Abstract
Salivary gland mucosa-associated lymphoid tissue (MALT) type lymphomas are typically indolent B-cell neoplasms that are often associated with Sjogren's syndrome. To better define the cell of origin and evaluate whether antigen receptor stimulation may be playing a role in tumor growth, the Ig heavy and light chain variable genes (VH and VL) expressed by five salivary gland MALT lymphomas were cloned and sequenced. Comparison to known germline sequences indicated that three of the lymphoma VH genes were derived from 51p1, a member of the VH1 family, while the other two used different VH gene segments from the VH3 family, 22-2B and HG19. All five of the VL genes belonged to the VkIII family, with three derived from Humkv325 and the other two from the Vg and Humkv328 genes. Numerous point mutations relative to the proposed germline genes were present in all of the lymphoma VH and VL genes. In addition, the VH and VL genes from each lymphoma showed intraclonal sequence heterogeneity indicative of ongoing somatic hypermutation. Because the process of Ig gene hypermutation is thought to occur at the germinal center stage of B-cell development, these findings suggest the MALT lymphoma cell of origin may be a germinal center B cell. Selection against mutations that result in replacement of amino acids suggested that Ig stimulation may be important for lymphoma growth. The possibility that antigen receptor stimulation may be involved in the growth of salivary gland MALT lymphomas is further suggested by the noted restricted use of VH and VL gene segments.
LYMPHOMAS OF mucosa-associated lymphoid tissue (MALT) type represent a distinct group of commonly occurring extranodal non-Hodgkin's lymphomas that can develop in a variety of locations.1,2 The majority are low-grade neoplasms, but transformation to high-grade MALT lymphoma occurs in some cases.3 Low-grade MALT lymphomas often remain localized at their sites of origin for many years in contrast to low-grade nodal base lymphomas, which are typically widely disseminated at diagnosis.4,5 Sites where MALT lymphomas frequently develop are also usually involved by an associated autoimmune disease or infection that precedes the lymphoma.5 For example, salivary gland MALT lymphomas are associated with myoepithelial sialadenitis (MESA), which is characteristic of Sjogren's syndrome,6,7 while MALT lymphomas of the thyroid gland and stomach are associated with Hashimoto's thyroiditis and Helicobacter pylori infection, respectively.8-10 It has been suggested that the growth of low-grade MALT lymphomas along with their propensity to remain localized may be dependent on antigen stimulation and/or help provided by the associated local immune reactions.5 This idea has gained considerable support recently from studies of gastric MALT lymphomas where it appears that most low-grade tumors can be eradicated upon the elimination of Helicobacter pylori using antibiotics.11 12
MALT lymphomas are thought to represent neoplasms of marginal zone B cells.13 Marginal zones are prominent features of reactive MALT as typified by Peyer's patches and are located outside of the follicular mantle areas extending to and infiltrating the epithelium, which is also a characteristic location where MALT lymphoma cells accumulate.14 In addition, MALT lymphomas have immunophenotypes that are similar to marginal zone B cells and are typically CD5−, CD10−, and IgM+ IgD−.13,14 Marginal zone B cells that express IgM appear to be enriched for early memory B cells, which are the direct progeny of germinal center cells.15 Because somatic hypermutation of Ig genes is thought to occur only in germinal center B cells,16-18 finding that gastric MALT lymphomas use mutated VH genes without intraclonal variation is consistent with a derivation from postgerminal center marginal zone memory B cells.19,20 However, a recent study has reported finding intraclonal variation in gastric MALT lymphoma VH genes suggesting the process of hypermutation is still ongoing in these neoplasms.21 Molecular studies of VH or VL genes expressed by MALT lymphomas that do not originate in the stomach or are not associated with Helicobacter pylori have yet to be reported.
In this study, we cloned and sequenced the VH and VL genes expressed by five salivary gland MALT lymphomas to better define the cell of origin and to also investigate whether direct antigen stimulation may be involved in the development or growth of these tumors. Information regarding direct antigen stimulation of lymphoma cells can be obtained from analyzing the distribution of somatic mutations within expressed Ig genes or frequencies that particular VH or VL gene segments are used.22-24 In addition to finding mutations in all of the VH and VL genes, we also found evidence that the process of Ig gene hypermutation is still ongoing in lymphoma cells. This suggests that the cell of origin may be a germinal center B cell, which then gives rise to the marginal zone-like cells that characterize MALT lymphomas. Moreover, ongoing hypermutation further suggests that antigen is directly stimulating lymphoma growth, which is also suggested by finding three of the lymphomas using the same VH and VL gene segments.
MATERIALS AND METHODS
Case information.The five low-grade salivary gland MALT lymphomas evaluated in this study represent cases from the University of Pittsburgh for which DNA or frozen cells were available for analysis and were not otherwise selected. All of the patients were females with ages ranging from 59 to 68 years and were biopsied to evaluate parotid gland enlargement. Three of the patients (HA, OR, RO) had a previous diagnosis of Sjogren's syndrome. Diagnoses of low-grade MALT lymphoma were based on histologic examination of the excised parotid gland tissue, which in all cases, showed typical features including residual MESA. Southern blot analysis demonstrated a monoclonal heavy chain gene rearrangement in four of four tested cases without T-cell receptor beta or Bcl-2 rearrangement. Flow cytometric immunophenotypic analysis performed on the same four cases showed CD19+ B cells that lacked expression of CD5 or CD10. However, B-cell monoclonality could not be established with certainty by flow analysis.
Preparation of DNA and cDNA.DNA was isolated from the parotid gland tissue using a standard method employing proteinase K and sodium dodecyl sulfate (SDS).25 RNA was isolated using RNA-Zol following the manufacturer's directions (Biotecx Laboratories, Houston, TX). cDNA was synthesized using RNA from approximately 2 × 106 lymphoma cells, random heximer primers (5 mmol/L), 40 U of RNAsin (Promega, Madison, WI) and 200 U of Superscript II reverse transcriptase using the suppliers buffer and directions (GIBCO-BRL Gaithersburg, MD).
Amplification of V genes.Rearranged VH and VL genes were amplified essentially as previously described26 in reactions that contained only one or two of the 5′ leader region primers for the indicated 6 VH and 6 VK families (Table 1). A corresponding 3′ J or C region primer was used depending on whether the template was DNA or cDNA, respectively. Individual primary polymerase chain reactions (PCR) reactions were performed in 50 μL total volumes and contained approximately 1/20 of cDNA or 1 μg of DNA and 5′ V and 3′ J or C primers each at a final concentration of 0.5 μm. Amplification for 30 to 37 cycles was performed using Taq polymerase with all other reagents as recommended by the model 480 thermocycler manufacturer (Perkin Elmer, Norwalk, CT). Cycling parameters were 20 seconds at 94°C (first cycle 2 minutes), 30 seconds at 55°C, and 30 seconds at 72°C (last cycle 5 minutes). Following amplification, approximately one fourth of the PCR products were electrophoresed in 1.5% agarose and visualized by ethidium bromide staining. Secondary amplification with the FW3-JH primer combination to assess clonality was performed in a similar fashion for 12 to 14 cycles using approximately 0.5 μL of the VH PCR products as templates. For better resolution of smaller sizes, these latter resultant products were electrophoresed in 8% acrylamide before staining with ethidium bromide.
Primers Used for V Gene Amplification
| Name . | Sequence (5′-3′) . |
|---|---|
| VH1 leader | CACCATGGACTGACCTGGA |
| VH2 leader | ATGGACATACTTTGTTCCAC |
| VH3 leader | CCATGGAGTTTGGGCTGAGC |
| VH4 leader | ATGAAACACCTGTGGTTCTT |
| VH5 leader | ATGGGGTCAACCGCCATCCT |
| VH6 leader | ATGTCTGTCTCCTTCCTCAT |
| FW3 | GACACGGC(C,T)(A,G)TGTATTACTG |
| JH | ACCTGAGGAGACGGTGACC |
| Cμ | GTCCATCCGCCCCAACCCTT |
| VK1 leader | ATGGACATGAGGGTCCCCGC |
| VK2 leader | ATGAGGCTCCCTGCTCAGCT |
| VK3 leader | ATGGAAACCCCAGC(G,T)CAGCT |
| VK4 leader | ATGGTGTTGCAGACCCAGGTC |
| VK5 leader | ATGGGGTCCCAGGTTCACCTC |
| VK6 leader | ATGTTGCCATCACAACTCATTG |
| JK124 | ACGTTTGAT(C,T)TCCA(C,G)CTTGGTC |
| JK3 | ACGTTTGATATCCACTTTGGTC |
| JK5 | ACGTTTAATCTCCAGTCGTGTC |
| CK | AGATGGCGGGAAGATGAAGAC |
| Name . | Sequence (5′-3′) . |
|---|---|
| VH1 leader | CACCATGGACTGACCTGGA |
| VH2 leader | ATGGACATACTTTGTTCCAC |
| VH3 leader | CCATGGAGTTTGGGCTGAGC |
| VH4 leader | ATGAAACACCTGTGGTTCTT |
| VH5 leader | ATGGGGTCAACCGCCATCCT |
| VH6 leader | ATGTCTGTCTCCTTCCTCAT |
| FW3 | GACACGGC(C,T)(A,G)TGTATTACTG |
| JH | ACCTGAGGAGACGGTGACC |
| Cμ | GTCCATCCGCCCCAACCCTT |
| VK1 leader | ATGGACATGAGGGTCCCCGC |
| VK2 leader | ATGAGGCTCCCTGCTCAGCT |
| VK3 leader | ATGGAAACCCCAGC(G,T)CAGCT |
| VK4 leader | ATGGTGTTGCAGACCCAGGTC |
| VK5 leader | ATGGGGTCCCAGGTTCACCTC |
| VK6 leader | ATGTTGCCATCACAACTCATTG |
| JK124 | ACGTTTGAT(C,T)TCCA(C,G)CTTGGTC |
| JK3 | ACGTTTGATATCCACTTTGGTC |
| JK5 | ACGTTTAATCTCCAGTCGTGTC |
| CK | AGATGGCGGGAAGATGAAGAC |
Cloning and sequencing of PCR products.Following electrophoresis and staining with ethidium bromide, appropriately sized PCR products were isolated from 1.5% agarose and further purified using Wizard DNA preps (Promega). Approximately one tenth of the resultant DNA was cloned using the pCR-Script cloning kit (Stratagene, La Jolla, CA), following the manufacturer's directions. Plasmid DNA was isolated from overnight cultures of randomly selected white colonies using Wizard Mini-preps (Promega). Diodeoxy sequencing was performed with Sequenase (United States Biochemical, Cleveland, OH) following the manufacturer's protocol for double stranded plasmid template using approximately one third of the isolated DNA. All clones were sequenced from both directions using the M13 forward and reverse primers. Sequence analysis was performed using the MacVector software (IBI, New Haven, CT).
Analysis of mutations.For n total mutations, the expected number of replacement (R) mutations in a complementarity-determining region (CDR) or framework region (FWR) was calculated according to the formula Rcdr = n × Acdr × Rfcdr where Acdr is the relative size of the CDR (or FWR) and Rfcdr is the frequency of R mutation for the CDR (or FWR).27 Rf values are typically different for CDRs and FWRs and can vary among different genes depending on the specific nucleotide sequence. Because mutations are either R or silent (S), the expected number of S mutations was obtained for the CDRs and FWRs using the same formula with Rf replaced by 1-Rf. The expected values assume that mutations distribute randomly throughout the variable (V) gene, which is not completely accurate28 and, therefore, should be considered as only approximations. The probability P that k number of R mutations occurred in the CDRs by chance alone was determined as previously described with a binomial model22 except the gene specific Rfcdr value was used instead of the more approximate value of 0.75.
RESULTS
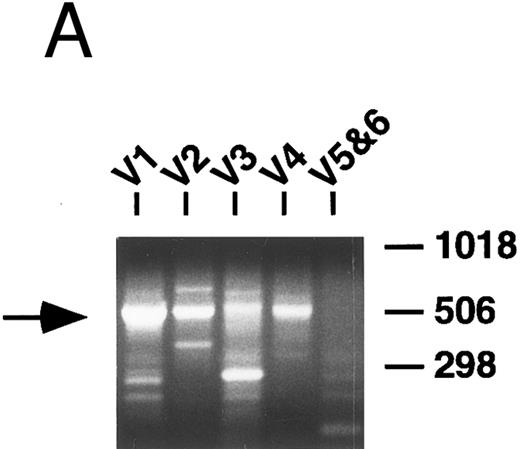
PCR analysis of V genes.Rearranged V genes were amplified from DNA or cDNA prepared from the lymphoma specimens using 5′ VH or VK leader primers in conjunction with an appropriate 3′ J or C region primer. Primers for the infrequently used VH5 and VH6, as well as VK5 and VK6 families, were combined in one reaction tube, while primers that amplify the other VH and VK familes were used in individual reactions. In each case, levels of appropriately sized PCR products from one of the five VH or VL amplification reactions appeared to be increased relative to the others following electrophoresis and staining with ethidium bromide (Fig 1A). These dominant products were subsequently cloned and sequenced and a unique repeat VH or VK sequence was obtained in each case confirming the presence of a monoclonal B-cell population as further detailed below.
PCR analysis of rearranged VH genes. Lymphoma DNA was amplified with six different VH leader primers and a consensus JH primer and the resultant products electrophoresed in 1.5% agarose and stained with ethidium bromide as shown in (A). Note the dominant band of the expected size (arrow) generated with the VH1-JH primer combination suggesting the lymphoma is using a VH1 family gene. To assess clonality, the VH PCR products and lymphoma DNA were further amplified with the FW3 and JH primers and the resultant products electrophoresed in 10% acrylamide and stained with ethidium bromide as shown in (B). Observe that the VH1 PCR products give a clonal band that comigrates with the clone in the lymphoma DNA (arrow), while the other PCR products give only polyclonal laders representing different sized V-D-J joints.
PCR analysis of rearranged VH genes. Lymphoma DNA was amplified with six different VH leader primers and a consensus JH primer and the resultant products electrophoresed in 1.5% agarose and stained with ethidium bromide as shown in (A). Note the dominant band of the expected size (arrow) generated with the VH1-JH primer combination suggesting the lymphoma is using a VH1 family gene. To assess clonality, the VH PCR products and lymphoma DNA were further amplified with the FW3 and JH primers and the resultant products electrophoresed in 10% acrylamide and stained with ethidium bromide as shown in (B). Observe that the VH1 PCR products give a clonal band that comigrates with the clone in the lymphoma DNA (arrow), while the other PCR products give only polyclonal laders representing different sized V-D-J joints.
Analysis of MALT Lymphoma VH and VL Genes
| Patient . | VH Genes . | VL Genes . | ||||||
|---|---|---|---|---|---|---|---|---|
| . | Family . | Tumor Seq/Clones Seq . | Germline . | Homology % (no. nuc)* . | Family . | Tumor Seq/Clones Seq . | Germline . | Homology % (no. nuc) . |
| HA | VH1 | 10/13 | 51p1 (DP10)† | 98 (7) | VkIII | 4/4 | Humkv325 (A27) | 97 (10) |
| SM | VH1 | 7/9 | 51p1 (DP10) | 93 (22) | VkIII | 4/5 | Humkv325 (A27) | 95 (16) |
| OR | VH1 | 7/7 | 51p1 (DP10) | 99 (4) | VkIII | 4/4 | Humkv325 (A27) | 97 (10) |
| RO | VH3 | 4/5 | 22-2B (DP35) | 90 (28) | VkIII | 3/3 | Vg (L6) | 94 (22) |
| SH | VH3 | 4/4 | HHG19 (DP54) | 97 (9) | VkIII | 3/3 | Humkv328 (L16) | 96 (14) |
| Patient . | VH Genes . | VL Genes . | ||||||
|---|---|---|---|---|---|---|---|---|
| . | Family . | Tumor Seq/Clones Seq . | Germline . | Homology % (no. nuc)* . | Family . | Tumor Seq/Clones Seq . | Germline . | Homology % (no. nuc) . |
| HA | VH1 | 10/13 | 51p1 (DP10)† | 98 (7) | VkIII | 4/4 | Humkv325 (A27) | 97 (10) |
| SM | VH1 | 7/9 | 51p1 (DP10) | 93 (22) | VkIII | 4/5 | Humkv325 (A27) | 95 (16) |
| OR | VH1 | 7/7 | 51p1 (DP10) | 99 (4) | VkIII | 4/4 | Humkv325 (A27) | 97 (10) |
| RO | VH3 | 4/5 | 22-2B (DP35) | 90 (28) | VkIII | 3/3 | Vg (L6) | 94 (22) |
| SH | VH3 | 4/4 | HHG19 (DP54) | 97 (9) | VkIII | 3/3 | Humkv328 (L16) | 96 (14) |
Abbreviations: Seq, sequence; nuc, nucleotide.
(% homologies and number of nucleotide differences from the start of FW1 in consensus sequence).
(Alternative name).
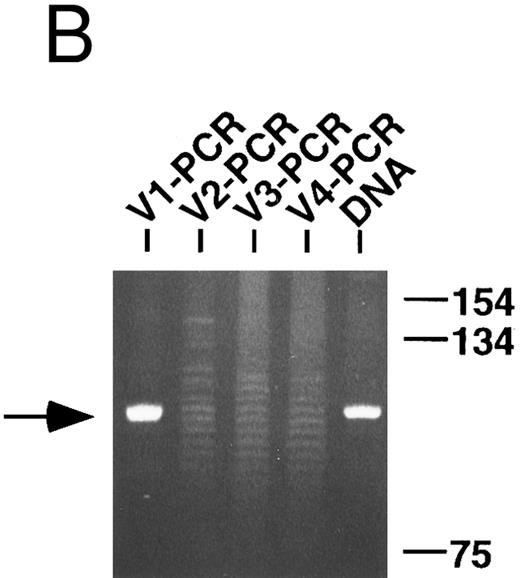
Because of the theoretical possibility that some of the specimens could be oligoclonal, the clonality of the VH PCR products that were not sequenced, as well as the lymphoma DNA (or cDNA), was also tested using a standard PCR technique.29 Observe in the representative example shown in Fig 1B that only the VH1 PCR products gave a dominant band indicative of a monoclonal population, while the other VH PCR products generated polyclonal ladders, which presumably represent specimen contamination by normal B cells. Also observe that the VH1 clone comigrates with the clone identified in the unmanipulated lymphoma DNA and, therefore, appears to be identical. By this type of analysis, all of the VH PCR products not cloned and sequenced were determined to be polyclonal and a single heavy chain clone was identified in four of the five lymphoma specimens. A heavy chain clone could not be identified in case RO with this technique presumably because of mismatches with the consensus 5′ FW3 primer (see below).
Sequence analysis of lymphoma V genes.Complete nucleotide sequences were obtained for at least three randomly selected clones of the VH and VL PCR products amplified from each case (Table 2). Thirteen VH clones from patient HA were analyzed because a lymph node, as well as parotid gland specimen, were evaluated. In most cases, all of the VH or VL gene sequences from a patient were clonally related having identical V-D-J joints or CDR3s, although occasional unrelated clones were also identified for some of the VH or VL genes. These unrelated clones were considered to represent PCR amplification of the background B cells. In addition, all of the consensus VH and VL genes appeared to represent expressed genes for functional Ig, as no stop codons were identified and all were in frame at the end of CDR3 or start of FWR4. Consensus sequences for each of the lymphoma VH and VL genes have been submitted to the GenBank data base (accession nos. U79581-U79590). Amino acid sequences deduced from the consensus nucleotide lymphoma V gene sequences are shown in Fig 2.
Deduced amino acid sequences of lymphoma V genes. Differences of the VH and VL consensus sequences from the most closely related germline genes are shown in (A) and (B), respectively, as upper case letters or are underlined in the J regions. The locations of silent mutations in the nucleic acid sequences are indicated with lower case letters.
Deduced amino acid sequences of lymphoma V genes. Differences of the VH and VL consensus sequences from the most closely related germline genes are shown in (A) and (B), respectively, as upper case letters or are underlined in the J regions. The locations of silent mutations in the nucleic acid sequences are indicated with lower case letters.
As anticipated from the PCR analysis, three of the lymphomas express VH genes derived from the VH1 family, while the other two use members of the VH3 family. Sequence analysis also confirmed that all of the lymphoma VL genes were members of the VkIII family. Candidate germline genes were assigned by searching Genebank or the VBASE directory.30 All three of the lymphoma VH1 genes were found to be most closely related the 51p1 germline gene having homology values of 93% or greater. One of the lymphomas expressing a VH3 family gene appeared to be derived from the 22-2B gene and the other from the HHG19 germline gene. Of the five light chain genes, three appeared to be derived from the Humkv325 germline gene, one from the Vg gene, and one from the Humkv328 gene. Note that all of the lymphoma Humkv325 VL genes are paired with a 51p1-derived VH gene.
Mutation analysis.All of the lymphoma VH and VL genes showed numerous single nucleotide differences from the candidate germline genes, which will hereafter be termed somatic mutations (see Discussion). The analysis to help determine whether the mutations in the consensus sequences were related to antigenic selection is summarized in Table 3. None of the lymphoma V genes demonstrated marked clustering of R mutations in the CDRs above the expected values that would be indicative of positive antigen selection.22,31 Moreover, the binomial model derived probabilities for obtaining the observed number of R mutations in the CDRs by chance alone were greater than .10 with only one exception where the P value was .05. The decreased number of R mutations from the expected values evident in most of the FWRs is typical for expressed Ig molecules and is thought to reflect negative selection in these areas to maintain Ig function.32
Distribution of Mutations in Consensus VH and VL Sequences
| Patient . | Region . | VH Genes . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Expected . | Observed . | P3-150 . | VL Genes . | |||||||
| . | . | R . | S . | R . | S . | . | Expected . | Observed . | P . | . | ||
| . | . | . | . | . | . | . | R . | S . | R . | S . | . | . |
| HA | CDR3-151 | 1 | 0 | 1 | 0 | .38 | 2 | 0 | 0 | 2 | .10 | |
| FWR | 4 | 1 | 1 | 5 | 5 | 2 | 3 | 5 | ||||
| SM | CDR | 4 | 1 | 2 | 5 | .14 | 3 | 1 | 2 | 4 | .22 | |
| FWR | 13 | 4 | 4 | 11 | 8 | 3 | 3 | 6 | ||||
| OR | CDR | 1 | 0 | 2 | 1 | .27 | 1 | 0 | 1 | 1 | .36 | |
| FWR | 2 | 1 | 0 | 1 | 4 | 1 | 4 | 1 | ||||
| RO | CDR | 6 | 1 | 7 | 6 | .14 | 4 | 1 | 7 | 4 | .05 | |
| FWR | 17 | 6 | 8 | 9 | 11 | 3 | 5 | 3 | ||||
| SH | CDR | 2 | 0 | 3 | 1 | .17 | 2 | 1 | 2 | 2 | .30 | |
| FWR | 5 | 2 | 4 | 1 | 6 | 2 | 5 | 2 | ||||
| Patient . | Region . | VH Genes . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Expected . | Observed . | P3-150 . | VL Genes . | |||||||
| . | . | R . | S . | R . | S . | . | Expected . | Observed . | P . | . | ||
| . | . | . | . | . | . | . | R . | S . | R . | S . | . | . |
| HA | CDR3-151 | 1 | 0 | 1 | 0 | .38 | 2 | 0 | 0 | 2 | .10 | |
| FWR | 4 | 1 | 1 | 5 | 5 | 2 | 3 | 5 | ||||
| SM | CDR | 4 | 1 | 2 | 5 | .14 | 3 | 1 | 2 | 4 | .22 | |
| FWR | 13 | 4 | 4 | 11 | 8 | 3 | 3 | 6 | ||||
| OR | CDR | 1 | 0 | 2 | 1 | .27 | 1 | 0 | 1 | 1 | .36 | |
| FWR | 2 | 1 | 0 | 1 | 4 | 1 | 4 | 1 | ||||
| RO | CDR | 6 | 1 | 7 | 6 | .14 | 4 | 1 | 7 | 4 | .05 | |
| FWR | 17 | 6 | 8 | 9 | 11 | 3 | 5 | 3 | ||||
| SH | CDR | 2 | 0 | 3 | 1 | .17 | 2 | 1 | 2 | 2 | .30 | |
| FWR | 5 | 2 | 4 | 1 | 6 | 2 | 5 | 2 | ||||
P is the probability for obtaining the observed number of R mutations in the CDRs by chance.
CDRs encoded by the VH or VL gene segments.
Intraclonal heterogeneity of V genes.All of the lymphoma V genes showed evidence of intraclonal heterogeneity although the five mutations identified in five VH clones from case OR were restricted to the leader region and intron and are not shown (Table 4, Fig 3). Some clones of a given V gene differed from the others by only one or two nucleotides, while other clones were identified that had three or four noncommon mutations, ie, mutations not present in all of the other related clones. In many cases, the noncommon mutations were shared among some of the clones of a given V gene, eg, see SMLC1 and SMLC3 (Table 4), or HALC2 and C3 (Fig 3). Similar to the consensus sequence mutations, which represent almost exclusively mutations that are shared among all of the clones of a given V gene, clustering of the noncommon R mutations to the CDR regions is also not evident (Table 4). Counting shared mutations only once, a total of 20 R and 27 S noncommon mutations were identified from the start of FWR1 in the 44 VH and VL clones evaluated with 5R and 5S mutations located in the CDRs and 15R and 22S mutations located in the FWRs.
Noncommon Mutations in VL and VH Clones
| Patient . | Clones4-150 . | Mutations4-151 . |
|---|---|---|
| HA | LC1 | G95A, A239G, G256A, C264t |
| LC2 | T188A, C282t | |
| LC3 | G130T, T188A, G256A, C282t | |
| LC4 | C96t G256A | |
| SM | LC1, 2 | G33t, C111t |
| LC3 | G33t, C111t, T257A, A272C | |
| LC4 | — | |
| OR | LC1 | A4G, C147g |
| LC2, 3 | A4G, C147a, T257G | |
| LC4 | C147g | |
| RO | LC1, | G183a, T218C |
| LC2 | A91G | |
| LC3 | A243g | |
| SH | LC1, 2, 3 | G270a |
| LC4 | T216c | |
| HA | HC1, 2, 3 | C204t |
| HC4 | G57a, C204t | |
| HC5, 6 | G3a C204t | |
| HC7 | G3a C204t C227G | |
| HC8, 9 | C204t C212T, G246a, G277A, G279c | |
| HC10 | — | |
| SM | HC1 | G39a C270t |
| HC2 | C255t | |
| HC3 | C252t | |
| HC4 | G113C, C177T, C247t | |
| HC5 | T161C | |
| HC6, 7 | — | |
| RO | HC1 | G254A |
| HC2 | C255t, C256t | |
| HC3, 4 | — | |
| SH | HC1 | C40T, T42g, C93t G279c |
| HC2 | A174T, C249A | |
| HC3 | C105t | |
| HC4 | — |
| Patient . | Clones4-150 . | Mutations4-151 . |
|---|---|---|
| HA | LC1 | G95A, A239G, G256A, C264t |
| LC2 | T188A, C282t | |
| LC3 | G130T, T188A, G256A, C282t | |
| LC4 | C96t G256A | |
| SM | LC1, 2 | G33t, C111t |
| LC3 | G33t, C111t, T257A, A272C | |
| LC4 | — | |
| OR | LC1 | A4G, C147g |
| LC2, 3 | A4G, C147a, T257G | |
| LC4 | C147g | |
| RO | LC1, | G183a, T218C |
| LC2 | A91G | |
| LC3 | A243g | |
| SH | LC1, 2, 3 | G270a |
| LC4 | T216c | |
| HA | HC1, 2, 3 | C204t |
| HC4 | G57a, C204t | |
| HC5, 6 | G3a C204t | |
| HC7 | G3a C204t C227G | |
| HC8, 9 | C204t C212T, G246a, G277A, G279c | |
| HC10 | — | |
| SM | HC1 | G39a C270t |
| HC2 | C255t | |
| HC3 | C252t | |
| HC4 | G113C, C177T, C247t | |
| HC5 | T161C | |
| HC6, 7 | — | |
| RO | HC1 | G254A |
| HC2 | C255t, C256t | |
| HC3, 4 | — | |
| SH | HC1 | C40T, T42g, C93t G279c |
| HC2 | A174T, C249A | |
| HC3 | C105t | |
| HC4 | — |
VL and VH clones are prefixed by L and H, respectively.
First letter and numbers give the mutated nucleotide and position from FW1. Underlined mutations are in CDRs. Lower and upper case second letters denote S and R mutations, respectively. Clones with only common mutations are also included and indicated by — .
Intraclonal heterogeneity of lymphoma V genes. The four VL and 10 VH clones sequenced from case HA are shown in (A) and (B), respectively, compared with the proposed germline genes. VH clones HC6, HC8, HC9, and HC10 were isolated from the lymph node specimen, while the others were derived from the parotid gland specimen. Replacement mutations are in upper case, while silent mutations are in lower case. Mutations not common to all of the VH or VL sequences are bolded and also listed in Table 4 along with similar mutations identified in the other lymphoma V genes. Individual clones referred to in the text are prefixed with the identifying patient letters, eg, HAHC10 refers to clone HC10 from patient HA.
Intraclonal heterogeneity of lymphoma V genes. The four VL and 10 VH clones sequenced from case HA are shown in (A) and (B), respectively, compared with the proposed germline genes. VH clones HC6, HC8, HC9, and HC10 were isolated from the lymph node specimen, while the others were derived from the parotid gland specimen. Replacement mutations are in upper case, while silent mutations are in lower case. Mutations not common to all of the VH or VL sequences are bolded and also listed in Table 4 along with similar mutations identified in the other lymphoma V genes. Individual clones referred to in the text are prefixed with the identifying patient letters, eg, HAHC10 refers to clone HC10 from patient HA.
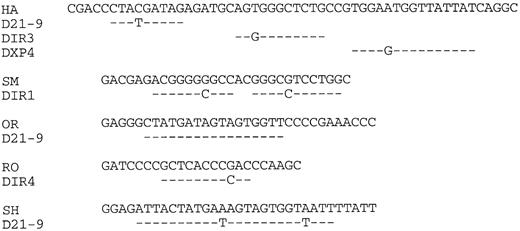
Lymphoma nucleotide sequences compared with germline D segments. The lymphoma VH CDR3 regions are shown up to but not including the JH segments indicated in Fig 2.
Lymphoma nucleotide sequences compared with germline D segments. The lymphoma VH CDR3 regions are shown up to but not including the JH segments indicated in Fig 2.
To help determine if some of the noncommon mutations could be due to Taq polymerase errors, VH genes were amplified in a similar manner from three different mantle cell lymphomas, which are B-cell neoplasms that typically do not show ongoing Ig gene hypermutation33 and a total of 20 randomly selected clones were sequenced split 5, 7, and 8 from the three cases. Only three differences were identified among related clones in the total of 9,270 nucleotides analyzed giving a Taq error rate of approximately .03% (not shown). This error rate is substantially less than the noncommon mutation rate in the MALT lymphoma VH or VL genes (Table 4).
VH CDR3 regions.Three of five lymphoma VH genes used JH3 (Fig 2). However, the small sample size does not allow the conclusion that this is significantly different from normal peripheral blood, which is dominated by JH4.34 Assignment of D segments to the VH CDR3s is shown in Fig 4. Because of uncertainty regarding the ability of DIR segments to undergo rearrangement,35 assignments of these elements to expressed genes should be considered tentative. It appears, nevertheless, that at least two of the five genes (OR, SH) are using D21-9. Lengths of the CDR3s averaged 15 amino acids, which is not significantly different from the normal value of 13.7 reported from an analysis of 325 unrelated VH genes.36
DISCUSSION
These studies demonstrate that the Ig VH and VL genes expressed by salivary gland MALT lymphomas harbor numerous point mutations relative to the most closely related germline genes. Indeed, for the five cases analyzed, the average VH and VL mutation frequencies were 4.6% and 4.2%, respectively. Because germline V genes were not isolated from these patients, it is possible that some of the nucleotide differences could represent individual polymorphisms as opposed to somatic mutations. However, sequence polymorphism among V genes is generally low and would, therefore, not be expected to account for more than a few of these changes.30 Moreover, in the specific case of the 51p1 derived lymphoma VH genes, polymorphism is particularly unlikely, as germline 51p1 genes have been extensively studied and none of the polymorphisms identified among 48 unrelated individuals match the sequence differences from 51p1 in these lymphoma genes.37 It is also possible that some of the lymphomas may be expressing previously uncharacterized germline genes, although again this is unlikely because most human VH and Vk genes have now been identified.30
In addition to highly mutated VH and VL genes, substantial intraclonal VH and/or VL gene diversity was also identified in each case indicating that the process of Ig gene hypermutation is still ongoing in lymphoma cells. Some of the nucleotide substitutions thought to occur in lymphoma cells because they were not present in all of the VH or VL gene clones from a given case, ie, the noncommon mutations, could theoretically represent Taq polymerase errors introduced during the amplification process. However, this explanation is unlikely to account for more than a few of the noncommon mutations because the ongoing mutation rate of approximately .6% in some of the V genes is an order of magnitude greater than our Taq error rate of .03%, which was calculated from analyses of mantle cell lymphoma VH gene clones. It is important to point out that our Taq error rate is close to other reported values38 and can also be derived by noting all seven VH clones analyzed from case OR were identical from before the start of FWR1. The VH gene from case OR is not anomalous with respect to ongoing mutation, however, as five different noncommon substitutions were identified in the leader region or intron in five of the clones. In addition to the high frequencies, many of the noncommon mutations were shared by different clones of a given V gene, which also indicates they are most likely of a lymphoma cell origin. Moreover, the vast majority of silent noncommon mutations that are not subjected to selection were C to T or G to A transitions, which strongly supports their derivation from the Ig mutator,39 as opposed to Taq polymerase, which most frequently generates T to C errors.40 Ongoing Ig gene hypermutation may be a general feature of MALT lymphomas that occur at sites besides the salivary gland, as intraclonal diversity of gastric MALT lymphoma VH genes has also been recently reported.21
Ongoing Ig gene hypermutation raises the possibility that MALT lymphomas may arise from germinal center B cells rather than marginal zone cells. In normal B-cell development, somatic hypermutation is thought to occur at the germinal center stage and may be further restricted to a subset of germinal center cells.16-18 Subsequent antigen selection of mutated germinal center B cells is the mechanism whereby high-affinity memory B cells and antibody producing plasma cells are generated.41,42 Somatic hypermutation of Ig genes in malignant B cells has, in past studies, also reflected the tumors' developmental stage in that pregerminal center cell neoplasms use unmutated germline V genes, while postgerminal center cell malignancies express V genes with numerous somatic mutations, but do not show evidence of ongoing mutation.33,43,44 Only follicular lymphomas, which are thought to be malignancies of germinal center cell B cells, have been shown to consistently have mutated V genes that also demonstrate intraclonal diversity indicative of an active Ig gene hypermutation mechanism.33,45 Further supporting a germinal center cell origin of these MALT lymphomas is the magnititude of intraclonal variation in some of the V genes, which is similar to what is seen in follicular lymphomas.22 46
Interestingly, MALT lymphomas were initially proposed to arise from germinal center B cells based on the tumors' histologic appearance.47,48 Germinal centers or secondary follicles are characteristic features of MALT lymphomas and a proportion of the follicles usually appear abnormal and/or neoplastic.49,50 In addition, the neoplastic cells that accumulate in MALT lymphomas morphologically resemble centrocytes or cleaved follicular center cells.1,2 Phenotypic analyses of the neoplastic cells, however, showed differences from germinal center cells and a closer resemblance to marginal zone B cells.13,14 Moreover, MALT lymphomas do not have translocations between chromosomes 14 and 18, which are characteristic of follicular small cleaved cell lymphomas.13,51 In keeping with a marginal zone cell origin, the abnormal or neoplastic follicles seen in MALT lymphomas have been explained by proposing the malignant centrocyte-like cells migrate into follicles that are initially reactive.48,50 This type of lymphoma cell migration termed “follicular colonization,” however, has not been directly observed and it is, therefore, possible that the centrocyte-like cells or their precursors arise in the follicles, but sometimes fail to migrate out.49 In keeping with a follicular center cell origin, the phenotypic differences identified between MALT lymphomas and germinal center B cells could be explained by further differentiation of the neoplastic precursor cells,14 or by transformation of a small subpopulation that differs phenotypically from the predominant type of germinal center cell.52 Differentiation of malignant germinal center cells could also explain the frequent presence of numerous monoclonal plasma cells seen in many MALT lymphomas along with the centrocyte-like cells because normal germinal center cells give rise to both plasma cells and memory B cells.42 53
An alternative interpretation is that salivary gland MALT lymphomas do, indeed, originate in postgerminal center marginal zone B cells and the ongoing Ig mutation reflects reentry into a germinal center environment. This is keeping with the view that Ig gene hypermutation, even in MALT lymphoma cells, requires the specialized germinal center environment and will not occur in marginal zones. As noted above, neoplastic cells are frequently found in MALT lymphoma-associated germinal centers, which may reflect follicular colonization.50 Reentry of normal B cells into germinal centers has been described for rat splenic marginal zone cells following antigenic stimulation42,54 suggesting that follicular colonization may have a normal counterpart. In addition, theoretical analyses of affinity maturation suggest that germinal center reentry may be followed by further rounds of Ig gene hypermutation.55 However, the basic issue of whether postgerminal center memory B cells can accumulate additional Ig gene mutations is highly controversial and the mechanism whereby this may occur is not understood.41,42 56
Replacement mutations that affect residues in the hypervariable or CDR loops can be positively or negatively selected by antigen, as these regions form the conventional antigen binding site. Mutations that have been positively selected lead to Ig molecules with higher affinity antigen binding properties and the B cell V genes often show clustering of R mutations in the CDRs.22,31 Mutations in the MALT lymphoma V genes studied here were not distributed in a manner that was especially suggestive of positive selection or affinity maturation. This was true for both the common mutations that were present in all clones of a given V gene as well as the noncommon mutations. However, positive selection can occur in the absence of unexpected R mutation clustering in the CDRs because single nucleotide substitutions in these areas can dramatically affect antibody affinity without necessarily altering an otherwise random distribution of mutations.57 58 It is, therefore, possible that positive selection could be responsible for some of the lymphoma V gene mutations, although the numbers of R mutations in the CDRs were close to the values expected from chance alone.
Although the mutation distributions in the consensus sequences were not suggestive of positive selection, the two lymphoma VH genes with the greatest number of mutations, and therefore potentially the most informative, showed evidence of negative selection. Without selective pressure, the expected ratios of R and S mutations within the CDRs of the SM and RO VH genes should be 3.6:1 and 4.3:1, respectively.27 Therefore, the observed R:S values of 2:5 and 7:6 for the SM and RO CDRs, respectively, which are substantially lower than the expected values, suggests there was selection against R mutations in the CDRs. Negative selection was also suggested when considering the noncommon mutations either in individual V genes or altogether, as the CDR R:S ratios were all less than or equal to 1, while the expected values should all be greater than 3.6.27 Negative selection may indicate that residues in the CDRs need to be conserved to bind antigens that are important for lymphoma cell survival.59 Negative selection of R mutations in FWRs is also thought to be required to maintain Ig function.32 Therefore, finding fewer R mutations than expected by chance in most of the FWRs, further suggests that Ig expression is important for lymphoma cell survival and is consistent with the notion of antigen related selection against R mutations in the CDRs.
Although only five cases were examined, our findings strongly suggest that the use of Ig V genes by salivary gland MALT lymphomas is highly biased or nonrandom. Recent estimates indicate that there are approximately 60 VH and 50 Vk gene segments in the average human genome that can potentially rearrange to form an expressed V gene.30,60 Therefore, the probability of selecting by chance alone three of five lymphomas that coexpress the 51p1 VH and Humkv325 VL gene segments is extremely low. In a recent study of normal peripheral blood B cells using single cell PCR, only one of 71 productive VH rearrangements was found to include a 51p1-like gene, which is close to the value of 1.7% (1/60) expected from random use.61 However, nonrandom 51p1 and Humkv325 expression has been described in other B-cell populations.62,63 For example, in individuals with the maximal number of four 51p1 related genes, an average of 9.5% of tonsillar IgD+ B cells were found to express the G6 idiotype, a marker for 51p1 expression, while in individuals with the more typical two copies of 51p1 related genes, 6.8% of the IgD+ B cells on average were G6 positive.62 Similiary, an average of 3.9% tonsillar B cells were found to express the 17.109 idiotype, a marker for Humkv325 gene expression, which is also higher than the value of 2% (1/50) expected for random use.63 However, less than 1% of tonsillar B cells were found to be positive for both G6 and 17.109 in this study, which suggests that coexpression of 51p1 with Humkv325 is still highly unusual. In addition, because the G6 and 17.109 idiotypes are predominately expressed on IgD+ B cells,62 63 the use of 51p1 and humkv325 by IgD− B cells, the subset that appears to give rise to MALT lymphomas, may be much lower. With these considerations taken into account, the observed biased use of VH and VL gene segments is, therefore, strongly suggestive of Ig-mediated selection of B cells and further supports antigen stimulation playing an important role in salivary gland lymphamagenesis.
Preferential use of the 51p1 and Humkv325 genes has also been reported for chronic lymphocytic leukemia (CLL), a low-grade malignancy of CD5 positive B cells. Recent estimates indicate that approximately 20% of CLLs have rearranged a 51p1 gene, and that approximately 20% of kappa positive CLLs express Humkv325.24,64 Unlike the salivary gland MALT lymphomas, however, the Ig V genes expressed by CLL are typically not mutated from germline.24 Despite this difference and the fact that CLL is a histologically, phenotypically, and clinically distinct B-cell neoplasm from MALT lymphoma, the use of similar VH and VL gene segments by CLL and salivary gland lymphoma suggests these entities may still may be related in terms of pathogenesis or antigen specificity.
In addition to this report, the notion that an antigen may be selecting B cells that express 51p1 and Humkv325 for malignant transformation in the salivary gland is also supported by studies of lymphoproliferations in patients with Sjogren's syndrome. It has been estimated that patients with Sjogren's syndrome have a 44-fold increased risk of developing lymphoma.65 Although the transition from reactive MESA to monoclonal lymphoma is not well understood, the number of B cells that express the Humkv325-related idiotype 17.109 and 51p1-related idiotype G6 appear to be increased in salivary gland tissue from patients with Sjogren's syndrome relative to controls.66-68 In addition, it has been reported that paraproteins in patients with Sjogren's syndrome frequently react with G6 and, therefore, are most likely encoded by 51p1.69 Although the antigen specificy of the salivary gland MALT lymphomas is unknown, paraproteins encoded by 51p1 and kv325 frequently display reactivity towards autoantigens, especially IgG.70 However, the autoreactive properties of 51p1 and kv325 encoded Ig expressed by CLL appear to critically depend on sequences in the CDR3 regions,71 which may be different in the salivary gland MALT lymphomas.
These studies extend analyses of lymphoma V genes to MALT lymphomas that arise at anatomical sites outside of the stomach. Because antigens that may be involved in the development of MALT lymphomas outside of the stomach will be different from Helicobacter pylori, certain aspects of MALT lymphoma biology may be site-specific. Although only a limited number of cases have been analyzed, the available evidence suggests that, unlike salivary gland lymphomas, gastric MALT lymphomas do not use a restricted set of VH gene segments.19-21 Moreover, only one of the 11 VH gastric MALT lymphoma VH genes characterized to date is derived from the 51p1 germline gene.19 Studies are in progress to determine whether salivary gland lymphomas do indeed bind a more limited set of antigens than gastric MALT lymphomas as the V gene restriction suggests and whether any of these antigens may be related to the antigenic trigger of MESA.
ACKNOWLEDGMENT
The authors thank Dr Joseph Locker for providing DNA, helpful discussions, and review of the manuscript.
Supported by the Pathology Education and Research Foundation (University of Pittsburgh) and Grant No. IRG-58-34 from the American Cancer Society.
Address reprint requests to David W. Bahler, MD, PhD, Department of Pathology NW 628, Montefiore University Hospital, 200 Lothrop St, Pittsburgh, PA 15213.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal