To the Editor:
Aplan et al1 recently reported a site-specific DNA cleavage induced by topoisomerase II (Topo II) inhibitors within the MLL gene breakpoint cluster region on chromosome 11q23. The finding was initially identified by Southern blot analysis of circulating blasts taken from a case of T-cell acute lymphoblastic leukemia (T-ALL) 16 hours after the induction of multiagent chemotherapy, including doxorubicin, a known inhibitor of Topo II. The investigators subsequently reproduced the same pattern of DNA fragmentation by in vitro culture of malignant cell lines with doxorubicin and etoposide, showing that cleavage was not restricted to the T-lymphoid lineage because it was observed in B-lymphoid, myeloid, and nonhematopoietic malignant cell lines and even in the peripheral blood lymphocytes of normal individuals. They suggested that Topo II inhibitor induced MLL cleavage correlated with sensitivity to epidophyllotoxins and that lymphoid lines may be more sensitive to such cleavage, because it was observed in 4 of 6 malignant T-cell lines and 3 of 4 B-cell lines, but in only 1 of 6 myeloid cell lines and 1 of 6 solid tumor cell lines. We show here that an identical MLL DNA cleavage occurs relatively frequently in de novo AML and that it is not restricted to cases analyzed after induction with Topo II inhibitors.
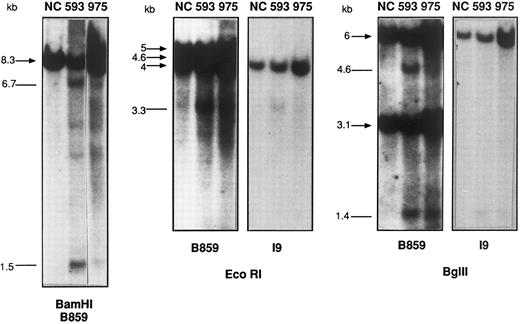
Southern blot analysis of BamHI-, EcoRI-, and Bgl II-digested DNA from patients no. 593 and 975, compared with a placenta negative control (NC). The sizes (in kilobases) of the germline bands are indicated by arrows and the rearranged bands as lines. The 2 faint ≈4- and ≈4.3-kb bands in the BamHI digest from patient no. 593 are probably due to star activity, because they are not seen in other digests.
Southern blot analysis of BamHI-, EcoRI-, and Bgl II-digested DNA from patients no. 593 and 975, compared with a placenta negative control (NC). The sizes (in kilobases) of the germline bands are indicated by arrows and the rearranged bands as lines. The 2 faint ≈4- and ≈4.3-kb bands in the BamHI digest from patient no. 593 are probably due to star activity, because they are not seen in other digests.
Having demonstrated that MLL rearrangements occur frequently in de novo acute myeloid leukemia (AML) of diverse French-American-British (FAB) subtypes,2 we have instituted routine screening for MLL rearrangement for patients included in the EORTC/GIMEMA AML 10 trial (No. 06931) by Southern blot hybridization of BamHI and HindIII-digested DNA with an MLL cDNA probe, B859, encompassing exons 5 through 113 (kindly provided by Guiseppe Cimino, La Sapienza, Rome, Italy). Briefly, this trial includes patients 15 to 60 years of age with untreated, newly diagnosed AML of all FAB subtypes other than M3. Seventy clinical centers from 9 countries submit patients to AML 10, but the current molecular analysis is restricted to data from the French centers, which represent approximately 10% of patients. Parallel molecular analyses are performed on Dutch, Belgian, and Italian patients. Patient recruitment for the French centers started in January 1994 and prospective molecular screening began in January 1996. All patients with cryopreserved material obtained between January 1994 and January 1996 were studied retrospectively. MLL status was assessed by Southern blotting in 62 of 96 (65%) patients from the French centers included in the AML 10 protocol between January 1994 and April 1996. Whereas 9 of 62 (15%) of patients were shown to have major MLL rearrangements, a further 3 (5%) patients showed minor, biallelic rearranged bands with both enzymes, with identically sized BamHI fragments to those identified by Aplan et al.1 The intensity of the rearranged bands corresponded to approximately 5% to 10% of the DNA (Fig 1). Further analysis of EcoRI and Bgl II digested DNA (Fig 1) from 2 of these 3 cases with the B859 cDNA and an intron 9 genomic probe, I9,2 showed that the MLL breakpoint localized to the 2-kb EcoRI-Bgl II genomic fragment containing exons 9 and 10 of the breakpoint cluster region, with the size of the rearranged fragments identified (Table 1) corresponding exactly to the breakpoint putatively localized to a Topo II consensus recognition sequence 1,470 bp 5′ of the BamHI site at the 3′ extremity of the breakpoint cluster region.1 Both rearranged bands were of equivalent intensity in patients no. 593 and 722 (≈10%; Fig 1 and data not shown), whereas the 5′ fragment was relatively weak in Bgl II digests from patient no. 975, with the 3′ fragment more clearly visible when probed with B859 (Fig 1). The amount of cleaved DNA was lower in this patient and no clear rearranged band could be discerned with the 300-bp genomic I9 probe. BamHI analysis of patient no. 593 showed 2 further minor rearrangements of approximately 4.3 and 4 kb (Fig 1), which are compatible with a second cleavage site, but these bands were not detected with other digests and may therefore represent star activity. The possible presence of a minor translocation leading to transcription of one of the common AML fusion transcripts was analyzed in patient no. 593, but RT-PCR analysis failed to show MLL-AF6, MLL-AF9, MLL-ENL, MLL-ELL, or MLL duplication (data not shown).
Clinical (A) and Molecular (B) Characteristics of AML Patients With MLL Cleavage
| A. Patient Characteristics . | |||||
|---|---|---|---|---|---|
| . | . | . | |||
| Patient No. . | Age/Sex . | FAB . | WBC . | % Age Blasts . | Cytogenetics . |
| 593 | 35/M | M5 | 104 | 96 | 46,XY [25] |
| 722 | 38/M | M4 | 160 | 80 | 46,XY[1]/47,XY, +8 [39] |
| 975 | 55/M | M4 | 23.5 | 42 | 46,XY [20] |
| B. Southern Blot Hybridization With the B859 cDNA and 19 Genomic Probes | |||||
| A. Patient Characteristics . | |||||
|---|---|---|---|---|---|
| . | . | . | |||
| Patient No. . | Age/Sex . | FAB . | WBC . | % Age Blasts . | Cytogenetics . |
| 593 | 35/M | M5 | 104 | 96 | 46,XY [25] |
| 722 | 38/M | M4 | 160 | 80 | 46,XY[1]/47,XY, +8 [39] |
| 975 | 55/M | M4 | 23.5 | 42 | 46,XY [20] |
| B. Southern Blot Hybridization With the B859 cDNA and 19 Genomic Probes | |||||
| Restriction Enzyme | Germline Fragments (kb) | Fragments Induced by Topo II Inhibitor (kb)* |
| BamHI | 8.3 | 6.7, 1.5 |
| HindIII | 15 | 12.5, 2.5 |
| BgI III | 3.1, 6 | 3.1, 4.6, 1.4 |
| EcoRI | 4.6, 5, 4 | 4.6, 5, (0.7), 3.3 |
| Restriction Enzyme | Germline Fragments (kb) | Fragments Induced by Topo II Inhibitor (kb)* |
| BamHI | 8.3 | 6.7, 1.5 |
| HindIII | 15 | 12.5, 2.5 |
| BgI III | 3.1, 6 | 3.1, 4.6, 1.4 |
| EcoRI | 4.6, 5, 4 | 4.6, 5, (0.7), 3.3 |
The 3′ fragment identified by the 19 genomic probe is underlined. The 0.7-kb fragment in EcoRI digests is unlikely to be detected with a cDNA probe in view of its size and limited exon content.
In contrast to the T-ALL reported by Aplan et al,1 samples from all 3 patients were taken before the induction of chemotherapy. Clinical and biologic details of the patients are shown in Table 1. All 3 were men and presented with monocytic M4/5 AML, in which karyotypic 11q23 abnormalities are found most frequently.4 Questioning did not show any prior malignancy or chemotherapy and there was no apparent exposure to toxic agents. Cytogenetic analysis was normal in 2 patients and the third demonstrated an isolated trisomy 8. The only finding of possible relevance was the presentation white blood cell (WBC) count, because both patients with easily detectable MLL cleavage had marked leukocytosis at diagnosis. The original case described by Aplan et al1 also showed a high presentation blast count, as is commonly found in T-ALL.
We therefore show that 5% of patients with AML demonstrate MLL cleavage in a minor proportion of blasts tested before the induction of chemotherapy. This observation has obvious significance for the interpretation of Southern blots in routine molecular diagnostic screening, when it is conceivable that a proportion of apparent MLL rearrangements may in fact be MLL cleavage. It is also interesting with regard to the known capacity of Topo II inhibitors to induce secondary leukemias with MLL rearrangement, as discussed by Aplan et al.1 It would obviously be interesting to determine whether MLL cleavage is more common in relapsed AML. Our data also suggest that MLL cleavage may occur preferentially in cases with a rapid cell doubling time. This intrinsic propensity may be further accentuated by treatment with Topo II inhibitors. Routine molecular screening of AML at diagnosis, as undertaken in the EORTC/GIMEMA AML 10 trial, will allow us to determine whether MLL cleavage has prognostic significance, particularly with regard to secondary malignancies, provided that this particular Southern blot profile is recognized.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal