcsmcrR1T1RZvp6sZSeSiF6L1MPk51eEB4p406m5ylMwUrgnf8d6zW8KELi-pGZQ3MSCkhiPuRw6MpBejhNpGMXIwwTiNsJIP5RFiLYdRayupB3CSDGSlgo6CRCQUCCzCO~I~IVtgxIuMKMgMz5hiwVDGHX5fjgNXxdYPH42uCTFnZNnj7NkG3h4G2ISGBm8j4XUl4gt7LfpTFmhI9CoVxQ6m8tkzbUXfR6eX3Wc3EKDRrLNMkUFntZeUQGLePsMOvVRpFm63uozMvvWAxsW8abA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal