Abstract
Recently, point mutations in the gene of the granulocyte colony-stimulating factor (G-CSF ) receptor have been reported in two patients with severe congenital neutropenia who developed acute myeloid leukemia (AML). We investigated the frequency of these specific G-CSF receptor mutations in patients with congenital neutropenia undergoing treatment with r-metHuG-CSF (Filgrastim) and the clinical relevance of these mutations. Nucleotides 2306 to 2561 including the critical region (nucleotides 2384-2429) from the intracellular domain of the G-CSF receptor gene were amplified by reverse transcriptase-polymerase chain reaction. Detection of point mutations was performed with specific restriction enzyme analysis, as well as sequencing of PCR products. Both genomic DNA and cDNA from neutrophils and mononuclear cells were analyzed from 28 patients with severe congenital neutropenia. Four of 28 patients with congenital neutropenia displayed a point mutation in the tested cytoplasmic region of the G-CSF receptor gene. The point mutations replace a glutamine codon by a stop codon of the G-CSF receptor gene. Among these four congenital neutropenia patients with a mutated G-CSF receptor, two developed AML. All four patients were investigated regularly and no correlation between occurrence of G-CSF receptor mutation and time or dose of r-metHuG-CSF treatment was found. No point mutations in the G-CSF receptor critical domain could be detected in cells from the other 24 congenital neutropenia patients. Furthermore, we tested six family members of the two patients with AML including mothers and fathers, one sister, and one brother who suffers from congenital neutropenia, as well. All family members displayed a normal G-CSF receptor gene. After the acquisition of the G-CSF receptor mutations, the congenital neutropenia patients continued to respond to G-CSF therapy with an increase in absolute neutrophils in the peripheral blood. We conclude that the point mutations in the critical region of the intracellular part of the G-CSF receptor occur spontaneously and are not inherited. From our data, we suggest that the described G-CSF receptor point mutations do not alter the response to treatment with r-metHuG-CSF and are not the cause of severe congenital neutropenia.
SEVERE CONGENITAL neutropenia or Kostmann's syndrome is characterized by a maturation arrest of neutrophil precursors at the level of promyelocytes or myelocytes in the bone marrow.1,2 The absolute neutrophil counts (ANC) are less than 200/μL in peripheral blood. Congenital neutropenia predisposes a patient to frequent and severe bacterial infections. Several clinical studies demonstrated that treatment of congenital neutropenia patients with recombinant human granulocyte colony-stimulating factor (G-CSF ) (r-metHuG-CSF, Filgrastim) results in an increase in circulating neutrophils and a reduction in infection-related events in more than 95% of the patients.3-6 G-CSF preferentially stimulates the growth and differentiation of neutrophil precursors and the function of neutrophils. Data from the International Registry for severe chronic neutropenia include 220 patients with severe congenital neutropenia undergoing treatment with r-metHuG-CSF.7 Between 1988 and 1995, 16 of these 220 congenital neutropenia patients developed myelodysplastic syndrome or acute myeloid leukemia (MDS/AML). None of the registered patients with cyclic (n = 78) or idiopathic (n = 122) neutropenia developed MDS/AML, suggesting that r-metHuG-CSF is not involved in leukemogenesis, and severe congenital neutropenia might represent a premalignant disorder of myelopoiesis.7
The pathophysiology of severe congenital neutropenia is still unknown. We could show that congenital neutropenia patients are capable of producing biologically active G-CSF.8 Scatchard analysis showed no alterations in the expression and binding affinity of G-CSF receptors on neutrophils from congenital neutropenia patients undergoing r-metHuG-CSF therapy as compared with normal cells.9 Defects in G-CSF–mediated signal transduction have been proposed and increased tyrosine phosphorylation and activation of the receptor-associated protein tyrosine kinase JAK2 could be shown in neutrophils from patients with congenital neutropenia.10 Recently, point mutations at nucleotide positions 2390 and 2429 in the intracellular region of the G-CSF receptor gene have been described and linked to the development of secondary AML in two patients with congenital neutropenia.11
We tested congenital neutropenia patients and their family members for the occurrence of point mutations in the critical region of the cytoplasmic domain of the G-CSF receptor gene (nucleotides 2384-2429) to answer the following questions: (1) What is the incidence of G-CSF receptor mutations in severe congenital neutropenia? (2) Are point mutations in the intracellular part of the G-CSF receptor inherited and, therefore, present from birth or are they spontaneous somatic mutations in the course of the disease? (3) Do point mutations abrogate or change the response to G-CSF in vivo? and (4) How frequent are G-CSF receptor mutations in patients who develop AML secondary to neutropenia and is there a potential role for these mutations in leukemogenesis?
MATERIALS AND METHODS
Patients.Between 1988 and 1990, 32 patients with severe congenital neutropenia were enrolled in our Phase I/II studies at the Medical School Hannover4 and are now on daily r-metHuG-CSF treatment for up to 8 years. All 32 patients are included in the 220 patients registered by the International Registry for severe chronic neutropenia.7 We tested myeloid cells from 28 patients who were clinically diagnosed as severe congenital neutropenia or Kostmann's syndrome according to the following criteria: ANC below 200/μL in the peripheral blood, maturation arrest at the promyelocyte level in the bone marrow, absence of antineutrophil antibodies, and onset of severe bacterial infections during the first 12 months of life. All patients being tested were treated with r-metHuG-CSF (Filgrastim) and they showed no signs of bacterial infections at the time when the study was performed.
Separation of myeloid cells.Neutrophils and mononuclear cells have been isolated from fresh heparinized blood by dextran sedimentation followed by Ficoll density centrifugation. Low-density mononuclear cells (mainly lymphocytes and monocytes) were obtained after Ficoll-separation and neutrophils residing in the pellet were isolated after hypotonic lysis of residual erythrocytes. More than 98% of the cells were viable as assayed by trypan blue dye exclusion. Mononuclear cells from bone marrow samples were isolated according to the above described protocol omitting the dextran sedimentation step.
RNA isolation.Total RNAs were extracted from the cells according to the single step isolation method12 using RNAzolB (WAK-Chemie Medical GmbH, Bad Homburg, Germany). A total of 1 to 3 × 107 cells were lysed with 1 mL RNAzolB. After addition of 0.1 mL of chloroform, the lysate was mixed thoroughly and incubated on ice for 5 minutes. Centrifugation of the lysate forms two phases. RNA remains exclusively in the upper aequeous phase and was precipitated with an equal volume of isopropanol and washed once with 70% ethanol. The air-dried RNA pellet was dissolved in water treated with diethylpyrocarbonate. RNA concentration was determined by measuring extinction at 260 nm.
Isolation of DNA.Genomic DNA was isolated from neutrophils and mononuclear cells using spin columns and buffers from the QIAamp tissue kit (Qiagen, Chatsworth, CA). A total of 106 to 107 cells were resuspended in 200 μL phosphate-buffered saline (PBS) and incubated with 25 μL Proteinase K (20 mg/mL) and 200 μL of buffer AL (lysis buffer) for 10 minutes at 70°C. After addition of 210 μL ethanol, the samples were applied to spin columns, washed three times with buffer AW (wash buffer), and eluted with 50 to 200 μL of 10 mmol/L Tris (pH 9.0). Concentration of DNA was determined by measuring extinction at 260 nm.
Polymerase chain reaction (PCR).RNA was transcribed into cDNA with reverse transcriptase (RT) (10 U/μg RNA, AMV reverse transcriptase, Promega, Madison, WI) using random primers (0.5 μg/μg RNA) in 50 mmol/L Tris-HCl, 50 mmol/L KCl, 10 mmol/L MgCl2 , 10 mmol/L dithiothreitol (DTT), 0.5 mmol/L spermidine, and 1 mmol/L deoxynucleoside triphosphates (dNTPs) at 37°C for 15 minutes and at 42°C for 45 minutes.13 14
PCR was performed with specific primers amplifying nucleotides 2306-2561 (Q1 and Q2) or 2306-2906 (Q1 and P2) of the cytoplasmic domain of the G-CSF receptor. The primer sequences were as follows: Q1: 5′-AACAGCTCAGAGACCTGTGGCCT-3′, Q2: 5′-CCAAGGGGCTGGCCTGGA-3′, P2: 5′-GGCCATTGGGTGGGGCTGGAT-3′. The numbering of the nucleotides refers to Fukunaga et al.15 The critical region of the G-CSF receptor is defined from nucleotide 2384-2429 and includes all mutations that have been reported, so far. Amplification of cDNA or DNA templates was performed in 25 μL of 10 mmol/L Tris-HCl, 50 mmol/L KCl, 1 mmol/L MgCl2 , 0.1 μmol/L primer, and 1 U of Taq Polymerase (Boehringer, Mannheim, Germany) for 25 cycles in a thermocycler (VariusV, Landgraf, Langenhagen, Germany). Each cycle consisted of denaturation for 40 seconds at 94°C, annealing for 60 seconds at 64°C, and extension for 60 seconds at 72°C. Fragments were analyzed on agarose gels stained with ethidium bromide.
Nonradioactive DNA sequencing.Sequencing of DNA was performed according to the dideoxynucleotide chain-termination method16 by cycle sequencing using DIG-labeled primer Q1 (sense) and Q2 (antisense) and the DIG Taq DNA sequencing kit from Boehringer. After direct blotting onto a membrane (GATC, Konstanz, Germany) and ultraviolet (UV) cross-linking, the DIG labeled fragments were detected with anti-DIG antibodies conjugated to alkaline phosphatase and chemiluminescent substrate. In a first screening, individual bands of PCR products were sequenced directly. To confirm the results, PCR products were ligated into a pCR-script SK(+) plasmid (Stratagene, La Jolla, CA) and cloned in Escherichia coli. After isolation of plasmid DNA containing the PCR product inserted into the multiple cloning site, individual clones were submitted to DNA sequencing.
Restriction enzyme analysis.PCR fragments generated with primers Q1 and P2 were digested for at least 2 hours with 2 U of the restriction endonuclease PvuII (Boehringer) and analyzed by agarose gel electrophoresis and ethidium bromide staining. PvuII recognizes the nucleotide sequence CAGCTG, which is destroyed by the point mutation at nucleotide position 2429.
RESULTS
Frequency of point mutations in the critical region of the G-CSF receptor and occurrence of AML.Point mutations in the G-CSF receptor cytoplasmic domain in neutrophils from two patients with congenital neutropenia have been described previously.11 17 To investigate the frequency of G-CSF receptor point mutations in patients with severe congenital neutropenia, we have tested neutrophils and mononuclear cells from 28 patients. The intracellular part of the G-CSF receptor including the critical region from nucleotide 2384 to 2429 was amplified from cDNA and genomic DNA of both neutrophils and mononuclear cells using specific primers in a PCR (Fig 1). PCR products were sequenced directly in both directions subsequent to cloning into a bluescript vector and transformation in E coli. Additionally, restriction enzyme analyses of PCR fragments were performed. The point mutation at nucleotide position 2429 destroys a PvuII restriction endonuclease recognition site. Using this technique, PCR products from neutropenic patients could be screened for this specific G-CSF receptor mutation before sequencing.
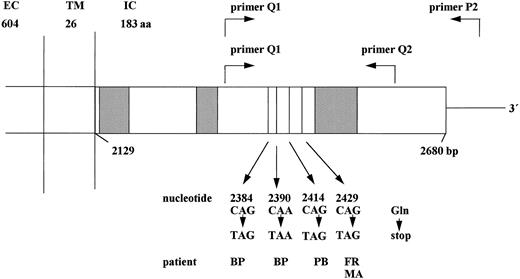
Schematic structure of the cytoplasmic domain of the G-CSF receptor gene. The cytoplasmic domain of the G-CSF receptor is shown including the critical region (nucleotides 2384-2429), which is amplified by primers Q1, Q2, and P2 in the RT-PCR reaction. The nucleotide positions given below indicate the point mutations detected in patients with congenital neutropenia. Hatched boxes indicate the conserved domains homologous to other members of the cytokine receptor family. EC, extracellular; TM, transmembrane; IC, intracellular domain; aa, amino acids; bp, base pairs.
Schematic structure of the cytoplasmic domain of the G-CSF receptor gene. The cytoplasmic domain of the G-CSF receptor is shown including the critical region (nucleotides 2384-2429), which is amplified by primers Q1, Q2, and P2 in the RT-PCR reaction. The nucleotide positions given below indicate the point mutations detected in patients with congenital neutropenia. Hatched boxes indicate the conserved domains homologous to other members of the cytokine receptor family. EC, extracellular; TM, transmembrane; IC, intracellular domain; aa, amino acids; bp, base pairs.
From the 28 congenital neutropenia patients analyzed, 24 did not show any mutation in the G-CSF receptor intracellular region from nucleotide position 2306 to 2561. Four congenital neutropenia patients displayed point mutations in the G-CSF receptor gene. These mutations could be detected at nucleotide position 2429 (patients FR and MA), or 2414 (patient PB), or 2384 and 2390 (patient BP) (Fig 1). All these mutations replace a C (cytosine) by a T (thymine), thereby introducing a stop-codon at amino acid position 731, 726, 718, or 716. Both direct sequencing of PCR products and sequencing of 20 (patients FR and MA) individual clones demonstrated that only one allele of the G-CSF receptor gene was affected by the point mutation. These point mutations were detectable both at the level of cDNA and of genomic DNA of the intracellular part of the G-CSF receptor in neutrophils and mononuclear blood and bone marrow cells. Intriguingly, the subgroup of the four congenital neutropenia patients with receptor mutations included those two patients who had developed secondary AML. The other two patients show no signs of MDS/AML at the present time.
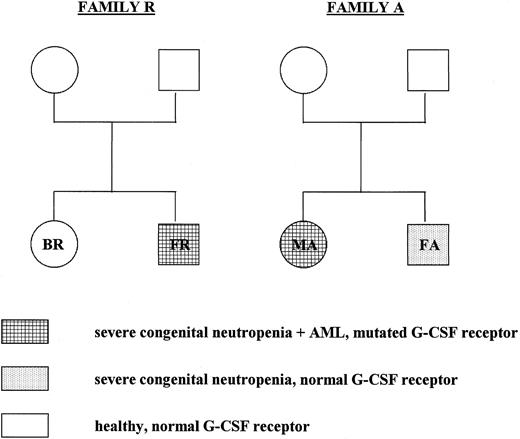
G-CSF receptor mutations are not inherited and occur at different stages of congenital neutropenia.Severe congenital neutropenia or Kostmann's syndrome has originally been reported as a recessive congenital disorder.1 2 We were interested whether the G-CSF receptor mutations were inherited or whether they occurred spontaneously. Therefore, we tested cells from six healthy family members of two congenital neutropenia patients with a mutated receptor. We analyzed both genomic DNA and cDNA isolated from neutrophils and mononuclear cells from the family members by restriction enzyme analysis, direct sequencing of PCR products, and sequencing of at least 20 individual clones of cloned PCR fragments. None of the healthy family members tested displayed any point mutation in the critical region of the G-CSF receptor cytoplasmic domain (Fig 2). The brother FA of patient MA suffers from congenital neutropenia, too, with no signs of MDS/AML. Restriction enzyme analysis, direct sequencing of PCR fragments, and sequencing of 32 individual clones demonstrated no G-CSF receptor mutation at the level of genomic DNA and cDNA isolated from neutrophils, blood mononuclear cells, and bone marrow cells from patient FA. Figures 3 and 4 show representative results of a restriction enzyme analysis and a direct sequencing reaction performed on cDNA isolated from neutrophils of the indicated patients and family members. Similar results were obtained with both cDNA and DNA analyzed from mononuclear cells from blood and bone marrow (data not shown).
Pedigree of the members of families A and R who have been tested for G-CSF receptor mutations. In both genomic DNA and cDNA isolated from neutrophils and mononuclear cells from the parents and siblings of patients FR and MA, the critical region in the cytoplasmic domain of the G-CSF receptor was analyzed by restriction enzyme analysis and DNA sequencing as described in the text.
Pedigree of the members of families A and R who have been tested for G-CSF receptor mutations. In both genomic DNA and cDNA isolated from neutrophils and mononuclear cells from the parents and siblings of patients FR and MA, the critical region in the cytoplasmic domain of the G-CSF receptor was analyzed by restriction enzyme analysis and DNA sequencing as described in the text.
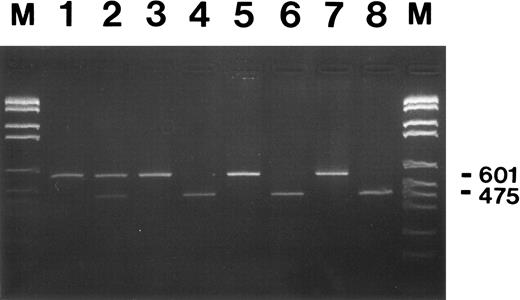
Restriction enzyme analysis of G-CSF receptor RT-PCR products from neutrophils of patient MA and her family members. Neutrophils from congenital neutropenia patient MA who developed secondary AML (lanes 1 and 2), her brother FA, also a congenital neutropenia patient, (lanes 3 and 4), their healthy mother (lanes 5 and 6), and father (lanes 7 and 8) were used for RNA isolation and subsequent RT-PCR analysis. PCR fragments amplified with primers Q1 and P2 are either shown undigested (lanes 1, 3, 5, and 7) or digested with PvuII (lanes 2, 4, 6, and 8) at 37°C overnight and after agarose gel electrophoresis and ethidium bromide staining. Numbers on the right indicate the undigested (601) and the digested (475) PCR fragments. The smaller digested fragment of 126-bp is not shown. DNA fragments lengths standard M.VI from Boehringer Mannheim was loaded in lanes M (2176, 1766, 1230, 1033, 653, 517, 453, 394, 298, 234 bp).
Restriction enzyme analysis of G-CSF receptor RT-PCR products from neutrophils of patient MA and her family members. Neutrophils from congenital neutropenia patient MA who developed secondary AML (lanes 1 and 2), her brother FA, also a congenital neutropenia patient, (lanes 3 and 4), their healthy mother (lanes 5 and 6), and father (lanes 7 and 8) were used for RNA isolation and subsequent RT-PCR analysis. PCR fragments amplified with primers Q1 and P2 are either shown undigested (lanes 1, 3, 5, and 7) or digested with PvuII (lanes 2, 4, 6, and 8) at 37°C overnight and after agarose gel electrophoresis and ethidium bromide staining. Numbers on the right indicate the undigested (601) and the digested (475) PCR fragments. The smaller digested fragment of 126-bp is not shown. DNA fragments lengths standard M.VI from Boehringer Mannheim was loaded in lanes M (2176, 1766, 1230, 1033, 653, 517, 453, 394, 298, 234 bp).
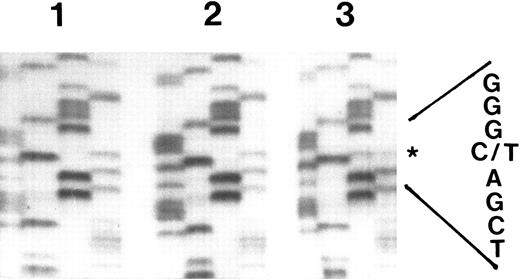
Nonradioactive direct sequencing of RT-PCR fragments of the G-CSF receptor cytoplasmic domain in myeloid cells from congenital neutropenia patients. G-CSF receptor cDNA from neutrophils of congenital neutropenia patient MA before (lane 2) and at the time point of diagnosis of AML (lane 3) and from her brother FA, who is also a congenital neutropenia patient, but who has not yet developed AML (lane 1), was amplified with primers Q1 and Q2 and sequenced directly. In each lane, the nucleotides are shown in the following order: GATC. The asterisk indicates the position of the point mutation.
Nonradioactive direct sequencing of RT-PCR fragments of the G-CSF receptor cytoplasmic domain in myeloid cells from congenital neutropenia patients. G-CSF receptor cDNA from neutrophils of congenital neutropenia patient MA before (lane 2) and at the time point of diagnosis of AML (lane 3) and from her brother FA, who is also a congenital neutropenia patient, but who has not yet developed AML (lane 1), was amplified with primers Q1 and Q2 and sequenced directly. In each lane, the nucleotides are shown in the following order: GATC. The asterisk indicates the position of the point mutation.
To determine the time point of occurrence of G-CSF receptor mutations in the course of the life of the patients, we tested their neutrophils and mononuclear cells at various time points. Again, PCR products generated from both genomic DNA and cDNA were investigated by restriction enzyme analysis, direct sequencing, and sequencing of individual clones. Interestingly, in patient MA, the point mutation could not be detected up to 3 months in neutrophils, mononuclear cells, or bone marrow cells before the diagnosis of AML at the age of 9 years when the mutated receptor was detectable in the blast cells (Figs 4 and 5). In patient FR who has been previously described in detail,11 the mutated G-CSF receptor was detectable in bone marrow cells before monosomy 7 was diagnosed at the age of 22 years. Patient BP displayed no G-CSF receptor point mutations in a sample of bone marrow mononuclear cells obtained in 1987 at the age of 8 years and before r-metHuG-CSF treatment. At a routine examination in 1995, two different point mutations could be detected in neutrophils and mononuclear cells, while the patient shows no signs of MDS/AML, so far. In neutrophils and blood mononuclear cells from patient PB, the mutated G-CSF receptor has been detected in samples since 1992 up to the present time. From patient PB, no samples are available that were taken at an earlier time point.
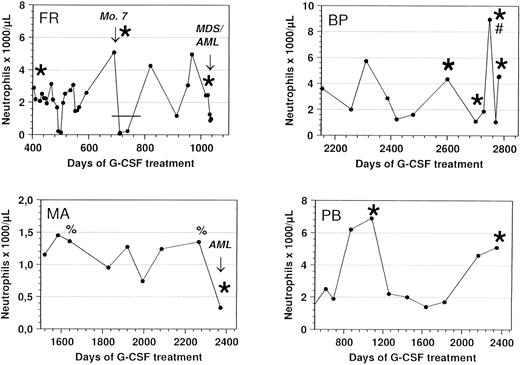
Response of congenital neutropenia patients with G-CSF receptor mutations to r-metHuG-CSF treatment. ANC of patients FR, MA, BP, and PB undergoing treatment with r-metHuG-CSF are depicted. The patients received a daily dose of 1 μg/kg/d (FR and BP), 3 μg/kg/d (PB), and 20 μg/kg/d (MA). In patient BP, the dose was increased up to 10 μg per day in March 1996 (#) to mobilize progenitor cells into the peripheral blood before leukaphoresis. Bone marrow mononuclear cells from patient BP taken before the start of G-CSF treatment in 1987 showed no G-CSF receptor mutation. In patient FR, G-CSF therapy was interrupted when monosomy 7 was diagnosed as indicated by the bar (-). Sequencing of the G-CSF receptor at different time points demonstrated either a normal (%) or a mutated (*) G-CSF receptor.
Response of congenital neutropenia patients with G-CSF receptor mutations to r-metHuG-CSF treatment. ANC of patients FR, MA, BP, and PB undergoing treatment with r-metHuG-CSF are depicted. The patients received a daily dose of 1 μg/kg/d (FR and BP), 3 μg/kg/d (PB), and 20 μg/kg/d (MA). In patient BP, the dose was increased up to 10 μg per day in March 1996 (#) to mobilize progenitor cells into the peripheral blood before leukaphoresis. Bone marrow mononuclear cells from patient BP taken before the start of G-CSF treatment in 1987 showed no G-CSF receptor mutation. In patient FR, G-CSF therapy was interrupted when monosomy 7 was diagnosed as indicated by the bar (-). Sequencing of the G-CSF receptor at different time points demonstrated either a normal (%) or a mutated (*) G-CSF receptor.
In vivo response to G-CSF of patients with G-CSF receptor mutations.G-CSF treatment was started in patient MA at the age of 2 years, patient FR at the age of 20 years, patient PB at the age of 7 years, and patient BP at the age of 10 years. During the course of treatment with r-metHuG-CSF, the congenital neutropenia patients who acquired the G-CSF receptor mutations showed no changes in their response to treatment with r-metHuG-CSF (Fig 5). The same doses of G-CSF were given before and after the detection of the receptor mutation. Patients FR and BP required G-CSF doses of 1 μg/kg/d, patient PB 3 μg/kg/d, and patient MA 20 μg/kg/d to maintain an effective ANC of more than 200/μL, which results in a decrease in the number of bacterial infections and in an improvement of the quality of life. In patient FR, treatment with r-metHuG-CSF was interrupted when monosomy 7 was diagnosed. Severe bacterial infections necessitated the reestablishment of r-metHuG-CSF therapy and resulted in an effective increase in ANC, although the G-CSF receptor mutation was already detectable (Fig 5). To summarize, these four patients received various doses of r-metHuG-CSF over different time periods without an alteration in their biological response to G-CSF.
DISCUSSION
Data from the International Registry for severe chronic neutropenia indicate that 16 of 220 congenital neutropenia patients developed secondary AML, while none of the patients with cyclic (n = 78) or idiopathic (n = 122) neutropenia developed AML.7 Recent reports of G-CSF receptor mutations in patients with congenital neutropenia11,17 raised the questions whether a point mutation within the G-CSF receptor gene is involved in the pathomechanism of congenital neutropenia or whether it is responsible for the development of leukemia secondary to congenital neutropenia. The mutations described so far, are located between amino acid position 716 and 731 (nucleotide positions 2384 and 2429, respectively) in the cytoplasmic domain of the G-CSF receptor, which we assume to represent a critical region for point mutations. Herein, we have demonstrated that point mutations in this critical region of the intracellular part of the G-CSF receptor occur in a subgroup of patients with congenital neutropenia. Four of 28 congenital neutropenia patients displayed such mutated G-CSF receptors, two of whom developed AML. Similar results have been reported by Touw et al.18 They demonstrated five of 25 cases with mutations in the G-CSF receptor cytoplasmic domain, four of whom developed AML. Because all patients with AML secondary to congenital neutropenia (n = 5) tested up to date by I.P. Touw and by us demonstrated G-CSF receptor mutations, these mutations are suspected to be involved in the pathogenesis of AML. The absence of these G-CSF receptor mutations in the majority of patients indicates that mechanisms other than G-CSF receptor mutations are the cause of severe congenital neutropenia.
Severe congenital neutropenia is a very rare disease that has been described as autosomal recessive inherited.1,2 Smith et al19 reported a family with the mother and three children suffering from severe chronic neutropenia suggesting a probable autosomal dominant pattern of inheritance. Among the children of this family were identical twins, with one of them developing MDS transforming into AML secondary to congenital neutropenia. Among the patients we have tested, there is a family with two congenital neutropenia patients (FA and MA). Sequencing of the critical G-CSF receptor cytoplasmic region showed no mutations in myeloid cells from the healthy parents, as well as from the patient FA (brother of MA) with congenital neutropenia. Only the leukemic cells from patient MA within this family demonstrated the G-CSF receptor mutation. In the second family, the healthy parents and sister of patient FR were tested for G-CSF receptor mutations, and all healthy family members displayed an intact cytoplasmic region of the G-CSF receptor. These data suggest that the G-CSF receptor mutation was not inherited from one or both of the parents and is not present in the siblings of the patients. The absence of G-CSF receptor mutation in the brother of patient MA, who suffers from congenital neutropenia, is the strongest proof that the described point mutations in the G-CSF receptor cytoplasmic domain are a spontaneous event in the course of the disease. Moreover, bone marrow cells taken from patient MA in 1988 and T-cell lines established from the same patient in 1990, did not show the mutated G-CSF receptor in both direct sequencing and sequencing of 34 individual clones. Our data suggest that in this familial form of congenital neutropenia, the G-CSF receptor defect is not the primary cause of the disease.
However, other genetic defects have been reported in patients with AML secondary to congenital neutropenia. In a subgroup of congenital neutropenia patients with MDS/AML, mutations in the ras gene have been reported to also be an important step in leukemogenesis.20 In 5 of 11 patients with MDS/AML secondary to congenital neutropenia, specific ras mutations could be detected. Most likely, ras mutations are a late event in leukemogenesis. Among the 4 patients we have investigated harboring the G-CSF receptor point mutation, patient FR was negative for ras mutations (patient number 5 in the publication of Kalra et al20 ), while the other 3 have not been tested. Cytogenetics were performed on all four patients with the mutated G-CSF receptor showing no chromosomal aberrations present in patients BP and PB, while patient FR displayed a monosomy 7 and patient MA showed the following karyotype: 46,XX/46,XX,der(1)t(1; 3)(p34; q23-25),add(5)(q35). Further studies showed increased tyrosine phosphorylation and activation of JAK2 in patient BP and slightly increased G-CSF serum levels in patients BP and FR. Increased JAK2 phosphorylation and G-CSF serum levels have been reported to be a common feature in patients with severe congenital neutropenia undergoing G-CSF therapy.8 10
In all patients with a mutated G-CSF receptor, the mutation presented only in one allele of the G-CSF receptor gene. Therefore, on the protein level, this would result in the expression of both forms of the G-CSF receptor protein forming homo and heterodimers on the cell surface. A model has been proposed with the truncated receptor protein exerting a dominant negative effect over the normal receptor protein with regard to the transduction of a maturation signal.11 Indeed, transfection experiments introducing the mutated G-CSF receptor gene into murine leukemic cell lines demonstrated that these cells proliferate in response to G-CSF, but do not differentiate as wild-type transfected cells do.11,17 Functional domains of the G-CSF receptor have been described and the C-terminal part that is truncated by the point mutations has been linked to the transduction of maturation signals.21-23 In congenital neutropenia patients, the situation is more complex. In this report, we demonstrate that after acquisition of the G-CSF receptor mutation, congenital neutropenia patients continue to respond to the same dose of r-metHuG-CSF with an increase in ANC similar to the one before the mutation was detectable. These data suggest that the neutrophilic progenitor cells expressing both the normal and the mutated G-CSF receptor alleles would still generate the same degree of proliferation and differentiation as before the mutation, despite the decrease in the formation of normal homodimeric receptor complexes. A second scenario could be hypothesized: There are two subpopulations of progenitor cells, one expressing the mutated and the second expressing the normal G-CSF receptor. The population of cells with the mutated receptor might have a selective advantage over other populations. However, the fact that the response to G-CSF is unchanged would argue against this hypothesis. Additionally, there is no increase in nondifferentiating myeloid cells in the bone marrow, which we would expect from the response of a subpopulation of cells expressing only the mutated receptor.
Taken together, our data suggest that point mutations in the critical region of the G-CSF receptor gene in congenital neutropenia patients occur spontaneously and are not the primary cause of neutropenia. The contribution of these mutations to leukemogenesis remains to be investigated.
Supported in part by the Madeleine-Bühler-Kinderkrebs-Stiftung (Fürth, Germany), Grant No. DFG We 942/4-1 from the Deutsche Forschungsgemeinschaft (Bonn, Germany), and Amgen (Thousand Oaks, CA).
Address reprint requests to K. Welte, MD, Pädiatrische Hämatologie und Onkologie, Medizinische Hochschule Hannover, D-30623 Hannover, Germany.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal