To the Editor:
The Rh blood group system is highly complex due to the occurrence of a large number of antigenic polymorphisms. Of these, the D category VI (DVI) is a well-recognized partial D antigen and is divided into two subtypes according to its association with C/c and E/e antigens.1 Type I refers to the DVIccEe association and type II the DVICCee association. It was suggested that the former is correlated with a deletion of exons 4 through 6 in the RhD gene and the latter with the formation of a RhD-CE-D hybrid gene retaining exons 4 through 6 of the RhCE gene.2 Here evidence is presented that DVI type I is defined by a novel RhD-CE-D hybrid gene but not an internally deleted RhD gene and that its CE portion contains exons 4 and 5 originating from a RhE allele.
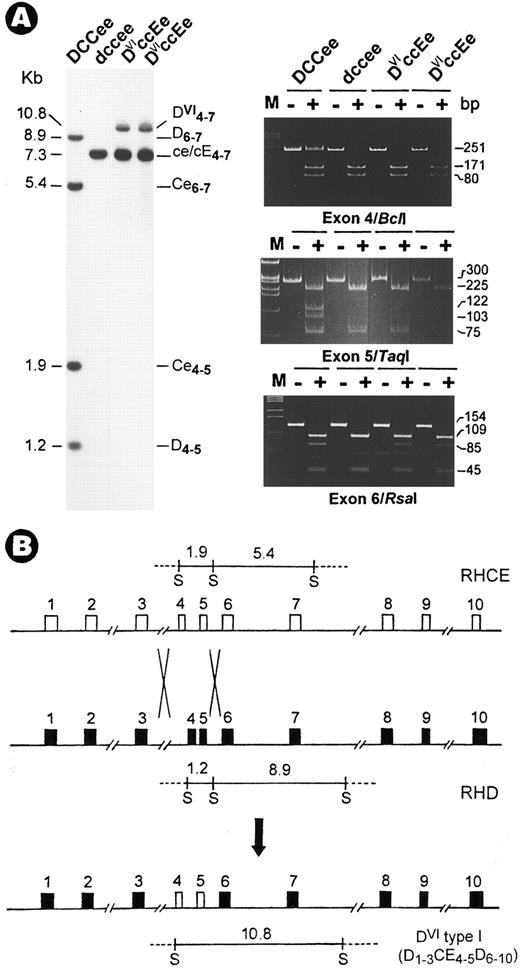
DVIccEe blood was obtained from a mother (T.S.) and her daughter (V.S.). Analysis of Rh gene structure and expression was performed as described.3 Haplotyping by Sph I RFLPs4 indicated a DVIcE/dce genotype for both subjects. Although no gross change was found in CE genes, the absence of two Sph I bands in the D gene was associated with the appearence of a 10.8-kb new fragment (Fig 1A). Next, exons retained in the new fragment were mapped and their identity was determined by polymerase chain reaction (PCR) coupled with diagnostic restriction digestion and DNA sequencing.3 The results showed that in both T.S. and V.S., exons 4 and 5 are CE-specific (Fig 1A, Bcl I and Taq I panels), whereas other exons including exon 6 (Fig 1A, Rsa I panel) are from both D and CE genes. Thus, the DVI type I variant is associated with the occurrence of a D-CE-D hybrid gene, but not a simple deletion of exons 4 to 6 in the RhD gene.2 This hybrid gene most likely originated from a genetic recombination event by which exons 4 and 5 of CE were transferred to replace the D counterpart (Fig 1B).
Identification of a novel RhD-CE-D gene associated with DVI type I variant. (A) Southern blot (left) and exon PCR assay (right). DNAs were digested with Sph I and hybridized with an Rh cDNA probe spanning exons 4 to 7. Lane 1, Rh-positive (DCCee); 2, Rh-negative (dccee); 3, DVIccEe (T.S.); and 4, DVIccEe (V.S.). The size (in kilobases), gene origin, and exon content of the various bands are indicated. The 10.8-kb new fragment is seen in T.S. and V.S. only. Exon PCR assay was performed exactly as described.3 Shown is a typical polyacrylamide gel electrophoresis analysis for exons 4, 5, and 6 PCR products digested with the restriction enzymes, Bcl I, Taq I, and Rsa I. M, φX174 DNA size markers; −, uncut; +, digested. The expected size (in base pairs) of various bands is indicated at the right margin. (B) Diagram for the origin of the DVI type I gene. The two crosses denote the sites for recombination by which a D-CE-D hybrid is formed along with the relocation of the Sph I fragments. In the hybrid gene, exons 4 and 5 from the RhCE gene are flanked by exons from the RhD gene.
Identification of a novel RhD-CE-D gene associated with DVI type I variant. (A) Southern blot (left) and exon PCR assay (right). DNAs were digested with Sph I and hybridized with an Rh cDNA probe spanning exons 4 to 7. Lane 1, Rh-positive (DCCee); 2, Rh-negative (dccee); 3, DVIccEe (T.S.); and 4, DVIccEe (V.S.). The size (in kilobases), gene origin, and exon content of the various bands are indicated. The 10.8-kb new fragment is seen in T.S. and V.S. only. Exon PCR assay was performed exactly as described.3 Shown is a typical polyacrylamide gel electrophoresis analysis for exons 4, 5, and 6 PCR products digested with the restriction enzymes, Bcl I, Taq I, and Rsa I. M, φX174 DNA size markers; −, uncut; +, digested. The expected size (in base pairs) of various bands is indicated at the right margin. (B) Diagram for the origin of the DVI type I gene. The two crosses denote the sites for recombination by which a D-CE-D hybrid is formed along with the relocation of the Sph I fragments. In the hybrid gene, exons 4 and 5 from the RhCE gene are flanked by exons from the RhD gene.
To further characterize this hybrid gene and its expression, Rh cDNAs were obtained using a novel approach that exploits the sequence difference between D and CE transcripts to avoid their coamplification.3 Surprisingly, rather than detecting a single truncated cDNA species,2 the D and CE cDNAs from T.S. and V.S. were found to be comparable to that of controls in size and amount (gels not shown). To establish the transcript structure, the nucleotide sequences of D and CE cDNAs from T.S. and V.S. were determined. In agreement with the results of genotyping and phenotyping, the sequence analysis showed the expression of three full-length transcripts. The CE cDNAs included both Rhce and RhcE forms as indicated by the coexistence of allelic differences.5 However, normal RhD transcript was not found in the erythroid cells of both DVI individuals. Instead, a hybrid transcript was identified which has the same arrangement of the proposed D-CE-D gene (Fig 1B).
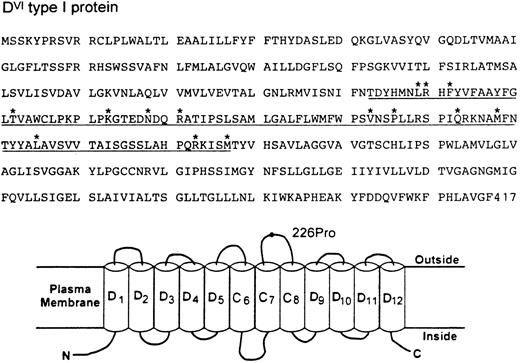
The DVI type I transcript maintains a normal open reading frame and thus should encode a protein with 417 amino acid residues (Fig 2). Immunoblotting with a D-specific monoclonal antibody confirmed the expression of this Rh protein in the erythrocyte membranes (data not shown). The deduced sequence differs from the normal D sequence by 14 amino acids from positions 169 through 267 (Fig 2). Sequence comparison also shows that the CE portion of the DVI type I protein must be from the RhE allele because a proline (Pro) occurs at position 226. Assessment of this protein by hydropathy plot6 suggests a model for its membrane disposition where the RhE portion would contribute to the formation of three membrane spans (C6-C8) plus the entire fourth extracellular loop (Fig 2). Whether the placement of a RhE portion in the context of D might elicit a new antigenicity is not known at present. Nevertheless, it is of interest to note that this partial D category exhibits a very weak D expression, yet there was no apparent reduction of its transcript or protein in the erythrocytes from both individuals. These observations, together with the definition of a new hybrid arrangement, suggest strongly that the amino acid sequence and its variations encoded by exons 4 and 5 of the RhD gene represent a minimal requirement for the exhibition of D antigen.
Deduced amino acid sequence (top) and topologic model for RhDVI type I protein (bottom). The CE portion of DVI type I protein encoded by exons 4 and 5 is underlined and its specific amino acid changes from positions 169-267 are marked by asterisks. (B) Hypothetical organization of DVI type I protein in the erythrocyte plasma membrane. Of the 12 potential membrane-spanning segments, the CE portion would form three passes (C6-C8) and the D portion would contribute to the remaining. The fourth extracellular loop is solely derived from the CE portion and its E allelic specificity is indicated by a 226Pro residue. N and C denote the amino and carboxyl terminal ends.
Deduced amino acid sequence (top) and topologic model for RhDVI type I protein (bottom). The CE portion of DVI type I protein encoded by exons 4 and 5 is underlined and its specific amino acid changes from positions 169-267 are marked by asterisks. (B) Hypothetical organization of DVI type I protein in the erythrocyte plasma membrane. Of the 12 potential membrane-spanning segments, the CE portion would form three passes (C6-C8) and the D portion would contribute to the remaining. The fourth extracellular loop is solely derived from the CE portion and its E allelic specificity is indicated by a 226Pro residue. N and C denote the amino and carboxyl terminal ends.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health Grant No. HL54459. I thank Drs Colvin Redman for reading the letter and Marion Reid for providing DVI blood samples and sharing unpublished observations. This work was first submitted to Blood as a regular paper on April 25, 1996, but instead a letter was solicited.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal