Abstract
High efficiency retroviral-mediated gene transfer to rhesus CD4+ peripheral blood lymphocytes (PBL) was accomplished using an optimized transduction protocol using a gibbon ape leukemia virus (GaLV) envelope-containing packaging cell line PG13. Engineered CD4+ PBL were administered to three nonmyeloablated animals in three or four separate infusions over 9 months. Polymerase chain reaction (PCR) demonstrated in vivo reconstitution of the genetically engineered CD4+ PBL at levels between 1% and 10% of the circulating leukocytes. This level of gene marking indicates that up to 30% of endogenous circulating CD4+ cells can be genetically engineered. The high levels of marked lymphocytes persist for the first 3 weeks following reinfusion then decline to ≤ 0.1% over the next 21 weeks. Lymph node (LN) biopsies were performed to determine if the engineered CD4+ lymphocytes could traffic to lymphoid tissues. Marked lymphocytes were detected in LN biopsies 100 days following reinfusion of the transduced cells. Expression of retroviral vector-derived sequences was detected by reverse transcriptase (RT)-PCR analysis from CD4-enriched lymphocytes that were activated by culturing in the presence of recombinant interleukin-2 (rIL-2). A humoral immune response to fetal bovine serum (FBS) was detected in all animals following the second administration of the culture expanded CD4+ lymphocytes. No antibody response was detected to the neomycin-resistance (NeoR) transgene, the murine retroviral group-specific antigen (gag), or GaLV envelope (env) proteins.
PERIPHERAL BLOOD LYMPHOCYTES (PBLs) are an attractive target for the introduction of genes into the lymphohematopoietic system for gene therapy. PBL have previously been successfully used for gene transfer of therapeutic genes in tissue culture, in preclinical protocols, and in clinical protocols for disorders of the immune system, such as adenosine deaminase (ADA) deficiency and the acquired immune deficiency syndrome (AIDS).1-6 Clinical results suggest that genetically engineered lymphocytes can contribute to reestablishing immune function in some immunodeficient patients.7 8 A limitation to the use of PBL as a target for gene therapy is that very little is known about their life span in vivo, the trafficking pattern of these cells, and the effects of in vivo expression of foreign genes in PBL.
In an early study evaluating the use of lymphocytes as targets for gene therapy, rhesus macaque PBL were transduced with retroviral vectors containing the human ADA gene and transduced cells were detected for up to 727 days following a single infusion.9 Several studies concerning the in vivo life span of human lymphocytes have recently been published. In human immunodeficiency virus (HIV)-1 infected patients, it was reported that the life span of PBL can vary widely from 15 days to 100 days.10,11 Long-term persistence of mature T cells (>18 months) has also been observed in a clinical trial in which Epstein-Barr virus (EBV)-specific cytotoxic T lymphocytes were administered to bone marrow transplant recipients.12 These conclusions, however, are based on studies involving PBL isolated from severely immunocompromised individuals and, therefore, may not accurately reflect the “true” life span of a lymphocyte. Studies of the life span of mature lymphocytes in cancer patients suggest that mature lymphocytes may persist as long as 2 years.13-16 Results of an ADA clinical gene therapy protocol further suggest that a mature T cell can have a protracted life span in vivo (>4 years).7-13 Taken together, all of these studies support the contention that certain lymphocytes have a significant life span in vivo.
A potential obstacle to the success of human gene therapy trials is the generation of an immune response to the genetically modified somatic cells. It was recently reported that HIV-1 infected patients receiving HIV-1 specific CD8+ T cells can develop cytotoxic T lymphocytes specific to a novel fusion protein encoded by the gene-marking vector.17 Moreover, the ex vivo transduction and growth of the cells in tissue culture systems can be associated with immune responses. In one ex vivo lymphocyte marking study in HIV-1 discordant identical twins, some patients developed an antibody response to the fetal bovine serum (FBS) used for cell culture (R. Walker, personal communication, March 1996). There have also been antibody responses observed to retroviral vector proteins in ADA patients receiving transduced lymphocytes (R. M. Blaese, personal communication, June 1996). All three of these reports have been in immunocompromised patients; it is, therefore, of interest to determine whether the same responses can be observed in immunocompetent individuals.
In this study, we sought to more fully define the in vivo survival kinetics, distribution, and expression pattern of CD4+ lymphocytes using a nonhuman primate animal model. A previously developed protocol was used to successfully transduce both human and nonhuman primate PBL using a gibbon ape leukemia virus (GaLV) envelope-containing vector.18 We report that, following multiple apheresis/transduction procedures, transduced CD4+ lymphocytes can accumulate up to 30% of endogenous circulating CD4+ cells, have a minimum life span of several months, and can traffic to the lymph nodes (LNs) of nonmyeloablated and nonimmunocompromised rhesus macaques. In addition, an antibody response to FBS was detected, but no antibody response was detected to either the NeoR gene product or viral antigens.
MATERIALS AND METHODS
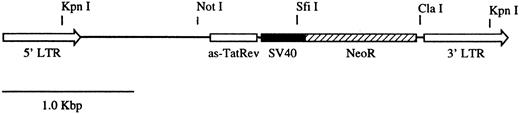
Cell culture and retroviral vectors.The packaging cell line used for these studies was PG13 (American Type Culture Collection, Rockville, MD, CRL-10686). PG13 cells were grown in supplemented Dulbecco's modified Eagle's medium (DMEM) as previously described.18 Cells were cultured at 37°C in a humidified 5% CO2 + 95% air incubator. The retroviral vector used for the marking study is denoted as GC-(anti–Tat/Rev)SNDC (Fig 1) and has previously been described.19 This vector was produced in the PG13 (GaLV) packaging cell line and had a titer measured on HeLa cells of 1.6 × 106 G418-resistant colony-forming units per milliliter.
Map of the retroviral vector GC-(anti–Tat/Rev) SNDC. The vector contains an antisense Tat/Rev gene that encompasses the entire Rev sequence and the second exon of the Tat gene. The antisense gene is expressed from a MoMLV 5′LTR. The vector also contains the neomycin resistance gene (NeoR) driven by the SV40 early promoter.
Map of the retroviral vector GC-(anti–Tat/Rev) SNDC. The vector contains an antisense Tat/Rev gene that encompasses the entire Rev sequence and the second exon of the Tat gene. The antisense gene is expressed from a MoMLV 5′LTR. The vector also contains the neomycin resistance gene (NeoR) driven by the SV40 early promoter.
CD4+ lymphocyte harvesting, culture, transduction, and reinfusion.Peripheral blood mononuclear cells (PBMC) were obtained from healthy rhesus macaques by a leukapheresis protocol using a modified CS3000 Plus Blood Cell Separator (Baxter Healthcare, Deerfield, IL). A detailed description of the apheresis protocol has previously been published.20 The MN cells were further purified using Ficoll-Paque (Pharmacia Biotech, Uppsala, Sweden) density centrifugation and cleared of contaminating red blood cells at 4°C with ACK red blood cell lysing buffer (Biofluids Inc, Rockville, MD). An enriched population of CD4+ lymphocytes was isolated by immunomagnetic depletion of CD8+ cells (Dynal, Great Neck, NY). As previously described, the CD4-enriched lymphocytes were seeded at 2 × 106 cells per mL in 6-well plates at 2 mL per well in recombinant human interleukin-2 (rhIL-2) supplemented AIM-V medium.18 Following a 24-hour incubation, the cells were transduced three times at 24-hour intervals with the antisense Tat/Rev retroviral vector at an moi of 1.0. As previously described, the transduction protocol consisted of two rounds of optimized retroviral transduction and a single round of standard transduction.18 The cells were expanded for 3 additional days and before reinfusion, resuspended in Hank's buffered saline solution (HBSS) containing 2% heat-inactivated FBS and 10 U/mL heparin and infused intravenously over 5 to 10 minutes into the rhesus macaque.
Analysis for retrovirus vector positive cells.The in vitro transduction efficiency of the retroviral vector preparation was evaluated by semiquantitative polymerase chain reaction (PCR) for the NeoR gene using DNA isolated from the lymphocytes immediately before reinfusion. The in vivo marking of PBL was determined at various time points by PCR analysis for the NeoR gene using genomic DNA isolated from PBMC. The trafficking of marked lymphocytes to the inguinal LNs by PCR analysis was performed using genomic DNA isolated from LN biopsies. Sorted CD4+ and CD8+ lymphocytes were isolated using a Coulter Elite flow cytometer (Coulter, Hialeah, FL) and evaluated by PCR for provirus. Total genomic DNA was isolated from all sources using the Nucleon II DNA isolation kit (ScotLab Bioscience, Shelton, CT). A total of 200 ng of genomic DNA was used per PCR reaction. The primer sequences used to amplify the NeoR gene were: 5′-GATAGAAGGCGATGCGCGCGAATCG-3′, and 5′-TCCATCATGGCTGATGCAATGCGCTGCGAATCG-3′. The PCR reactions were performed under the following conditions: initial denaturation at 94°C for 2 minutes; followed by 30 cycles of amplification with denaturation (94°C for 30 seconds), annealing (65°C for 1 minute), elongation (72°C for 30 seconds); concluding with an incubation at 72°C for 5 minutes followed by cooling to 4°C. The genomic DNA was also used to PCR the β-actin gene using the following PCR primers: 5′-CATTGTGATGGACTCCGGAGACGG-3′, and 5′-CATCTCCTGCTCGAAGTCTAGAGC-3′. The PCR reaction for β-actin was performed as follows: initial denaturation at 94°C for 2 minutes; followed by 30 cycles of amplification with denaturation (94°C for 30 seconds), annealing (58°C for 1 minute), elongation (72°C for 1.5 minutes); concluding with an incubation at 72°C for 5 minutes and then cooled to 4°C. The PCR products were detected by Southern hybridization with 32P-labeled oligonucleotides as follows: NeoR (5′-CCAGGCTCAAGGCGCGCATGC-3′) and β-actin (5′-CATCTCCTGCTC-GAAGTCTAGAGC-3′). The gene marking efficiency was quantitated by conversion of radioactivity to photo-stimulated luminescence using a PhosphoImager (Molecular Dynamics, Sunnyvale, CA) and direct comparison between the β-actin control samples and the neomycin phosphotransferase PCR results.
Analysis of expression of antisense sequences following reinfusion.Expression of the antisense TAT/REV was analyzed by a reverse transcriptase (RT)-PCR assay. CD4-enriched lymphocytes were isolated from 20 mL of PB by Ficoll-Hypaque density gradient centrifugation (BioWhittaker, Walkersville, MD), and cultured for up to 7 days as described above. On days 0, 3, and 7, equivalent aliquots of cells were removed and total cellular RNA was isolated with TRIzol reagent (Life Technologies, Grand Island, NY). A total of 500 ng of total cellular RNA was reverse transcribed at 42°C for 60 minutes with AMV-RT (Promega, Madison, WI). One tenth of the reverse transcription product was subjected to PCR for the antisense TAT/REV sequences using primers (5′-ATCTCCTATGGCAGGAAGA-AGCGG-3′, 5′-TGTGGCATTGAGCAAGCTAACAG3′).
Western blotting.Western blot analysis for the retroviral vector was performed by concentrating the retroviral supernatant. To this end, 200 mL of freshly isolated PG13 packaging cell line supernatant was centrifuged in an SW41 rotor at 41K for 4 hours at 4°C. The retroviral vector pellets were resuspended in a total volume of 200 μL of phosphate-buffered saline (PBS). The concentrated retrovirus preparation was electrophoresed on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel and electroblotted to Hybond-ECL (Amersham, Arlington Heights, IL) for Western analysis. Plasma samples from the rhesus macaques were diluted 1:500 and 1:1,000 in PBS. The plasma from a rhesus macaque exposed to replication-competent retrovirus21 was used as a positive control and was diluted 1:1,000. Plasma from a naive animal was used as a negative control. A rabbit antimonkey whole serum antibody (Accurate Chemical, Westbury, NY) was used as the secondary antibody at a 1:1,000 dilution in PBS, and a goat antirabbit IgG (H + L) labeled with peroxidase was used for detection by the ECL system (Amersham, Arlington Heights, IL).
Western blots for the neomycin phosphotransferase type II protein (NPTII) were performed by electrophoresis of 0.5 mg of purified NPTII (5 Prime - 3 Prime, Boulder, CO) on a 10% SDS polyacrylamide gel followed by detection with the same antibodies as discussed above. The positive control in this experiment was serum from an animal that had previously been immunized with purified NPTII protein and had high titer NPTII antibodies (H. Muslow, unpublished observations, May 1996). Both preimmune plasma from the experimental animals and plasma from naive animals were used as negative controls for this assay.
Ouchterloney immunodiffusion assay.Immunodiffusion assays were performed for NPTII, FBS, and bovine serum albumin (BSA) using the titering T plates (Inova Diagnostics, San Diego, CA). A total of 1 mg of purified NPTII, 2.5 mg of purified BSA, or 50 μL of undiluted FBS were incubated in the presence of 50 μL of plasma from the transduced or naive animals. The immunoprecipitate was allowed to form for 3 days at room temperature. The plates were read by visualization of the immunoprecipitated band.
ELISA for NPTII antibodies.96-well plates were coated with purified NPTII protein by addition of 100 ng/well of NPTII protein in HCO3 buffer (pH 9.6). Control wells were coated with 1% human serum albumin (HSA) in HCO3 buffer (pH 9.6). After binding, the plates were washed three times in PBS + 0.05% Tween20, and blocked with PBS + 1% HSA for 1 hour at 37°C. The plates were again washed three times in PBS + 0.05% Tween20 before incubation with dilutions of plasma from the transduced animals, the positive control animal, or a naive animal (dilutions ranged from 1:100 to 1:12,800 in PBS + 1% HSA). The plasma samples were incubated at 37°C for 1 hour. The plates were washed four times in PBS + 0.05% Tween20 and incubated with a 1:2,000 dilution of mouse antihuman IgG (H + L) alkaline phosphatase (Southern Biotechnology, Birmingham, AL) in PBS + 1% HSA at 37°C for 1 hour. The plates were washed four times in PBS + 1% HSA before incubation with the diethanolamine + PNPP color development substrate solution (Pierce Chemical, Rockford, IL). The enzyme-linked immunosorbent assay (ELISA) plates were read at 405 nm in an E-max plate reader (Molecular Dynamics, Sunnyvale, CA).
RESULTS
In vivo reconstitution of transduced CD4+ cells.To address the issues of the persistence and distribution of mature CD4+ lymphocytes in vivo, gene marking studies were performed using autologous CD4+ lymphocytes in three normal rhesus macaques. The gene marking studies were initiated using an antisense HIV-1 gene therapy vector GC-(anti-Tat/Rev)SNDC (Fig 1). The vector contains a long terminal repeat (LTR) promoted antisense Tat/Rev gene and an internal SV40 virus early region promoter that drives the expression of the NeoR gene. Lymphocytes were isolated from rhesus macaques using a modified small chamber leukapheresis protocol20 and enriched for CD4+ cells by immunomagnetic depletion of CD8+ cells. After a 24-hour incubation in IL-2–supplemented medium, lymphocytes were transduced using an optimized retroviral-mediated transduction protocol.18 After the transductions, the lymphocytes were expanded for 3 days in the presence of rhIL-2, washed in PBS, and then reinfused into the animals. Greater than 90% of the cells reinfused expressed CD4 by flow cytometric analysis. The CD4+ lymphocyte transduction protocol was repeated as many as four times for each of the three animals.
As shown in Table 1, between 0.8 × 108 and 6.2 × 108 (mean, 3.8 × 108) CD4-enriched cells were reinfused into each animal following culture. This represented a cell reinfusion dose of 1.4 × 107 to 9.2 × 107 cells/kg (mean, 6.6 × 107 cells /kg). The dose of reinfused cells is roughly equal to the total number of circulating CD4+ lymphocytes before any treatment. Semiquantitative PCR analysis determined an in vitro transduction efficiency for the neomycin phosphotransferase gene between 12% and 35% (mean, 24%). Therefore, the estimated number of transduced CD4+ lymphocytes reintroduced into the animals ranged from 0.2 × 108 to 1.7 × 108 cells.
Transduction and Administration of CD4-Enriched PBL
| Animal . | Apheresis/Transduction (days between Aph/Trdx) . | Total No. of CD4+ Cells Reinfused . | No. Cells/Kg . | % Trdx . |
|---|---|---|---|---|
| RQ1114 | No. 1 | 3.5 × 108 | 5.6 × 107 | 20 |
| No. 2 (28 days) | 1.4 × 108 | 2.2 × 107 | 33 | |
| No. 3 (172 days) | 6.2 × 108 | 8.8 × 107 | 28 | |
| No. 4 (83 days) | 5.0 × 108 | 6.1× 107 | ND | |
| RQ900 | No. 1 | 0.8 × 108 | 1.4 × 107 | 30 |
| No. 2 (35 days) | 5.2 × 108 | 9.1 × 107 | 14 | |
| No. 3 (126 days) | 5.6 × 108 | 9.2 × 107 | 12 | |
| No. 4 (106 days) | 4.2 × 108 | 7.0 × 107 | ND | |
| RQ876 | No. 1 | 4.2 × 108 | 8.9 × 107 | 35 |
| No. 2 (30 days) | 3.7 × 108 | 7.6 × 107 | 25 | |
| No. 3 (148 days) | 3.3 × 108 | 5.8 × 107 | ND |
| Animal . | Apheresis/Transduction (days between Aph/Trdx) . | Total No. of CD4+ Cells Reinfused . | No. Cells/Kg . | % Trdx . |
|---|---|---|---|---|
| RQ1114 | No. 1 | 3.5 × 108 | 5.6 × 107 | 20 |
| No. 2 (28 days) | 1.4 × 108 | 2.2 × 107 | 33 | |
| No. 3 (172 days) | 6.2 × 108 | 8.8 × 107 | 28 | |
| No. 4 (83 days) | 5.0 × 108 | 6.1× 107 | ND | |
| RQ900 | No. 1 | 0.8 × 108 | 1.4 × 107 | 30 |
| No. 2 (35 days) | 5.2 × 108 | 9.1 × 107 | 14 | |
| No. 3 (126 days) | 5.6 × 108 | 9.2 × 107 | 12 | |
| No. 4 (106 days) | 4.2 × 108 | 7.0 × 107 | ND | |
| RQ876 | No. 1 | 4.2 × 108 | 8.9 × 107 | 35 |
| No. 2 (30 days) | 3.7 × 108 | 7.6 × 107 | 25 | |
| No. 3 (148 days) | 3.3 × 108 | 5.8 × 107 | ND |
Abbreviations: Aph, apheresis; Trdx, transduction; ND, not determined.
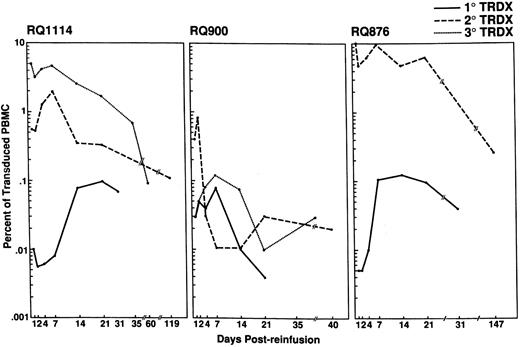
Following each reinfusion, PB samples were drawn at regular intervals for semiquantitative PCR analysis. Results of the in vivo detection of transduced lymphocytes are shown in Figs 2 and 3A. Following the initial infusion, all three of the experimental animals demonstrated the presence of transduced CD4+ lymphocytes in the range of 0.01% to 0.1% of PB mononuclear (MN) cells over the first 4 weeks. The percentage of transduced cells increased during the first 1 to 2 weeks, then stabilized in two of the animals (RQ1114 and RQ876), and gradually declined in the remaining animal (RQ900). Upon subsequent reinfusions of transduced cells, the total percentage of transduced cells increased. In two of the animals (RQ876 and RQ1114), we were able to reach in vivo levels of marked cells ranging between 5% and 10% immediately following reinfusion of the lymphocytes. Levels of 1% to 10% transduced lymphocytes were maintained for at least 3 weeks in both of these animals. Animal RQ900 had a similar accumulation of lymphocytes at each infusion, but did not reach a level near 1% until the third infusion. All animals demonstrated a reduction in the detectable number of transduced cells over time (Fig 2). By 8 weeks, a steady state level between 0.01% and 0.3% was achieved. This plateau was maintained over a period of 21 weeks in two of the animals (RQ876 and RQ1114).
Detection and persistence of transduced CD4+ lymphocytes in the PB. Following each reinfusion of transduced lymphocytes, peripheral blood samples were isolated at regular intervals and used to isolate PBMC. Semiquantitative PCR for the NeoR gene was performed on the genomic DNA isolated from the PBMC. The NeoR PCR product was detected by Southern blotting and hybridization with a NeoR-specific oligonucleotide. All samples were normalized to PCR reactions performed using the β-actin gene. The percent transduction was determined by comparison of the normalized NeoR signal from the experimental samples to a NeoR(+) standard curve prepared by isolating genomic DNA from predetermined percentages of G1N vector transduced SupT1 cells and nontransduced rhesus PBL. The percentage of Neo(+) cells in the standard curve were 10%, 1%, 0.1%, and 0.01%. The percentage of transduced cells in each animal following each reinfusion is plotted on a single graph per animal and the number of transduced PBL for each infusion is shown in the upper right corner of each plot.
Detection and persistence of transduced CD4+ lymphocytes in the PB. Following each reinfusion of transduced lymphocytes, peripheral blood samples were isolated at regular intervals and used to isolate PBMC. Semiquantitative PCR for the NeoR gene was performed on the genomic DNA isolated from the PBMC. The NeoR PCR product was detected by Southern blotting and hybridization with a NeoR-specific oligonucleotide. All samples were normalized to PCR reactions performed using the β-actin gene. The percent transduction was determined by comparison of the normalized NeoR signal from the experimental samples to a NeoR(+) standard curve prepared by isolating genomic DNA from predetermined percentages of G1N vector transduced SupT1 cells and nontransduced rhesus PBL. The percentage of Neo(+) cells in the standard curve were 10%, 1%, 0.1%, and 0.01%. The percentage of transduced cells in each animal following each reinfusion is plotted on a single graph per animal and the number of transduced PBL for each infusion is shown in the upper right corner of each plot.
(A) Detection of the transduced lymphocytes in vivo. Genomic DNA was isolated from the PBMC from RQ876 following the second infusion and RQ900 and RQ1114 following the third infusion of transduced cells were analyzed semiquantitative PCR for the NeoR gene. The samples labeled Pre represent the percentage of gene marked cells present in the animal immediately before reinfusion of the transduced lymphocytes. All samples were normalized to PCR reactions performed using the β-actin gene. The percentage of NeoR(+) cells in the standard curve were 10%, 1%, 0.1%, and 0.01%. (B) NeoR marking of fractionated PBL. PBL were isolated from RQ876 120 days following the second infusion of transduced CD4-enriched lymphocytes, and from RQ816, a negative control animal. The PBMC were sorted into CD4+ and CD8+ cell populations by fluorescence activated cell sorting (FACS). The purity of the separated cell subpopulations used for genomic DNA isolation exceeded 97%. Direct PCR with [32P]-dCTP (deoxycytosine triphosphate) was performed for the detection of the NeoR gene. The standard curve for the PCR was generated by isolating genomic DNA from a G418 resistant population of LSGN vector (glucocerebrosidase/NeoR vector) transduced cells. The genomic DNA was diluted 1:1, 1:5, 1:25, 1:125, and 1:625 for preparation of the standard curve. (C) Trafficking of the transduced CD4 lymphocytes to the LNs. At various times following the second reinfusion of transduced cells (indicated as D-21, -100, or -87), inguinal LN biopsies and PB were isolated from all of the animals. Genomic DNA was isolated from the samples and used for direct PCR for the NeoR gene in the presence of [32P]-dCTP. The NeoR(+) standards of 10%, 1%, 0.1%, and 0.01% are shown.
(A) Detection of the transduced lymphocytes in vivo. Genomic DNA was isolated from the PBMC from RQ876 following the second infusion and RQ900 and RQ1114 following the third infusion of transduced cells were analyzed semiquantitative PCR for the NeoR gene. The samples labeled Pre represent the percentage of gene marked cells present in the animal immediately before reinfusion of the transduced lymphocytes. All samples were normalized to PCR reactions performed using the β-actin gene. The percentage of NeoR(+) cells in the standard curve were 10%, 1%, 0.1%, and 0.01%. (B) NeoR marking of fractionated PBL. PBL were isolated from RQ876 120 days following the second infusion of transduced CD4-enriched lymphocytes, and from RQ816, a negative control animal. The PBMC were sorted into CD4+ and CD8+ cell populations by fluorescence activated cell sorting (FACS). The purity of the separated cell subpopulations used for genomic DNA isolation exceeded 97%. Direct PCR with [32P]-dCTP (deoxycytosine triphosphate) was performed for the detection of the NeoR gene. The standard curve for the PCR was generated by isolating genomic DNA from a G418 resistant population of LSGN vector (glucocerebrosidase/NeoR vector) transduced cells. The genomic DNA was diluted 1:1, 1:5, 1:25, 1:125, and 1:625 for preparation of the standard curve. (C) Trafficking of the transduced CD4 lymphocytes to the LNs. At various times following the second reinfusion of transduced cells (indicated as D-21, -100, or -87), inguinal LN biopsies and PB were isolated from all of the animals. Genomic DNA was isolated from the samples and used for direct PCR for the NeoR gene in the presence of [32P]-dCTP. The NeoR(+) standards of 10%, 1%, 0.1%, and 0.01% are shown.
Distribution of PBL and trafficking to LN.As the original lymphocyte population is only enriched for CD4+ cells, the original population of cells used for transduction also may contain monocytes, B lymphoctyes, and low levels of CD8+ lymphocytes. To ensure that our population of gene marked cells represented only CD4+ cells, CD4+ and CD8+ lymphocytes from one animal (RQ876) were purified by cell sorting and analyzed by PCR. As shown in Fig 3B, the NeoR gene could only be detected in the CD4+ cells from RQ876 120 days following reinfusion.
LNs from HIV-1 infected individuals have previously been reported to be the predominant site for virus replication.22 As viral burden increases, the normal architecture of LNs are disrupted with a significant loss of CD4+ cells.23 If HIV-1–directed gene therapies are to be efficacious, it may be necessary for protected lymphocytes to traffic to the LNs of infected patients. Biopsies of inguinal LNs were obtained from all animals between 21 and 100 days following either the second (RQ876) or third (RQ900 and RQ1114) infusion of transduced cells. NeoR marked cells were detected in the LNs of all three animals. The level of marked cells in the LNs were similar to PB in RQ876 and RQ1114, but lower than the PB signal in animal RQ900 (Fig 3C).
Expression of retroviral sequences in vivo.Expression of the LTR-promoted antisense TAT/REV and the NeoR genes was determined by RT-PCR analysis of RNA from the PBMC isolated at various times following reinfusion of marked cells. The CD4-enriched cells were cultured up to 7 days in the presence of rhIL-2. Total cellular RNA was isolated from aliquots of lymphocytes following 0, 3, and 7 days in culture. As shown in Fig 4, expression of the antisense TAT/REV RNA was detectable in two animals (RQ876 and RQ1114) after 3 days of culture of the CD4+ cells and in all three animals by 7 days of culture. The expression of the NeoR gene was detectable in the lymphocytes of all animals after 3 days of culture in the presence of rhIL-2 (data not shown).
Expression of the Tat/Rev antisense gene. At various times following the second reinfusion, PBMC were isolated from the PB of all of the animals. A CD4-enriched cell population was generated by the immunomagnetic depletion of CD8+ cells and placed into culture at 1 × 106 cells/mL in AIM-V + 10% FBS + 200 IU/mL rIL-2. On days 0, 3, and 7 postculture, equal aliquots of cells were removed and total cellular RNA was isolated. RT-PCR reactions were performed using 500 ng of RNA and [32P]-dCTP for detection of the antisense Tat/Rev RNA. Reactions minus the RT were performed as negative controls for this assay. All samples were analyzed on an 8% polyacrylamide gel.
Expression of the Tat/Rev antisense gene. At various times following the second reinfusion, PBMC were isolated from the PB of all of the animals. A CD4-enriched cell population was generated by the immunomagnetic depletion of CD8+ cells and placed into culture at 1 × 106 cells/mL in AIM-V + 10% FBS + 200 IU/mL rIL-2. On days 0, 3, and 7 postculture, equal aliquots of cells were removed and total cellular RNA was isolated. RT-PCR reactions were performed using 500 ng of RNA and [32P]-dCTP for detection of the antisense Tat/Rev RNA. Reactions minus the RT were performed as negative controls for this assay. All samples were analyzed on an 8% polyacrylamide gel.
Immune response.To investigate the potential immune response to components of the gene therapy protocol, plasma was isolated from all of the animals and used to analyze the development of antibodies to the retroviral vector, FBS, BSA, and NPTII.
The retroviral vector produced by the PG13 packaging cell line contains the Moloney murine leukemia virus (MoMLV) GAG and POL proteins (p30, p65, p180), and the GaLV ENV protein (gp70).24 We investigated the generation of antiviral antibodies to the PG13 (GaLV) retroviral vector by Western blot analysis of concentrated retroviral supernatant using plasma isolated after at least two infusions of marked cells. The positive control for this assay was plasma isolated from an animal previously exposed to replication competent MoMLV retrovirus (RCR).21 None of the transduced animals mounted an antibody response to any of the PG13 retroviral vector proteins, while the animal exposed to RCR had a significant immune response to the p30 protein (Table 2).
Immune Response in Apheresed/Transduced Animals
| Animal . | Retroviral Vector (PG13)* . | NPTII*†‡ . | Fetal Bovine Serum*† . | Bovine Serum Albumin* . |
|---|---|---|---|---|
| RQ900 | (−) | (−) | (+) | (−) |
| RQ876 | (−) | (−) | (+++) | (−) |
| RQ1114 | (−) | (−) | (+) | (−) |
| Animal . | Retroviral Vector (PG13)* . | NPTII*†‡ . | Fetal Bovine Serum*† . | Bovine Serum Albumin* . |
|---|---|---|---|---|
| RQ900 | (−) | (−) | (+) | (−) |
| RQ876 | (−) | (−) | (+++) | (−) |
| RQ1114 | (−) | (−) | (+) | (−) |
Western blot.
Ouchterloney Immunodiffusion.
ELISA.
We investigated the formation of antibodies to FBS by performing Ouchterloney immunodiffusion assays with plasma from the transduced animals. The results indicate that all three animals mounted an immune response to the FBS used either for culturing the cells during the ex vivo transduction period and/or the FBS present in the buffer used during the reinfusion of the cells (Table 2). Plasma isolated from the animals before the gene transfer protocol and from untreated animals failed to form an immunoprecipitate. One animal, RQ876, appeared to have had a much more vigorous reaction to FBS than the other experimental animals. Since greater than 80% of the protein in FBS is BSA, we studied the possibility that the antibodies detected against FBS were in fact generated against the BSA. Ouchterloney immunodiffusion assays were performed with purified BSA. All of the transplanted animals failed to form an immunoprecipitate with BSA, indicating the major antigen found in FBS was a component other than BSA.
The majority of retroviral vectors used to date in clinical trials contain the NeoR selectable marker gene. The expression of this prokaryotic protein in patient cells may be immunogenic. To analyze this, we performed ELISAs using purified NPTII protein (Table 2). Although the NeoR gene expression was detected in engineered lymphocytes, plasma from the transduced animals failed to react with the purified NPTII protein in a range of plasma dilutions from 1:100 to 1:12,800. The positive control plasma for this assay was isolated from a rhesus macaque that was previously immunized on four occasions with the purified NPTII protein in adjuvant. This animal's serum gave a strong reaction to NPTII at a 1:20,000 dilution. In addition to an ELISA, we performed both Ouchterloney immunodiffusions and Western blots with the purified NPTII protein, and again, all of our transduced animals failed to show an antibody response to NPTII after multiple infusions of transduced cells.
DISCUSSION
This report describes high efficiency in vivo reconstitution in nonmyeloablated animals following ex vivo retroviral-mediated gene transfer into rhesus CD4-enriched lymphocytes. The retroviral vector supernatant used in the ex vivo transduction procedure was produced using the PG13 packaging cell line.24 The PG13 packaging cells express a hybrid retroviral genome in which the Moloney murine leukemia virus gag and pol genes are expressed by one expression vector, while the gibbon ape leukemia virus (GaLV) env gene is expressed by another expression vector. We have previously demonstrated that the GaLV envelope-containing retroviruses generated by this packaging cell line could efficiently transduce rhesus and human PBL. This report is the first to successfully use PG13 retroviral vector transduced cells in vivo. Results from this study demonstrate that high levels of transduced CD4+ lymphocytes can accumulate in the circulation by repeated administration and persist there for several months. Up to 3 weeks following reinfusion, genetically engineered CD4+ lymphocytes were detected in 1% to 10% of PBMC, representing approximately 3% to 30% of the total number of circulating CD4+ cells. These cells were also found to have the capacity to traffic to lymphoid tissues, as retroviral vector was detected in all lymph node biopsies.
All animals tolerated multiple reinfusions of transduced CD4-enriched PBL with no adverse reactions. The number of CD4+ cells reinfused into these animals represented a quantity of cells that was equivalent to or exceeded the total number of endogenous circulating CD4+ cells. Our results demonstrate that the ex vivo genetic modification and reinfusion of large quantities of CD4+ cells can be accomplished both safely and effectively.
In two of the three transplanted animals (RQ876 and RQ1114), the percentage of transduced cells in vivo reached between 5% and 10%, for up to 3 weeks following reinfusion of the animals. Such levels of marking are rare and are in part due to the high efficiency retroviral transduction protocol used. The third animal (RQ900) required three infusions to reach a 1% gene marking level. For all animals, the percentage of transduced cells decreased over time before reaching a steady state level of 0.01% to 0.1% marked cells. This low level of transduced cells continued to be detected for up to 21 weeks following reinfusion. This decrease in the percentage of transduced cells may reflect the extravasation of transduced cells into the surrounding tissues, the trapping of the transduced cells in the reticuloendothelial system, the development of an undetected immune response to the transduced cells (for example, a cell-mediated immune response), the production of endogenous unmarked lymphocytes, and/or the natural turnover of these cells in vivo. These results are nearly identical to those obtained in the clinical studies that involved HIV-1 infected twins (R. Walker, unpublished observations, July 1996). The persistence of the marked cells for an extended period of time in healthy animals suggests that the immune status of the recipient may not play a major role in the survival kinetics of the transduced cells.
The detection of both the retroviral vector antisense Tat/Rev and NeoR mRNA indicates that CD4+ lymphocytes retain the ability to express the transduced vector sequences when appropriately stimulated in culture. It has previously been demonstrated in other experimental systems that expression of retroviral vectors can be extinguished upon reinfusion of the cells into the host.25,26 In our study, primary CD4+ cells had to be cultured ex vivo in the presence of rhIL-2 before any vector derived mRNA could be detected. We speculate that the CD4+ lymphocytes do not express significant levels of vector-derived mRNA in vivo as the cells return to their natural resting state after reinfusion, due to a lack of extracellular stimulation in normal rhesus macaques. As evidence of this phenomenon, it has previously been demonstrated in HIV-1 infected individuals that a pool of resting lymphocytes exists that are incapable of expressing HIV-1 retroviral sequences.27 In contrast, it has recently been demonstrated that primary lymphocytes transduced with retroviral vectors expressing the adenosine deaminase gene can give persistent vector mRNA expression over prolonged periods of time.28 It is postulated, however, that an ADA-deficient patient's transduced lymphocytes have a selective growth advantage over cells that fail to express ADA. Taken together, these data, along with our observations, suggest that primary lymphocytes retain the ability to efficiently express vector sequences in vivo, but they require some when they are in a state of activation to do so.
Our data, showing an immune response to FBS, when combined with the unpublished observations of R. Walker in the HIV-1 twin marking study, indicate that the techniques being used to culture or reinfuse lymphocytes should limit exposure to the antigenic component or components found in FBS. As no antibody response was identified to BSA in our experiment, the antigenic source for this immune response remains unclear. To minimize an antibody response to FBS, either an appropriate serum-free media or autologous serum during the ex vivo culture period and upon reinfusion of the transduced cells should be used. The use of autologous serum appears to permit the efficient transduction and expansion of human PBL (L. Muul and R.M. Blaese, personal communication, June 1996).
Unlike FBS, no humoral immune response to the selectable marker NPTII was detected. Our data indicates that even after multiple applications of large numbers of retrovirus marked CD4+ lymphocytes, in which NeoR mRNA expression was detected by RT-PCR; the animals failed to generate a detectable antibody response to NPTII. The lack of a detectable anti-NPTII immune response is also consistent with our finding that the number of circulating gene marked CD4+ cells, and their persistence, did not diminish with subsequent third and fourth infusions. This is consistent with previous studies in which antibody responses to NPTII were not detected after reinfusion of CD34+-derived cells or ADA-deficient PBL.8,29 We have not ruled out, however, the possibility of a cell-mediated immune response to NPTII. Such an immune response has previously been observed by Riddell et al17 to the selectable marker hygromyan-thimidine kinase.
There are several mechanisms to explain the lack of an antibody response to NPTII. First, NPTII may be intrinsically nonimmunogenic. This, however, is unlikely, as it is possible to generate a potent immune response in animals immunized with purified NPTII protein supplemented with adjuvant (R. M. Blaese, unpublished observations, April 1996). A more plausible reason for the failure to develop an immune response to NPTII is that NPTII is an immunogenic protein, but it is not presented effectively in the context of class II major histocompatability complex (MHC) on antigen-presenting cells. A third possibility is that mature lymphocytes may not express significant amounts of the NPTII until they have been activated by external stimuli. Expression of the LTR-promoted Tat/Rev was not detectable in lymphocytes analyzed immediately following removal from the animal (at these time points, the level of circulating transduced cells was 0.03%). RT-PCR analysis confirmed that lymphocytes were capable of expressing vector derived antisense or NeoR sequences only upon stimulation with rhIL-2.
The results of an HIV-1 human clinical trial lends credence to the use of mature lymphocytes as targets for HIV-1 gene therapy.30 In this study, CD4-enriched lymphocytes were isolated from three HIV-1 infected patients and plasmids expressing either a transdominant form of the HIV-1 Rev protein or a deletion mutant control were introduced into the lymphocytes by gold microparticle bombardment. After selection in G418 for 8 days, the cells were reintroduced into the patients and assessed for toxicity, gene expression, and survival of the genetically modified cells. It was determined that cells expressing the transdominant negative form of the Rev protein survived preferentially in the patients. These results also suggest that the high levels of stably transduced lymphocytes achieved in our study would greatly increase the likelihood of detecting a therapeutic benefit in HIV-1 infected individuals.
In summary, our data demonstrate that high efficiency gene transfer to CD4+ PBL in vivo can be achieved with a retroviral vector containing a GaLV env protein. In addition, this animal model may prove useful in the testing of gene therapies for hematopoietic cell-based disorders, such as AIDS.
ACKNOWLEDGMENT
The authors would like to thank Dr Harry Muslow for providing the reagents for the NPTII ELISA, Martha Kirby for performing the cell sorting, and Brian Agricola, Barrington Thompson, and Earl West for their assistance in caring for the animals. The authors would also like to thank Drs Michael Blaese, David Nelson, and Neal Young for critical review of the manuscript.
Address reprint requests to Robert E. Donahue, VMD, Hematology Branch, National Heart, Lung, and Blood Institute, 5 Research Court, Rockville, MD 20850.

![Fig. 3. (A) Detection of the transduced lymphocytes in vivo. Genomic DNA was isolated from the PBMC from RQ876 following the second infusion and RQ900 and RQ1114 following the third infusion of transduced cells were analyzed semiquantitative PCR for the NeoR gene. The samples labeled Pre represent the percentage of gene marked cells present in the animal immediately before reinfusion of the transduced lymphocytes. All samples were normalized to PCR reactions performed using the β-actin gene. The percentage of NeoR(+) cells in the standard curve were 10%, 1%, 0.1%, and 0.01%. (B) NeoR marking of fractionated PBL. PBL were isolated from RQ876 120 days following the second infusion of transduced CD4-enriched lymphocytes, and from RQ816, a negative control animal. The PBMC were sorted into CD4+ and CD8+ cell populations by fluorescence activated cell sorting (FACS). The purity of the separated cell subpopulations used for genomic DNA isolation exceeded 97%. Direct PCR with [32P]-dCTP (deoxycytosine triphosphate) was performed for the detection of the NeoR gene. The standard curve for the PCR was generated by isolating genomic DNA from a G418 resistant population of LSGN vector (glucocerebrosidase/NeoR vector) transduced cells. The genomic DNA was diluted 1:1, 1:5, 1:25, 1:125, and 1:625 for preparation of the standard curve. (C) Trafficking of the transduced CD4 lymphocytes to the LNs. At various times following the second reinfusion of transduced cells (indicated as D-21, -100, or -87), inguinal LN biopsies and PB were isolated from all of the animals. Genomic DNA was isolated from the samples and used for direct PCR for the NeoR gene in the presence of [32P]-dCTP. The NeoR(+) standards of 10%, 1%, 0.1%, and 0.01% are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/6/10.1182_blood.v89.6.1987/3/m_bl_0012f3.jpeg?Expires=1767843532&Signature=MkOwHzrRKqAxLvhIzv4xtdCQo5zWFvrJCQ85uyqfkQk3tXnR70MlR6f9JAza4aWA-3N7nj~YnCK1HTB6esC-nSxhvzy4EqVclAkDh7JmCQJjEZ6MIhY6PWjZgP9O2NCtO0cBHX-vY7x3NaDYlAORlXOc8yftaGh56KoD6f-K1aGrHv36JSxLDGhx75z9COI32tWlQzoDjalVtHtd6cYdn1d9ZFAjX42eEsAgxaMUzYu7oay1ntcIF9g7z-BkALQT43Aqm8Wts-4CsRW5qogZZGvgJ4UxjNs4qIoEz8nWjaB9Cs5IW75B5-RNnN3V9jDG7pPuISpD-gR1dk6CKX0mvw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Expression of the Tat/Rev antisense gene. At various times following the second reinfusion, PBMC were isolated from the PB of all of the animals. A CD4-enriched cell population was generated by the immunomagnetic depletion of CD8+ cells and placed into culture at 1 × 106 cells/mL in AIM-V + 10% FBS + 200 IU/mL rIL-2. On days 0, 3, and 7 postculture, equal aliquots of cells were removed and total cellular RNA was isolated. RT-PCR reactions were performed using 500 ng of RNA and [32P]-dCTP for detection of the antisense Tat/Rev RNA. Reactions minus the RT were performed as negative controls for this assay. All samples were analyzed on an 8% polyacrylamide gel.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/6/10.1182_blood.v89.6.1987/3/m_bl_0012f4.jpeg?Expires=1767843532&Signature=P3nehhzG9AQ0WW30Fc9vw7y1aLENswQYg4JvYAi3UT4CEXII5T4wqt5K-Msb41G7jqrpRH6INuN~gxNEvRNdHTZq~Ut8DLSSLsaAuhooQ7WNp2w1tPzqAVQN~WrApzvq7JDhBBB15vgfsUbYzUoJg4McMZ4~WF76iSgd~QRH~aMBiS66X0QwaRnm0On1FhGH0uIyjDnirrFzOxIQRh~uOBjZmK~yTpE-qp~MmobT7y~XcLLENppp6L0LJV7qKepBWTvF-uqMl4LMQbLIDgLCsWbOJEw1ox38FBPtqe9-0wn5MaTv8uyyW4sWgt9u4DELQ764nAdaLbLrWBSDHnrd1Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal