Abstract
The retinoblastoma tumor-suppressor gene, RB, has been implicated in tumor suppression, in regulation of the cell cycle, and in mediating cell differentiation. RB is necessary for hematopoiesis in mice, and aberrant RB-expression is associated with the progress and prognosis of leukemia. We have used antisense oligonucleotides, established clones stably expressing an antisense RB construct, and also established clones over expressing the retinoblastoma protein (pRb) to study the role of RB expression in monocytic differentiation induced by all-trans retinoic acid (ATRA) or 1-α-25-dihyroxycholecalciferol (Vit D3) in the monoblastic cell line U-937 and erythroid differentiation induced by transforming growth factor β1 (TGFβ1) and hemin in the erythroleukemic cell line K562. A reduction in pRb production in antisense RB-transfected U-937 clones was shown. Antisense oligonucleotides as well as expression of the antisense RB construct suppressed differentiation responses to ATRA or Vit D3, as judged by the capability to reduce nitro blue tetrazolium, by the appearance of monocyte-related cell surface antigens and by morphologic criteria. K562 cells showed decreased differentiation response to TGFβ1, but not to hemin, when incubated with antisense oligonucleotides. U-937 antisense RB-transfected cells were also suppressed in their ability to upregulate levels of hypophosphorylated pRb when induced to differentiate. Although U-937 cells incubated with antisense oligonucleotides and clones expressing the antisense RB construct were hampered in their ability to differentiate on incubation with ATRA or Vit D3, the induced G0/G1-accumulation was similar to differentiating control cells treated with ATRA or Vit D3. Intriguingly, U-937 clones overexpressing RB were also inhibited in their differentiation response to ATRA or Vit D3 but not inhibited in their ability to respond with G0/G1 accumulation when induced with these substances. The results indicate that pRb plays a role in induced differentiation of U-937 cells as well as K562 cells involving mechanisms that, at least partially, are distinct from those inducing G1 accumulation.
THE RETINOBLASTOMA gene, RB, a prototype tumor-suppressor gene, has been identified as the recessive gene whose loss of function confers susceptibility to development of the rare childhood eye tumor retinoblastoma as well as other neoplasias including sarcomas, small-cell carcinoma of the lung, and breast cancer.1-5 The tumor-suppressor properties of this gene have been further sustained by the observation that introduction of a single copy of the gene into different tumor cells lacking it leads to complete or partial suppression of their tumorigenic potential.6-8 The product of this in vertebrates ubiquitous gene, the retinoblastoma protein, pRb, is a nuclear 110-kD phosphoprotein9 whose function is closely related to cell-cycle control.10 The activity of pRb depends on its degree of phosphorylation, showing cell-cycle–dependent changes due to the activity of cyclin-dependent kinases and phosphatases.11-14 The hypophosphorylated form of pRb is the active one in restraining growth. It probably maintains cells in quiescence by its ability to bind to transcription factors of the E2F family, whose target genes encode proteins (c-myc, dihydrofolate reductase, thymidine kinase, and DNA polymerase α) necessary for progression of the cell cycle from G1- to S-phase.15-18 Loss of pRb function leads to liberation of E2F and subsequent loss of restriction at the G1-S transition, increasing the probability of uncontrolled cell division.19 The significance of this cell-cycle restriction mechanism is underlined by the observation that viral oncoproteins from certain tumor-inducing DNA viruses sequester and inactivate hypophosphorylated pRb of the infected cell, giving the virus access to a DNA-synthesising machinery without restrain to exploit.20,21 However, pRb is not only involved in cell proliferation shown as cell cycle regulation and suppression of tumor formation. Increasing amounts of evidence indicate that pRb is involved in the process of cell differentiation. Functional inactivation of both RB alleles in transgenic mice is lethal and the mutant embryos die after 14 to 15 days of gestation, showing histologic evidence of defective hematopoietic and neuronal differentiation22-25 and abnormal lens development.26 Furthermore, it has been shown that pRb is crucial for the establishment and maintenance of the terminally differentiated phenotype of muscle cells.27,28 Moreover, differentiation of normal human hematopoietic cells and leukemic cells has in vitro been correlated with activation of pRb by hypophosphorylation.11,29 Also, recent findings point out a role for the retinoblastoma gene in normal adult hematopoiesis; high levels of RB mRNA are induced and sustained during erythroid differentiation.30
In normal hematopoiesis, a regulated recruitment of stem cells leads to controlled clonal expansion along specific lineages. This results in mature functional populations of different blood cells, most of which have a relatively short life span. The proliferative capacity of these cells decreases as the degree of differentiation increases. This mass production of cells is regulated by factors produced in response to the demand of the organism. The ability of a cell to respond adequately to various proliferation and differentiation signals depends on its expression of specific genes, such as proto-oncogenes and tumor-suppressor genes. However, expression of these genes may be aberrant. For example, fusion of gene segments by chromosomal translocation may lead to inappropriate activation of an unaltered proto-oncogene or, sometimes, expression of a mutant mitogenic fusion protein. Such translocations are frequently observed in hematopoietic malignancies.31 Furthermore, tumor-suppressor genes may be inactivated by mechanisms such as deletions and mutations. Structural abnormalities of the RB gene and absent protein expression have been observed in the evolution of human acute leukemia, particularly in those of monocytic phenotype and in lymphoid leukemia, and in blast crisis of chronic myelogenous leukemia.32-36
The accumulation of immature nonfunctional malignant cells is a characteristic feature of acute leukemia. This accumulation is believed to be due to the inability of these cells to proceed towards terminal differentiation and to impaired programmed cell death, rendering them far longer life than normal blood cells. However, the inability of the transformed leukemic cell to fulfil its original commitment to differentiation can sometimes be abrogated with various agents. In vitro, all-trans retinoic acid (ATRA) or 1-α-25-dihyroxycholecalciferol (Vit D3), for example, induce myeloid differentiation in several leukemic cell lines, and the human monocytic leukemic cell line U-937, used in the present study, is one of them.37-39 Furthermore, substances such as hemin and transforming growth factor β1 (TGFβ1) have been shown to induce signs of erythroid differentiation in K562 cells.40,41 In vivo, administration of ATRA to patients with acute promyelocytic leukemia results in differentiation of the leukemic cells and even complete remission.42 Unfortunately, differentiation therapy in other forms of leukemia has not yet been very successful.
To reach a deeper understanding of the mechanisms of induced hematopoietic differentiation, we decided to test the hypothesis that tumor-suppressor genes are necessary for induction of maturation in leukemic cells. In this study, we show that suppression of pRb production in the leukemic cell line U-937, by incubation with antisense oligonucleotides or transfection with an antisense construct, reduces the ability of these cells to differentiate on incubation with ATRA or Vit D3. Antisense RB-transfected U-937 cells failed to produce detectable levels of hypophosphorylated pRb on induction of differentiation. However, the suppressed production of pRb did not uncouple the restraining effect on G1-S transition induced by ATRA or Vit D3 in U-937 cells. Furthermore, another leukemic cell line, K562, was found to be suppressed in its capacity for TGFβ1-induced erythroid differentiation when incubated with antisense Rb oligonucleotides. Intriguingly, forced overexpression of the human RB gene in U-937 also resulted in a differentiation block to ATRA as well as Vit D3, with a corresponding G0/G1 accumulation. The results indicate that the retinoblastoma protein is involved in induced differentiation of U-937 and K562 cells by ATRA or Vit D3 and TGFβ1, respectively, and that G1 arrest could be necessary, but not sufficient, for pRb-mediated induction of differentiation of leukemic cells.
MATERIALS AND METHODS
Incubation of cells.The leukemic cell lines U-93737 and K56240 were cultured in RPMI 1640 medium (GIBCO Ltd, Paisley, UK) supplemented with 10% heat inactivated fetal calf serum (GIBCO Ltd). Cells were grown in 5% CO2 at 37°C in a fully humidified atmosphere. Exponentially growing cells were used for all experiments. For determination of growth rate, cells were diluted at an initial concentration of 0.2 × 106 cells/mL in complete medium and aliquots were removed daily for determination of cell concentration and viability by trypan blue exclusion. In experiments with induction of differentiation, cells at an initial concentration of 0.2 × 106 cells/mL were incubated in complete medium with ATRA (Sigma Chemical Co, St Louis, MO), Vit D3 (a generous gift from Roche, Basel, Switzerland), or TGFβ1 (Genzyme, Cambridge, MA) for 4 days at the indicated concentrations, after which differentiation in U-937 cells was assessed by nitro blue tetrazolium (NBT) reduction test, flow cytometric analysis of cell surface antigens, and morphologic analysis; differentiation in K562 cells was assessed by benzidine staining.
Antisense RB oligonucleotides.The sequence of the 21-base–long phosphorothioate-modified antisense oligodeoxynucleotide (synthesised by Bio Molecular Resource Facility, Lund University, Lund, Sweden) read 5′ GGG GGT TTT GGG CGG CAT GAC 3′. The corresponding phosphorothioate-modified sense oligonucleotide was used as negative control. Wild-type U-937 or K562 cells were preincubated in complete medium with oligonucleotides at a concentration of 50 μmol/L for 24 hours before induction of differentiation by the addition of ATRA at 1 μmol/L or Vit D3 at 0.1 μmol/L (U-937) or TGFβ1 at 25 ng/mL (K562). After 4 days of incubation with inducer and in the continuous presence of oligonucleotide, the cells were counted and the extent of differentiation was determined.
Antisense RB cDNA vector construct.The antisense cDNA fragment for the human retinoblastoma gene was created by polymerase chain reaction (PCR) amplification (Perkin Elmer Cetus DNA Thermal Cycler 480; Perkin Elmer Cetus, Norwalk, CT) of the cDNA for the human retinoblastoma protein (generously provided by Dr S. Friend, Charlsetown, MA) using the upstream primer 5′ GAC TTC AGA AGC TTG GCG TCA TGC CGC CCA AAA CCC CC 3′ and the downstream primer 5′ CTG AAG TCG GTA CCA CAT AAT GCA GTA AAA TCA GGT 3′ (Scandinavian Gene Synthesis AB, Falkenberg, Sweden). The amplified product, approximately 200 bp of length, spanning from −9 to +194, including the restriction enzyme sites HindIII and Kpn I (in bold above), was cloned into the mammalian expression vector pCEP4 (In Vitrogen; British Biotechnology, Oxon, UK) in an antisense orientation, creating the expression vector pCEP4/antiRb. The pCEP4 vector is driven by a viral promoter, cytomegalovirus (CMV), providing high-level and constitutive expression. The plasmid pCEP4 alone was used to obtain control clones (mock-transfected clones).
RB cDNA vector construct.The complete human cDNA (a generous gift from Dr S. Friend) was cloned into the mammalian expression vector pCDNA3 (In Vitrogen), both previously digested with Kpn I and HindIII, creating the expression vector pCDNA3/HRbC. The plasmid pCDNA3 alone was used to obtain control clones (mock-transfected clones).
Transfection procedure.Plasmid DNA was introduced into cells by electroporation essentially as described.43 Briefly, 20 μg of plasmid was added to U-937 cells (8 × 106) and electroporation was performed with electrical settings at 1.6 kV and 25 μF (pCEP4) or 0.32 kV and 960 μF (pCDNA3), using the Bio-Rad Electroporation Apparatus (Bio-Rad, Melville, NY). After 48 hours of incubation in complete medium, 5,000 cells per well were seeded in 96-well plates with hygromycin B (1,200 U/mL; Sigma) or geniticin (500 μg/mL; Boehringer Mannheim, Mannheim, Germany) for selection of recombinant clones expressing the hygromycin B resistance gene carried by pCEP4 or the neomycin resistance gene carried by pCDNA3. After 4 weeks, individual clones growing in the presence of hygromycin B or geniticin were expanded to mass cultures.
Biosynthetic labeling and fluorography.Biosynthetic labeling of newly synthesised proteins was obtained by incubation of cells with 35S-methionine/35S-cysteine at 25 μCi/mL (Promix; Amersham, Amersham, UK) as previously described.43 Briefly, cells were incubated in methionine- and cysteine-free RPMI medium supplemented with 10% dialyzed fetal calf serum (GIBCO) for 30 minutes to deplete the intracellular pools of methionine and cysteine. Subsequently, cells were incubated for 2 hours at 37°C in identical medium supplemented with 35S-methionine/35S-cysteine to obtain radioactive labeling of newly synthesised proteins. Labeled pRb was immunoprecipitated and subjected to sodium dodecyl sulfate-polyacrylamide gradient gel electrophoresis (SDS-PAGE) and subsequent fluorography essentially as described.43 Briefly, a cell lysate was prepared by suspension of cells in lysis buffer (50 mmol/L Tris-HCl, pH 8.0, 0.15 mol/L NaCl, 5 mmol/L EDTA, and 0.5% NP-40), followed by removal of DNA by centrifugation at 37,500g for 1 hour. The radioactivity of each supernatant was measured by use of a beta counter, and equal amounts of radioactivity were then subjected to specific immunoprecipitation using 10 μg of a monoclonal anti-pRb antibody. For anti-Rb/pCEP4 transfectants Ab-1, clone C36E, E. Harlow (Cold Spring Harbor, Oncogene Science Inc, Uniondale, NY) was used. For HRbC/pCDNA3 transfectants, the monoclonal anti-pRb antibody sc-102 (Santa Cruz, Santa Cruz, CA) was used. The immunoprecipitates were analysed on a 7% to 20% SDS-PAGE, after which fluorography was performed by exposure of Hyperfilm MP (Amersham) at −70°C for 1 to 3 days to the dried gel, which had been previously treated with Amplify (Amersham). An anti-p53 antibody (Ab-3, PAb 240; Oncogene Science Inc) was used as a negative control.
Western blot analysis.Cell lysates were prepared by suspension of 10 × 106 cells in lysis buffer (50 mmol/L Tris-HCl, pH 8.0, 0.15 mol/L NaCl, 5 mmol/L EDTA, and 0.5% NP-40), followed by removal of DNA by centrifugation at 37,500g for 1 hour and then subjected to specific immunoprecipitation using 10 μg of a monoclonal anti-pRb antibody (sc-102; Santa Cruz). The immunoprecipitates were loaded onto a 7% SDS-PAGE. After electroblotting, a monoclonal anti-pRb antibody (sc-102) was used to probe the blotted membrane for pRb. Bands were visualized by using a horseradish peroxidase-coupled goat antimouse antibody (Bio-Rad, Richmond, CA). A monoclonal anti-p53 antibody (Ab-3, PAb 240; Oncogene Science Inc) was used as a negative control.
Assessment of differentiation by the NBT reduction test.Cells destined for the NBT reduction test were resuspended at a concentration of 1 to 2 × 106 cells/mL in complete medium containing 0.075% (wt/vol) NBT (Sigma) and 0.15 μg/mL phorbol-12-myristate-13-acetate (PMA; Sigma) for 25 minutes at 37°C. Cytospin slides were prepared and stained with May-Grünwald-Giemsa. The percentage of NBT-reducing cells, containing black formazan deposits, was determined by counting 200 cells. Viability before the NBT reduction test, as judged by trypan blue exclusion, was always greater than 90%.
Assessment of differentiation by flow cytometric analysis of cell surface antigens.Cells were washed in phosphate-buffered saline (PBS) and resuspended to 5 to 10 × 107 cells/mL. Fifty microliters of the cell suspension was incubated with 5 μL of the following monoclonal antibodies in microtiter wells: control IgG1-fluorescein isothiocyanate (FITC)/IgG1-phycoerythrin (PE), CD14-PE, CD11c-PE, and CD11b-PE (DAKO A/S, Glostrup, Denmark) for 10 minutes at room temperature under constant agitation. The cells were washed three times and fixed in 1% paraformaldehyde before flow cytometric analysis (FACScan; Becton Dickinson, San Jose, CA). Ten thousand cells were collected for each antibody. Dead cells and debris were excluded from analysis by gating before the calculation of the percentage of positive cells.
Assessment of differentiation by staining for hemoglobin.K562 wild-type cells were incubated for 4 days with substances known to induce erythroid differentiation, after which hemoglobin was stained by the benzidine method. After being washed two times with PBS, 100 μL of 0.9% NaCl was added to suspend 0.5 × 106 cells. To 1 mL of 0.2% tetramethyl-benzidine (T-2885; Sigma) in 0.5 mol/L HAc, 20 μL of 30% H2O2 was then added. Of this benzidine reagent solution, 50 μL was added to the homogenized cell suspension. The cell suspension was then left at room temperature for 30 minutes, protected from light, and occasionally agitated. Cells were subsequently diluted in 200 μL 0.9% NaCl and 200 cells were counted to determine the percentage of benzidine-positive cells, visualized as cells containing blue crystals.
Assessment of differentiation by morphologic analysis.Morphology was investigated by light microscopy. See the legend to Fig 3.
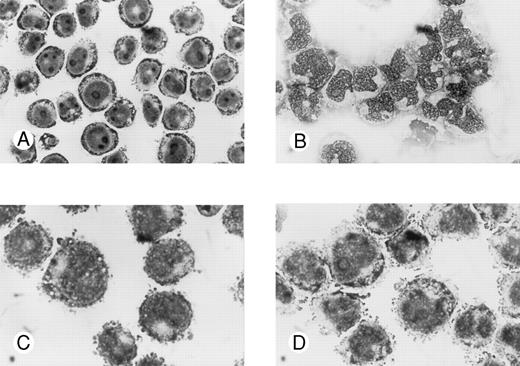
Effects of oligonucleotides on morphologic changes on induced differentiation of wild-type U-937 cells. Cells at an initial concentration of 0.2 × 106 cells/mL were preincubated for 24 hours with oligonucleotides at a concentration of 50 μmol/L. Cells were then induced with ATRA at 1 μmol/L. After 4 days of induction in the continuous presence of antisense Rb oligonucleotides or oligo buffer, cytospin slides were prepared and stained with May-Grünwald-Giemsa. Simultaneously, noninduced cells were incubated with antisense Rb oligonucleotides or oligo buffer and used as controls. Morphology was investigated by light microscopy. Cells incubated with oligo buffer only (A); with ATRA and oligo buffer (B); with antisense Rb oligonucleotides only (C); or with ATRA and antisense Rb oligonucleotides (D).
Effects of oligonucleotides on morphologic changes on induced differentiation of wild-type U-937 cells. Cells at an initial concentration of 0.2 × 106 cells/mL were preincubated for 24 hours with oligonucleotides at a concentration of 50 μmol/L. Cells were then induced with ATRA at 1 μmol/L. After 4 days of induction in the continuous presence of antisense Rb oligonucleotides or oligo buffer, cytospin slides were prepared and stained with May-Grünwald-Giemsa. Simultaneously, noninduced cells were incubated with antisense Rb oligonucleotides or oligo buffer and used as controls. Morphology was investigated by light microscopy. Cells incubated with oligo buffer only (A); with ATRA and oligo buffer (B); with antisense Rb oligonucleotides only (C); or with ATRA and antisense Rb oligonucleotides (D).
Determination of cell cycle distribution by flow cytometric DNA analysis.Staining of nuclei and flow cytometric analysis were performed essentially as previously described.44 45 Briefly, to achieve staining of nuclei, cells were washed in PBS and a nuclear isolation medium containing propidium iodide was added. Samples were incubated in darkness for 5 minutes at room temperature. The samples were then kept at 4°C for at least 15 minutes before flow cytometric analysis was performed in an Ortho-Cytoron Absolute (Ortho Diagnostic Systems, Raritan, NJ) equipped with a 15-mW argon ion laser. The laser line at 488 nm was used for excitation of propidium iodide and the fluorescence beyond 620 nm was detected. Up to 20,000 nuclei per sample were analyzed. Processor signals were digitized and sorted into frequency distributions (DNA histograms) with a resolution of 256 units. Cell cycle phase distribution, ie, the percentages of G0+G1, S, and G2+M nuclei of the analyzed cell population, was determined by applying Multi Cycle (Phoenix Flow Systems, Tucson, AZ) on the DNA histograms. The DNA histograms were corrected for contribution of nucleic debris.
RESULTS
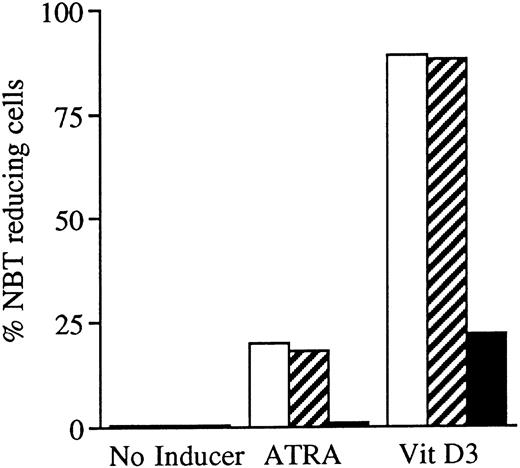
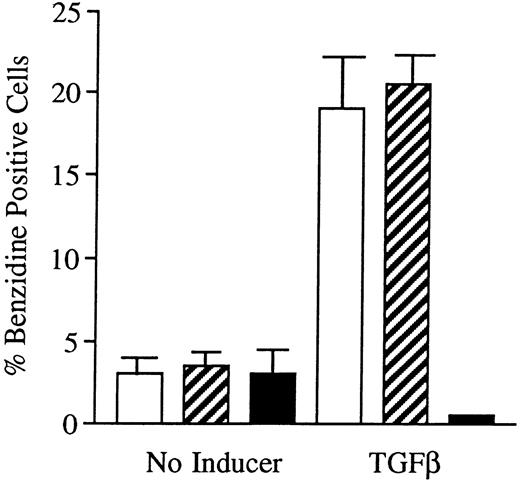
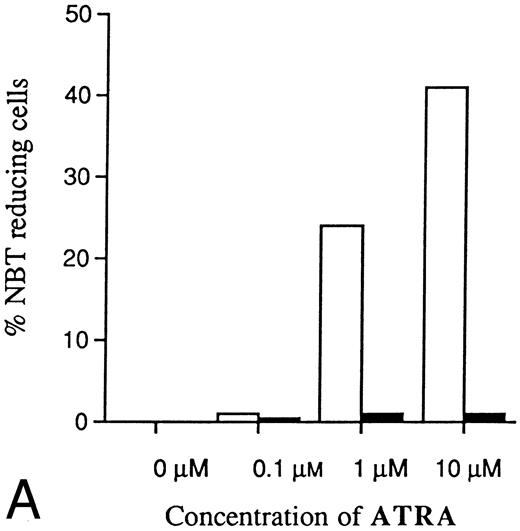
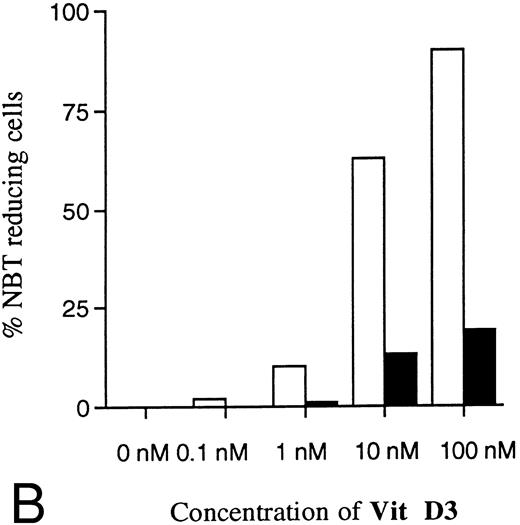
Incubation of wild-type U-937 or K562 cells with antisense RB oligonucleotides resulted in reduced capability to differentiate on induction with ATRA, Vit D3, or TGFβ1, respectively.Incubation of wild-type U-937 or K562 cells with antisense RB oligonucleotides at 50 μmol/L, as described in the Materials and Methods, reduced the capacity of induced differentiation as judged by the NBT reduction test or benzidine staining test, respectively. U-937 wild-type cells incubated with antisense oligonucleotides almost totally failed to differentiate upon exposure to ATRA, as did K562 cells incubated with antisense RB oligos on exposure to TGFβ1 (Figs 1 and 2). In U-937 cells, this reduced sensitivity to induction of differentiation was observed both with ATRA at 1 μmol/L and Vit D3 at 0.1 μmol/L (Fig 1). In K562 cells, it was observed with TGFβ1 (Fig 2) but not with hemin (data not shown), both previously described inducers of erythroid differentiation in these cells.40 41 The effect of the antisense RB oligonucleotide was specific, in as much as no response was observed in control cells incubated with sense RB oligonucletides at 50 μmol/L. The effect of the antisense RB oligonucleotide showed dose dependence with effects detected from 10 μmol/L (data not shown). The oligonucleotides did not affect cell viability, which was always greater than 90% (data not shown). The oligonucleotides also did not affect proliferation rates (data not shown).
Effects of oligonucleotides on ATRA or Vit D3-induced monocytic differentiation in wild-type U-937 cells assayed by the cells' capacity to reduce NBT. Cells at an initial concentration of 0.2 × 106 cells/mL were preincubated for 24 hours with phosphorothioate-modified oligonucleotides at a concentration of 50 μmol/L. Cells were then induced with ATRA or Vit D3 at 1 μmol/L and 0.1 μmol/L, respectively. After 4 days of induction in the continuous presence of (▪) antisense RB oligonucleotides, (▨) sense RB oligonucleotides, or (□) oligo buffer, respectively, cells were subjected to NBT reduction test. As controls, noninduced cells were incubated with antisense RB oligonucleotides, sense RB oligonucleotides, or oligo buffer, respectively. The percentage of NBT reducing cells was determined by counting 200 cells. Results are shown from one representative experiment.
Effects of oligonucleotides on ATRA or Vit D3-induced monocytic differentiation in wild-type U-937 cells assayed by the cells' capacity to reduce NBT. Cells at an initial concentration of 0.2 × 106 cells/mL were preincubated for 24 hours with phosphorothioate-modified oligonucleotides at a concentration of 50 μmol/L. Cells were then induced with ATRA or Vit D3 at 1 μmol/L and 0.1 μmol/L, respectively. After 4 days of induction in the continuous presence of (▪) antisense RB oligonucleotides, (▨) sense RB oligonucleotides, or (□) oligo buffer, respectively, cells were subjected to NBT reduction test. As controls, noninduced cells were incubated with antisense RB oligonucleotides, sense RB oligonucleotides, or oligo buffer, respectively. The percentage of NBT reducing cells was determined by counting 200 cells. Results are shown from one representative experiment.
Effects of oligonucleotides on TGFβ1-induced erythroid differentiation in wild-type K562 cells assayed by hemoglobin staining. Cells at an initial concentration of 0.2 × 106 cells/mL were preincubated for 24 hours with oligonucleotides at a concentration of 50 μmol/L. Cells were then induced with TGFβ1 (25 ng/mL). After 4 days of induction in the continuous presence of (▪) antisense Rb oligonucleotides, (▨) sense Rb oligonucleotides, or (□) buffer, respectively, cells were subjected to hemoglobin staining by the benzidine method, as described in the Materials and Methods. Simultaneously, noninduced cells were incubated with antisense Rb oligonucleotides, sense Rb oligonucleotides, or buffer, respectively, as controls. Two hundred cells from each incubation were counted and the percentage of benzidine-positive cells was determined. Results shown represent the mean values from three separate experiments, bars ± SEM.
Effects of oligonucleotides on TGFβ1-induced erythroid differentiation in wild-type K562 cells assayed by hemoglobin staining. Cells at an initial concentration of 0.2 × 106 cells/mL were preincubated for 24 hours with oligonucleotides at a concentration of 50 μmol/L. Cells were then induced with TGFβ1 (25 ng/mL). After 4 days of induction in the continuous presence of (▪) antisense Rb oligonucleotides, (▨) sense Rb oligonucleotides, or (□) buffer, respectively, cells were subjected to hemoglobin staining by the benzidine method, as described in the Materials and Methods. Simultaneously, noninduced cells were incubated with antisense Rb oligonucleotides, sense Rb oligonucleotides, or buffer, respectively, as controls. Two hundred cells from each incubation were counted and the percentage of benzidine-positive cells was determined. Results shown represent the mean values from three separate experiments, bars ± SEM.
Morphologic signs of differentiation of wild-type U-937 cells after induction with ATRA were inhibited by antisense RB oligonucleotides but not by sense RB oligonucleotides.Untreated wild-type U-937 cells appeared to be immature, with a big round nucleus with nucleoli and a low cytoplasm/nucleus ratio (Fig 3A). After 4 days of induction with ATRA, cells appeared more mature with more lobulated nuclei, fewer nucleoli, and a higher cytoplasma/nucleus ratio (Fig 3B). Antisense or sense RB oligonucleotides alone did not affect the morphology of the cells (Fig 3C and data not shown). However, antisense RB oligonucleotides were able to suppress the ATRA-induced morphologic signs of differentiation (Fig 3D). The capacity to suppress the differentiation response was specific, in as much as the corresponding sense oligonucleotides did not interfere with ATRA-induced differentiation (data not shown).
Establishment and characterization of U-937 clones stably expressing antisense RB.To further investigate and sustain the finding of a reduced capability to differentiate in response to ATRA or Vit D3 in U-937 cells incubated with antisense RB oligonucleotides, we decided to establish antisense RB expressing clones to reduce the endogenous production of pRb. This would allow us to more conveniently study the capability of retinoblastoma protein production and to correlate this to the capacity for induced differentiation and to cell cycle responses. Transfection of U-937 cells with the pCEP4/antiRB construct, as described in the Materials and Methods, resulted in several clones, from which eight individual clones were randomly picked, designated A1 through A8. Six clones from transfection with pCEP4 alone (mock-transfected controls) were isolated and designated M1 through M6. To determine the capability of the transfected clones to produce the retinoblastoma protein, biosynthetic labeling, immunoprecipitation, SDS-PAGE, and fluorography were performed, as described in the Materials and Methods. Wild-type U-937 cells are known to produce pRb.32 Not very surprisingly, therefore, mock-transfected clones (M1, M2, M3, M4, M5, and M6) showed distinct bands of biosynthetically labeled immunoprecipitated pRb upon fluorography (Fig 4). On the other hand, six of eight clones transfected with the antisense RB construct (A1, A2, A5, A6, A7, and A8) showed, when compared with the mock-transfected clones, corresponding bands of weaker intensity. The remaining two antisense clones (A3 and A4) showed bands of moderate intensity (Fig 4). Thus, the expression of the antisense PCEP/anti-RB construct suppressed the synthesis of pRb in six of eight clones, whereas the inhibition was less evident in the remaining two (A3 and A4).
Biosynthetic labeling of pRb. Indicated U-937 clones were biosynthetically labeled for 2 hours with 35S-methionine/cysteine and labeled proteins were subjected to immunoprecipitation with an anti-pRb antibody or a negative control antibody followed by SDS-PAGE and fluorography, as described in the Materials and Methods. Molecular weights are indicated to the left (in kilodaltons). Immunoprecipitated pRb is indicated with an arrow.
Biosynthetic labeling of pRb. Indicated U-937 clones were biosynthetically labeled for 2 hours with 35S-methionine/cysteine and labeled proteins were subjected to immunoprecipitation with an anti-pRb antibody or a negative control antibody followed by SDS-PAGE and fluorography, as described in the Materials and Methods. Molecular weights are indicated to the left (in kilodaltons). Immunoprecipitated pRb is indicated with an arrow.
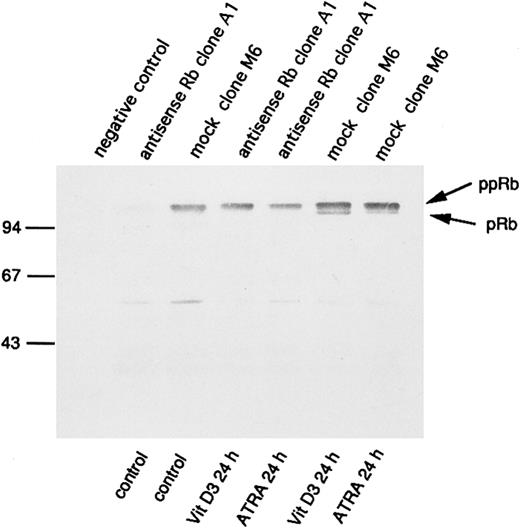
Increased expression and hypophosphorylation of pRb has been implicated in processes of differentiation. Therefore, we wanted to investigate the consequences of antisense RB expression for pRb production also during incubation of U-937 cells with agents capable of inducing differentiation, such as ATRA or Vit D3. For this purpose, we selected the clones M6 and A1 for incubation with and without ATRA or Vit D3 at concentrations of 1 μmol/L and 0.1 μmol/L, respectively. pRb levels and its degree of phosphorylation were determined by immunoprecipitation followed by 7% SDS-PAGE and Western blot analysis, as described in the Materials and Methods. As seen in Fig 5, a reduced amount of pRb in antisense-transfected cells was still evident 24 hours after the onset of induction of differentiation, both with ATRA or Vit D3. Furthermore, levels of pRb were generally increased on induction of differentiation, both in antisense-transfected cells and mock-transfected cells (Fig 5). Notably, only mock-transfected cells showed levels of hypophosphorylated pRb on incubation with ATRA or Vit D3, whereas the antisense RB-transfected cells did not (Fig 5). To determine the effect of the transfected antisense RB construct on the ability of de novo synthesis of pRb on induction of differentiation, biosynthetic labeling, followed by 7% SDS-PAGE and fluorography, was performed as described in the Materials and Methods. The pattern was almost identical to Fig 5, with upregulation of pRb synthesis and appearance of hypophosphorylated pRb in differentiating mock-transfected cells, whereas antisense-transfected cells did not show any hypophosphorylation of pRb on induction of differentiation (data not shown).
Changes in levels of the retinoblastoma protein and its degree of phosphorylation on induction of differentiation. Indicated U-937 clones (antisense Rb clone A1 and mock clone M6) were induced with ATRA (1 μmol/L) or Vit D3 (0.1 μmol/L). At 0 and at 24 hours, cells were harvested and subjected to protein extraction followed by specific immunoprecipitation with an antiretinoblastoma protein antibody, 7% SDS-PAGE, and Western blot analysis, as described in the Materials and Methods. Molecular weights are indicated to the left (in kilodaltons). Immunoprecipitated pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated).
Changes in levels of the retinoblastoma protein and its degree of phosphorylation on induction of differentiation. Indicated U-937 clones (antisense Rb clone A1 and mock clone M6) were induced with ATRA (1 μmol/L) or Vit D3 (0.1 μmol/L). At 0 and at 24 hours, cells were harvested and subjected to protein extraction followed by specific immunoprecipitation with an antiretinoblastoma protein antibody, 7% SDS-PAGE, and Western blot analysis, as described in the Materials and Methods. Molecular weights are indicated to the left (in kilodaltons). Immunoprecipitated pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated).
The suppression of retinoblastoma protein production in antisense transfected clones resulted in decreased sensitivity to induction of differentiation by ATRA or Vit D3.To determine the effect of the suppressed pRb production as well as the absence of detectable levels of hypophosphorylated pRb in induced antisense-transfected cells on inducibility of differentiation, incubations with ATRA or Vit D3 were performed as described in the Materials and Methods. U-937 cells are known to respond with maturation into monocyte-like cells by these agents.38 39 A marked difference in maturation responses to ATRA, as judged by NBT reduction capability, was observed between antisense- and mock-transfected clones (Fig 6A). All mock-transfected clones showed, after 4 days of incubation with ATRA at 1 μmol/L, a percentage of formazan-containing cells ranging from 19% to 39%, whereas six of eight clones transfected with the PCEP4/anti-Rb construct (A1, A2, A5, A6, A7, and A8) almost totally failed to differentiate on the same treatment. Fewer than 1% of cells from these latter clones were capable of reducing NBT. As seen in Fig 4, these nonresponding clones are identical to those showing bands of low intensity on fluorography after immunoprecipitation of biosynthetically labeled pRb. Thus, the repression of pRb synthesis conferred a reduced capacity to respond to ATRA with differentiation as judged by NBT reduction capability. Moreover, a correlation between the degree of pRb expression and differentiation inducibility was observed; antisense RB clones A3 and A4 showed bands on fluorography of moderate intensity (Fig 4) and these two clones were less inhibited in their response of differentiation to ATRA than the other antisense RB clones (Fig 6A), showing bands of low intensity. When incubated with Vit D3 at 0.1 μmol/L for 4 days, all mock-transfected clones, except for M1, showed a high percentage of NBT-reducing cells ranging from 90% to 98%. Similar to what was seen with ATRA, clones transfected with antisense RB showed a lower percentage of NBT-reducing cells, ranging from 23% to 67% (Fig 6B). These results show that the extent of differentiation of U-937 cells induced by Vit D3 was also reduced by expression of the pCEP4/anti-RB construct, although the inhibition was less pronounced than with ATRA.
Effects of ATRA or Vit D3 on NBT-reducing capability of antisense RB-transfected and mock-transfected clones. Cells at an initial concentration of 0.2 × 106 cells/mL were incubated with (A) ATRA (1 μmol/L) or (B) Vit D3 (0.1 μmol/L) for 4 days followed by an NBT-reducing test. Two hundred cells from each clone were counted on a cytospin slide and the percentage of cells containing reduced NBT was determined. Results shown are (A) from one representative experiment and (B) mean values from three separate experiments, bars ± SEM.
Effects of ATRA or Vit D3 on NBT-reducing capability of antisense RB-transfected and mock-transfected clones. Cells at an initial concentration of 0.2 × 106 cells/mL were incubated with (A) ATRA (1 μmol/L) or (B) Vit D3 (0.1 μmol/L) for 4 days followed by an NBT-reducing test. Two hundred cells from each clone were counted on a cytospin slide and the percentage of cells containing reduced NBT was determined. Results shown are (A) from one representative experiment and (B) mean values from three separate experiments, bars ± SEM.
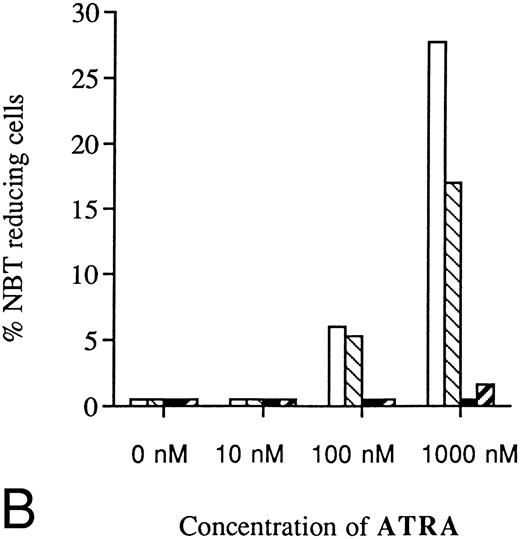
Two clones (A1 and M6) were selected for further experiments with different concentrations of ATRA or Vit D3. As shown in Fig 7A, the resistance to induction of differentiation by ATRA was even more evident with higher concentrations. Because of toxic effects, higher concentrations of ATRA than 10 μmol/L were not possible to use. Thus, the differentiation block induced by antisense RB expression was not possible to overcome with increased amounts of ATRA. Higher concentrations of Vit D3 than 0.1 μmol/L could not be used because of toxicity, but Fig 7B shows that the reduced response was most evident with the highest concentration possibly used, as was the case for ATRA. Control cells incubated without ATRA or Vit D3 showed no ability at all to reduce NBT (Fig 7A and B).
Dose-dependence of induced differentiation by ATRA or Vit D3 in antisense RB-transfected and mock-transfected U-937 cells. Cells (0.2 × 106 /mL) of (▪) the antisense transfected clone A1 and (□) mock-transfected clone M6 were incubated with ATRA (A) at concentrations ranging from 0 to 10 μmol/L or Vit D3 (B) at concentrations ranging from 0 to 100 nmol/L for 4 days followed by an NBT-reducing test. It was not possible to use higher concentrations of inducer due to toxic effects. Two hundred cells from each clone were counted and the percentage of cells containing reduced NBT was determined. Results shown are from one representative experiment.
Dose-dependence of induced differentiation by ATRA or Vit D3 in antisense RB-transfected and mock-transfected U-937 cells. Cells (0.2 × 106 /mL) of (▪) the antisense transfected clone A1 and (□) mock-transfected clone M6 were incubated with ATRA (A) at concentrations ranging from 0 to 10 μmol/L or Vit D3 (B) at concentrations ranging from 0 to 100 nmol/L for 4 days followed by an NBT-reducing test. It was not possible to use higher concentrations of inducer due to toxic effects. Two hundred cells from each clone were counted and the percentage of cells containing reduced NBT was determined. Results shown are from one representative experiment.
Antisense RB-transfected cells showed impaired inducibility of monocyte-related cell surface antigens as compared with control cells on induction with ATRA or Vit D3.The capacity to reduce NBT is a phenotypic characteristic of mature myeloid cells and can be considered as a functional marker of differentiation. To see whether other differentiation-induced phenotypic changes also were affected by antisense RB expression, the appearance of the cell surface antigens CD14, CD11b, and CD11c was monitored by flow cytometric analysis after treatment with or without Vit D3 or ATRA, as described in the Materials and Methods. CD14 is a receptor for the complex of lipopolysacharides (LPS) and LPS binding protein.46 The expression of CD14 on U-937 cells can be induced by Vit D3 or PMA.47 CD11b and CD11c constitute the α chains of CD11b/CD18 (Mac-1) and CD11c/CD18 (p150, 95), respectively, both members of the β2 integrin family of leukocyte adhesion molecules,48 and can be induced by Vit D3, ATRA, or PMA on U-937 cells.47
As shown in Table 1, the mock-transfected control cells (M6) responded with induction of all three surface antigens in response to ATRA or Vit D3. This increase correlated to acquired capacity to reduce NBT. However, in the antisense RB-transfected cells (A1), the induction of CD14 and CD11c by ATRA was strongly inhibited. The level of CD11b was very low in the antisense-transfected cells, although it was most easily inducible with ATRA in antisense- and mock-transfected cells. Similarly to the results from induction of NBT-reducing capability, the inhibition of induced differentiation in antisense RB-transfected cells, as judged by cell surface markers, was less prominent on incubation with Vit D3 than with ATRA. A diminished response in antisense RB-transfected cells on incubation with Vit D3 was only seen for CD11c (Table 1). The effect of antisense RB on Vit D3-induced appearance of surface antigens did not fully parallel the effect on NBT-reductive capacity. Thus, the results from ATRA-induced appearance of cell surface antigens confirmed those from NBT reduction experiments that cells with impaired capacity to synthesise pRb showed decreased tendency for differentiation, whereas the data on antisense RB effects on Vit D3-induced appearance of surface antigens were less convincing.
Appearance of Cell Surface Antigens Correlated to NBT Positivity
| . | Mock Clone M6 . | Antisense Rb Clone A1 . | ||||
|---|---|---|---|---|---|---|
| . | Control . | ATRA . | Vit D3 . | Control . | ATRA . | Vit D3 . |
| CD14 | 12 | 22 | 100 | 26 | 6 | 100 |
| CD11c | 0.5 | 94 | 98 | 2 | 38 | 78 |
| CD11b | 49 | 100 | 98 | 3 | 96 | 100 |
| NBT | 0 | 36 | 98 | 0 | 1 | 23 |
| . | Mock Clone M6 . | Antisense Rb Clone A1 . | ||||
|---|---|---|---|---|---|---|
| . | Control . | ATRA . | Vit D3 . | Control . | ATRA . | Vit D3 . |
| CD14 | 12 | 22 | 100 | 26 | 6 | 100 |
| CD11c | 0.5 | 94 | 98 | 2 | 38 | 78 |
| CD11b | 49 | 100 | 98 | 3 | 96 | 100 |
| NBT | 0 | 36 | 98 | 0 | 1 | 23 |
Cells were incubated at an initial concentration of 200,000 cells/mL and induced with ATRA at 1 μmol/L or Vit D3 at 0.1 μmol/L. After 4 days, the cells were subjected to analysis of surface antigens as well as NBT reduction test. Values shown are the percentage of cells expressing surface antigens or reducing NBT determined as described in the Materials and Methods. Results are shown from one representative experiment.
The cell cycle distribution in U-937 cells transfected with an antisense RB construct or incubated with antisense Rb oligonucleotides did not differ from that of control cells on incubation with ATRA or Vit D3.The retinoblastoma protein is involved in cell cycle regulation and is capable of executing a potent brake in the G1-phase of the cell cycle. Antisense RB-transfected clones, mock-transfected clones, and wild-type U-937 cells incubated with or without oligonucleotides all showed a similar growth rate and viability (data not shown). The viability was constantly greater than 90% (data not shown). Thus, neither the transfected antisense RB construct nor the oligonucleotides at concentrations used conferred any apparent effect on growth and viability.
Growth inhibition by ATRA or Vit D3 in antisense RB-transfected U-937 cells as compared with mock-transfected cells. Cells (0.2 × 106 /mL) from antisense RB clone A1 and mock clone M6 were incubated with Vit D3 at 0.1 μmol/L or ATRA at 1 μmol/L for 4 days. Aliquots were removed and counted daily. Simultaneously, noninduced cells were incubated as controls. Results shown are from one representative experiment. (□) M6; (▵) M6 + ATRA; (○) M6 + Vit D3; (▪) A1; (▴) A1 + ATRA; (•) A1 + Vit D3.
Growth inhibition by ATRA or Vit D3 in antisense RB-transfected U-937 cells as compared with mock-transfected cells. Cells (0.2 × 106 /mL) from antisense RB clone A1 and mock clone M6 were incubated with Vit D3 at 0.1 μmol/L or ATRA at 1 μmol/L for 4 days. Aliquots were removed and counted daily. Simultaneously, noninduced cells were incubated as controls. Results shown are from one representative experiment. (□) M6; (▵) M6 + ATRA; (○) M6 + Vit D3; (▪) A1; (▴) A1 + ATRA; (•) A1 + Vit D3.
Antisense RB Oligonucleotides Suppressed Differentiation But Not G0/G1 Accumulation on Incubation of Wild-Type U-937 Cells With ATRA or Vit D3
| Day . | . | No Inducer . | Vit D3 . | ATRA . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Buffer . | s-Rb . | a-Rb . | Buffer . | s-Rb . | a-Rb . | Buffer . | s-Rb . | a-Rb . |
| 0 | G0/G1 | 57 | 56 | 64 | 54 | 59 | 60 | 56 | 60 | 58 |
| 4 | G0/G1 | 78 | 78 | 74 | 96 | 97 | 98 | 96 | 95 | 85 |
| 4 | NBT | 0 | 0 | 0 | 86 | 83 | 26 | 39 | 42 | 1 |
| Day . | . | No Inducer . | Vit D3 . | ATRA . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Buffer . | s-Rb . | a-Rb . | Buffer . | s-Rb . | a-Rb . | Buffer . | s-Rb . | a-Rb . |
| 0 | G0/G1 | 57 | 56 | 64 | 54 | 59 | 60 | 56 | 60 | 58 |
| 4 | G0/G1 | 78 | 78 | 74 | 96 | 97 | 98 | 96 | 95 | 85 |
| 4 | NBT | 0 | 0 | 0 | 86 | 83 | 26 | 39 | 42 | 1 |
Cells were preincubated for 24 hours with buffer, sense RB oligos (s-Rb; 50 μmol/L), and antisense RB oligos (a-Rb; 50 μmol/L); respectively. The incubated cells were then induced with ATRA of 1 μmol/L or Vit D3 at 0.1 μmol/L at a concentration of 200,000 cells/mL. After 4 days, cells were again subjected to determination of cell cycle phase distribution by flow cytometric analysis as well as an NBT reduction test. Values shown are the percentages of cells in G0/G1 or reducing NBT. Results are from one representative experiment.
Antisense Rb-Transfected Cells Showed Decreased Ability to Differentiate But Not to Accumulate in G0/G1 as Compared With Mock-Transfected Cells on Incubation With ATRA or Vit D3
| Day . | . | Mock Clone M6 . | Antisense Rb Clone A1 . | ||||
|---|---|---|---|---|---|---|---|
| . | . | Control . | ATRA . | Vit D3 . | Control . | ATRA . | Vit D3 . |
| 0 | G0/G1 | 54 | 55 | ||||
| 4 | G0/G1 | 65 | 97 | 97 | 75 | 92 | 99 |
| 4 | NBT | 0 | 29 | 98 | 0 | 0 | 27 |
| Day . | . | Mock Clone M6 . | Antisense Rb Clone A1 . | ||||
|---|---|---|---|---|---|---|---|
| . | . | Control . | ATRA . | Vit D3 . | Control . | ATRA . | Vit D3 . |
| 0 | G0/G1 | 54 | 55 | ||||
| 4 | G0/G1 | 65 | 97 | 97 | 75 | 92 | 99 |
| 4 | NBT | 0 | 29 | 98 | 0 | 0 | 27 |
Cells were incubated at a concentration of 200,000 cells/mL and induced with ATRA at 1 mmol/L or Vit D3 at 0.1 μmol/L. After 4 days, cells were again subjected to determination of cell cycle phase distribution by flow cytometric analysis as well as an NBT reduction test. Values shown are the percentages of cells in G0/G1 or reducing NBT. Results are from one representative experiment.
During terminal cell differentiation, there is a characteristic accumulation of cells in G0/G1, parallelled or followed by cessation of growth. We therefore wanted to investigate the consequences of the reduced capacity to synthesise pRb on proliferation and on cell cycle phase distribution, with or without the presence of inducers of differentiation. Figure 8 shows the proliferation of U-937 mock-transfected control cells (clone M6) and of antisense-transfected cells (clone A1) treated with or without ATRA or Vit D3. A similar reduced growth rate on incubation with ATRA or Vit D3 was observed in antisense RB clones as well as mock-transfected clones when compared with uninduced control cells (Fig 8). Thus, in contrast to the difference in tendency for differentiation, the effects on proliferation rate was similar for control cells and antisense RB-expressing cells, with or without induction of differentiation (Fig 8). The viability of all cells was unaffected at the concentrations of inducer used, ATRA at 1 μmol/L or Vit D3 at 0.1 μmol/L, and was always greater than 90% (data not shown). In accordance with these data on cell proliferation, the cell cycle distribution, assayed by use of flow cytometric analysis, was similar for control cells and antisense RB-expressing cells, with or without differentiation inducers (Table 2). At the onset of induction, the cell cycle phase distribution was similar in the antisense-transfected cells and mock-transfected cells. After 4 days with inducers, virtually all mock-transfected cells as well as antisense RB-transfected cells were arrested in G0/G1. However, the fraction of NBT-positive cells was significantly lower in the antisense-transfected clone (Table 2). Thus, on induction with ATRA or Vit D3, the differentiation response was impaired in antisense RB-transfected cells, whereas proliferation rate and cell cycle distribution were similarly affected in both antisense- and mock-transfected cells.
To further sustain the occurence of a G1 accumulation similar to that of terminal differentiation in cells being suppressed in their ability to differentiate, we incubated wild-type U-937 cells with antisense RB oligonucleotides previously found to be able to block differentiation (Figs 1 and 2). Wild-type U-937 cells were induced with ATRA or Vit D3 for 4 days in the continuous presence of antisense RB oligos, using sense RB oligos and oligo buffer as controls. Although these cells were inhibited in their ability to differentiate, they showed a similar G0/G1 accumulation as control cells that did differentiate (Table 3). The oligonucleotides did not affect cell viability, which was always greater than 90% (data not shown). The oligonucleotides also did not affect proliferation rates (data not shown). The reduced proliferation rates on induction of differentiation were very similar with or without oligonucleotides (data not shown), which is analogous to the data in Fig 8. Thus, simply arresting cells in G0/G1 is not sufficient to induce differentiation of leukemic U-937 cells.
U-937 clones stably overexpressing pRb showed decreased sensitivity to induction of differentiation by ATRA or Vit D3.The present finding that suppression of Rb expression led to inhibited capacity for induced differentiation prompted us to investigate if overexpression of pRb would facilitate induction of differentiation. For this purpose, U-937 wild-type cells were transfected with the pCDNA3/HRbC construct as described in the Materials and Methods. This resulted in two RB clones, B3 and B4, both stably overexpressing pRb as judged by biosynthetic labeling and specific immunoprecipitation followed by fluorography. Intriguingly, these two clones were almost completely inhibited in their differentiation response to Vit D3 or ATRA, whereas the mock-transfected clones responded with differentiation in the expected manner, as judged by NBT reduction (Fig 9A and B). The reduced sensitivity to induced differentiation in clones overexpressing pRb was not due to unability to respond with hypophosphorylation of pRb on induction of differentiation (Fig 10), indicating that the observed differentiation block depends on other mechanisms than the absence of hypophosphorylated Rb.
Inhibited induction of differentiation by ATRA or Vit D3 in RB-transfected U-937 cells as compared with mock-transfected cells. Cells (0.2 × 106 /mL) of the RB-transfected clones (▪) B3 and (▨) B4, both stably overexpressing pRb, as well as mock-transfected clones (□) M2 and (▧) M6 were incubated with ATRA (A) or Vit D3 (B) at concentrations ranging from 0 to 1,000 nmol/L 4 days followed by an NBT-reducing test. Two hundred cells from each clone were counted and the percentage of cells containing reduced NBT was determined. Results shown are mean values from three separate experiments.
Inhibited induction of differentiation by ATRA or Vit D3 in RB-transfected U-937 cells as compared with mock-transfected cells. Cells (0.2 × 106 /mL) of the RB-transfected clones (▪) B3 and (▨) B4, both stably overexpressing pRb, as well as mock-transfected clones (□) M2 and (▧) M6 were incubated with ATRA (A) or Vit D3 (B) at concentrations ranging from 0 to 1,000 nmol/L 4 days followed by an NBT-reducing test. Two hundred cells from each clone were counted and the percentage of cells containing reduced NBT was determined. Results shown are mean values from three separate experiments.
Production of the retinoblastoma protein and changes in its degree of phosphorylation on induction of differentiation in Rb-transfected clones. Indicated U-937 clones (RB transfectants B3 and B4 and mock-transfectant M2) were induced with Vit D3 (0.1 μmol/L). At 0 and 24 hours, cells were harvested and indicated clones were biosynthetically labeled for 2 hours with 35S-methionine/cysteine. Labeled proteins were subjected to immunoprecipitation with an anti-pRb antibody followed by 7% SDS-PAGE and fluorography, as described in the Materials and Methods. Immunoprecipitated pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated).
Production of the retinoblastoma protein and changes in its degree of phosphorylation on induction of differentiation in Rb-transfected clones. Indicated U-937 clones (RB transfectants B3 and B4 and mock-transfectant M2) were induced with Vit D3 (0.1 μmol/L). At 0 and 24 hours, cells were harvested and indicated clones were biosynthetically labeled for 2 hours with 35S-methionine/cysteine. Labeled proteins were subjected to immunoprecipitation with an anti-pRb antibody followed by 7% SDS-PAGE and fluorography, as described in the Materials and Methods. Immunoprecipitated pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated).
The cell cycle distribution in U-937 cells overexpressing pRb did not differ from that of control cells on incubation with ATRA or Vit D3.RB-transfected U-937 clones overexpressing pRb accumulated in G0/G1 to a similar extent as mock-transfected clones on induction of differentiation with ATRA or Vit D3 (Table 4). Thus, although the cells overexpressing pRb were significantly inhibited in their differentiation response (Fig 9A and B), they still responded with a G0/G1 accumulation when incubated with ATRA or Vit D3 (Table 4). This result further sustained the interpretation of the cell cycle data on antisense clones/cells (Tables 2 and 3) that G0/G1 arrest per se is not sufficient for induction of differentiation by ATRA or Vit D3.
Rb-Transfected U-937 Cells Accumulated in G0/G1 to a Similar Extent as Mock-Transfected Clones Upon Incubation With ATRA or Vit D3
| Day . | . | Mock Clones (M2; M6) . | Rb Clones (B3; B4) . | ||||
|---|---|---|---|---|---|---|---|
| . | . | Control . | ATRA . | Vit D3 . | Control . | ATRA . | Vit D3 . |
| 4 | G0/G1 | 60; 58 | 94; 96 | 96; 97 | 50; 71 | 90; 97 | 96; 98 |
| Day . | . | Mock Clones (M2; M6) . | Rb Clones (B3; B4) . | ||||
|---|---|---|---|---|---|---|---|
| . | . | Control . | ATRA . | Vit D3 . | Control . | ATRA . | Vit D3 . |
| 4 | G0/G1 | 60; 58 | 94; 96 | 96; 97 | 50; 71 | 90; 97 | 96; 98 |
Cells from two mock-transfected clones (M2; M6) and two RB-transfected clones (B3; B4) were incubated at a concentration of 200,000 cells/mL and induced with ATRA at 1 μmol/L or Vit D3 at 0.1 μmol/L. After 4 days, cells were subjected to determination of cell cycle phase distribution by flow cytometric analysis. Values shown are the percentages of cells of the two mock-transfected and the two RB-transfected clones in G0/G1. Results are from one representative experiment.
DISCUSSION
Differentiation depends on RB gene expression.The aim of this study was to investigate if the retinoblastoma gene product is involved in the induction of differentiation of leukemic cells. The initial finding that U-937 cells as well as K562 cells, when incubated with antisense RB oligonucleotides, become hampered in their capability for induced differentiation by ATRA or Vit D3 and TGFβ1, respectively, prompted us to further investigate and sustain these results. For this purpose, clones of the monoblastic cell line U-937, which were transfected with an antisense cDNA construct for the human retinoblastoma gene, were established. The ability of the transfected clones to produce the retinoblastoma protein and its degree of phosphorylation were characterized as well as their responsiveness to differentiation induced by ATRA or Vit D3. Furthermore, cell cycle phase distribution was assayed. We have shown that several of the clones transfected with the antisense RB construct were suppressed in their ability to produce the retinoblastoma protein. Moreover, we have been able to show that this suppressed capacity of retinoblastoma protein production correlates with a reduced susceptibility to induction of differentiation, which in turn correlates with an inability to upregulate detectable levels of hypophosphorylated pRb. Despite the strong inhibitory effects on the differentiation induction exerted by antisense RB oligonucleotides, no appreciable effect on the expression of pRb was noted (data not shown). The reason for the absence of apparent effects on the protein level is unclear, but subtle changes of pRb below the detection limit may be critical for dynamic processes. Distinct effects of antisense oligonucleotides without remarkabe effects on the corresponding protein level have been reported previously.49 A major concern in using antisense stratagies in modifying gene expression is selectivity, a problem that can lead to side effects, because related target sequences may also be inhibited.50 However, both antisense-transfected clones, expressing low levels of the retinoblastoma protein, as well as wild-type U-937 and wild-type K562 cells incubated with antisense RB oligonucleotides showed less sensitivity to induction of differentiation with ATRA or Vit D3 and TGFβ1, respectively, than did controls. The results from these experiments taken together, using two different methods both aiming at reducing expression of pRb, one of which applied to two different cell lines, indicate that the observed effects are due to reduced pRb expression. Nevertheless, it cannot be ruled out that the observed effects of the antisense oligos are not sequence-specific.
To further explore the involvement of the RB gene during induction of differentiation, clones of U-937 cells overexpressing pRb after transfection with the human RB cDNA were established. Intriguingly, these clones behaved similarly to antisense-transfected clones, showing reduced sensitivity to induction of differentiation to ATRA or Vit D3 as judged by NBT positivity. In contrast to this inhibited differentiation response to ATRA and Vit D3, the RB overexpressing clones were able to respond with hypophosphorylation of pRb as well as G0/G1 accumulation on incubation with ATRA or Vit D3, which will be discussed below.
Our results showing that induction of differentiation depends on the expression of the retinoblastoma protein are consistent with several previous independent lines of evidence. By using transgenic mice devoid of pRb, it has been shown that pRb is crucial for neuronal and hematopoietic differentiation22-25 and for the establishment and maintenance of the terminally differentiated phenotype of muscle cells.27,28,51 Levels of pRb increase upon differentiation of basal keratinocytes and colonic crypt cells.52 Moreover, it has been shown that exposure to ATRA in the embryonic carcinoma cell line P19 induces differentiation into neurons with a subsequent increase in the levels of pRb.53 Furthermore, differentiation of normal human hematopoietic cells or leukemic cells in vitro has been correlated with activation of pRb by hypophosphorylation and with upregulation of pRb expression.11,29 30
In the present study, antisense RB-targeted U-937 or K562 cells did not differ in growth or viability as compared with nontargeted cells. Deletion of the RB gene in transgenic mice is lethal and confers widespread apoptotic neuronal cell death.22-25 This apoptotic reaction resulting from RB inactivation is believed to depend on the function of another tumor-suppressor gene, p53.26,54 A speculative interpretation of the unaffected viability of cell lines used in the present study is provided by the fact that U-937 cells and K562 cells are devoid of detectable levels of normal p53 protein.43 55 Also, the antisense targeted cells cannot be considered totally defective in retinoblastoma protein production. Therefore, another speculative explanation might be that the antisense targeted cells in the present study produce enough pRb for survival, but too little for unaffected ability of induced differentiation. Finally, during establishment of the antisense RB-transfected U-937 clones, the process of selection might have made these clones independent of pRb for survival.
How are differentiation responses mediated by the retinoblastoma protein?Cell differentiation and cell proliferation are mutually exclusive phenomena in terminally differentiating cell types such as myoblasts and neurons. Results from studies on myogenesis have shown that, in myoblasts, proteins of the Rb family are required for induced expression of tissue-specific genes, whereas inactivation of pRb is required for cell cycle progression and DNA duplication.27,28 It has been suggested that the decision of a cell to differentiate is made during the G1-phase of the cell cycle56 and that the phosphorylation state of pRb decides whether the cell will enter the cell cycle or stay out of it and commit to the pathway of terminal differentiation. Active hypophosphorylated pRb induces cell cycle arrest in the G1-phase.10 Hypothetically, the pRb-mediated cell cycle arrest could provide the cell opportunity to express its tissue-specific genes for differentiation, if an adequate inducing signal for differentiation is present. In other words, an arrest of the cell cycle in the G1-phase may increase the probability for differentiation. If the cell succeeds in reaching a higher or a terminally differentiated state, hypophosphorylated pRb is able to sustain the achieved phenotype, by arresting the cell at G1 phase or, if the terminal state has been reached, trapping the cell in G0.28 However, if the differentiation mediating property of pRb only depends on its cell cycle restriction potential, a correlation between the accumulation of cells in G0/G1 and the differentiation response would be expected in the present study. But this was not the case. The results from the cell cycle experiments showed that, despite the reduced capacity to differentiate on induction with ATRA or Vit D3 seen in the antisense RB-expressing cells, wild-type cells incubated with antisense RB oligos, and pRb overexpressing cells, the pattern of G0/G1 accumulation was almost identical to that of the induced differentiating control cells (Tables 2, 3, and 4). Thus, pRb seems to have functions other than those directly related to cell cycle arrest. It has been shown that pRb is able to interact with several specific transcription factors such as MyoD, GATA1, and NF-IL6, interactions that have been shown necessary for murine terminal muscle differentiation,27,51 human adult erythropoiesis,30 and monocytic differentiation of U-937 cells,57 respectively. Hypothetically, the G0/G1-accumulation observed in the induced antisense RB cells apparently devoid of hypophospforylated pRb could be due to action of other proteins, perhaps pRb-related proteins, such as p107 and p130. pRb-related proteins have previously been shown to be capable of binding E2F58,59 but unable to sustain terminal muscle cell differentiation.28 Another possible explanation of the G0/G1 accumulation induced by ATRA or Vit D3 in antisense cells may be that they produce enough pRb to induce cell cycle arrest but not enough to mediate signals for terminal differentiation, which might request associations between pRb and various myeloid transcription factors. In the present study, the most pronounced differences in maturation responses between U-937 antisense RB clones and mock-transfected clones were observed after incubation with ATRA. Also, in K562 cells, the ability of antisense RB oligonucleotides to inhibit differentiation was selective, because the maturation response to TGFβ1, but not to hemin, was blocked. The reason for this is unclear. It could be that different inducers of differentiation use distinct transcription factors, some of which rely more on pRb than others. This could also serve as a hypothetical explanation for the observed discrepancy in Table 1, in which antisense RB seems to be more potent in suppressing NBT positivity than the appearance of surface antigens. Also, most hypothetically, this discrepancy may be due to a sequential appearance of Vit D3-induced surface antigens and NBT positivity in which the induced appeareance of surface antigens preceeds the NBT positivity and the antisense mechanism is acting in between.
The present data on cell cycle distribution indicate that G1 arrest might be necessary, but it is not sufficient to induce terminal differentiation of leukemic cells. This would be in line with recent findings indicating that G1 prolongation is necessary but not sufficient for erythropoietin-induced erythroid differentiation in a murine interleukin-3–dependent cell line.60 Also, it has recently been reported that induced differentiation of Friend virus-transformed murine erythroleukemic cells requires pRb-mediated G1 prolongation, but that a G1 prolongation per se does not induce terminal differentiation.61
The present finding that overexpression of RB in U-937 cells resulted in inhibited ability to respond with differentiation to ATRA and Vit D3 (Fig 9A and B) could not be correlated with inability of the cells to hypophosphorylate pRb on such induction of differentiation (Fig 10). Even though these pRb overexpressing clones were hampered in their capacity to respond with differentiation as compared with controls, they accumulated in G0/G1 to a similar extent as did controls when incubated with ATRA or Vit D3 (Table 4). Thus, both inhibition and overexpression of pRb inhibits induced differentiation. These apparently contradictory results might be addressed in view of recent discoveries of pRb function. It has been proposed that the biologic activity of pRb depends on its ability to assemble specific protein complexes.62 Hypothetically, during conditions of forced overexpression or forced suppression of pRb expression, as in this study, a critical and optimal level of pRb needed for proper assembly of specific protein complexes favoring expression of tissue-specific genes is out of reach. Also, hypothetically, it may be that, during the process of selection of clones, in this study with either a stable overexpression or a suppressed expression of pRb, the capacity of induced differentiation is affected.
To conclude, our data indicate that induction of differentiation by ATRA or Vit D3 in the leukemic cell line U937 and by TGFβ1 in the leukemic cell line K562 involves pRb by mechanisms other than those merely inducing G1 accumulation.
ACKNOWLEDGMENT
The authors thank Gerd Häggbom for help with photographs on cytospin preparations.
Supported by the Swedish Cancer Society, the Georg Danielsson Foundation, the Gunnar, Arvid and Elisabeth Nilsson Foundation, the Thelma Zoéga Foundation, the John Persson Foundation, the John and Augusta Persson Foundation, Funds of Lunds Sjukvårdsdistrikt, Swedish Society for Medical Research, the Tobias Foundation and the Medical Faculty of Lund.
Address reprint requests to Gösta Bergh, MD, Research Dept. 2, E-block, University Hospital, S-221 85 Lund, Sweden.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal