Abstract
We investigated the DNA of 29 unrelated pyruvate kinase (PK) deficiency (PKD) patients from Central Europe with hereditary nonspherocytic hemolytic anemia for mutations in the PK-L/R gene. Among 58 potentially affected alleles, 53 mutations were identified, of which 17 were different from each other. Of these 17 mutations, 13 were single-nucleotide (nt) substitutions resulting in amino acid exchanges, G787A (Gly263-Arg), G994A (Gly332-Ser), G1006T (Ala336-Ser), G1010A (Arg337-Gln), A1081G (Asn361-Asp), G1127T (Ser376-Ile), G1174A (Ala392-Thr), G1281T (Glu427-Asp), C1454T (Ser485-Phe), C1456T (Arg486-Trp), G1493A (Arg498-His), G1529A (Arg510-Gln), and C1594T (Arg532-Trp); 1 in-frame triplet deletion, 1060delAAG (delLys354); 1 in-frame triplet insertion, 1203insAGC (insSer after Cys401); 1 splicesite mutation, 101-1G-A; and 1 frameshift deletion, 628delGT. Six mutations, 628delGT, G787A, G1010A, G1127T, G1281T, and C1454T, are described for the first time. To test the hypothesis of a single origin of the most common PK mutation in the European population, G1529A, we investigated all patients at four polymorphic sites in the PK-L/R gene: C/A at nt 1705, C/T at nt 1992, the (ATT)n microsatellite in intron J, and a polymorphism (T)10/(T)19 in intron I. Nine patients homozygous for mutation G1529A were consistent in all four markers. In the group of patients homozygous for mutation G1529A, the hematologic parameters and clinical manifestations have been studied in detail. Although having an identical mutation in the PK-L/R gene, the patients are affected differently. Their appearance ranges from a very mild compensated hemolysis to a severe anemia. Possible molecular explanations are discussed.
PYRUVATE KINASE (PK) deficiency (PKD) is the most common type of hereditary nonspherocytic hemolytic anemia caused by an enzyme defect of the glycolytic pathway. It was first described in 1961,1 and by 1989 more than 400 cases had been reported.2 Protein properties like thermal stability, susceptibility to degradation by proteases, or electrophoretic mobility had been used to characterize mutant enzymes.3-5 These methods produced contradictory results due to partial instability of mutant enzymes, difficulties in judging hybrid enzymes of compound heterozygotes, or general differences in the energy metabolism of patients of different ages and under different schemes of drug treatment.3 4
By cloning the human L-type PK cDNA,6 it became possible to analyze PK mutations directly at the DNA level. In 1991, the first two point mutations causing PKD were described.7,8 Since then, the largest groups of PKD patients have been investigated by Baronciani and Beutler9-13 in the United States, Kanno et al14-20 in Japan, Bianchi et al21,22 in Italy, and our group.23,24 In our previous report, we presented a preliminary study of 12 PKD patients analyzed by a polymerase chain reaction (PCR) based genomic sequencing strategy.24 Here, we describe the complete mutation analysis of 29 PKD patients from Central Europe representing the largest geographically homogeneous group studied thus far.
SUBJECTS AND METHODS
Patients.Twenty-nine unrelated patients with hereditary nonspherocytic hemolytic anemia of German, English, Czech, and Slovak origin were investigated.
In most of the cases, neonatal jaundice, anemia, and partly reticulocytosis were present at birth or in early childhood. Some patients remained chronically icteric. Patients no. 2, 12, 21, 23, and 24 have been splenectomized. All patients showed a decreased level of hemoglobin ranging from 5 to 12 g/dL (normal: males, 14.5 to 19.0 g/dL; females, 11.4 to 16.4 g/dL). Patients no. 5, 11, 14, and 16 showed an abnormal enzyme-kinetic behavior (high Km-PEP variants, ie, variants with a high Michaelis constant for phosphoenol pyruvate). The activity of the enzyme in patient no. 16 (high Km-PEP variant) was only 200% (Table 1) when assayed under conditions of substrate saturation. All the patients had hemolytic crises and were transfused as newborns.
Genetic and Hematologic Characteristics of Patients With PKD
| Patient No. . | Reticulocytes (%) . | Residual PK Activity (%) . | Mutations . | Amino Acid Substitutions . | Nationality . | (T)n in Intron I . | (ATT)n in Intron J . | C/A 1705 . | C/T 1992 . |
|---|---|---|---|---|---|---|---|---|---|
| . | . | . | (alleles 1 and 2) . | . | . | . | . | . | . |
| 1 | 60-80 | 6 | G1529A/1203insAGC | Arg510-Gln/ins Ser after Cys401 | German | 10/19 | 14/12 | C/A | C/T |
| 2 | 25 | 10 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 3 | 20-40 | 14 | C1594T/G994A | Arg532-Trp/Gly332-Ser | Slovak | 10/10 | 17/17 | C/C | C/C |
| 4 | 53-80 | 21 | Splicing 101-1G-A/G1493A | Splice-site mutation/Arg498-His | Czech | 10/19 | 15/13 | C/A | C/T |
| 5 | 4-5 | 87 | G1006T/? | Ala336-Ser/? | Czech | 10/10 | 14/14 | C/C | C/C |
| 6 | 60-90 | 12 | C1594T/1060delAAG | Arg532-Trp/del Lys354 | Czech | 10/10 | 17/15 | C/C | C/C |
| 7 | 8-10 | 24 | C1456T/A1081G | Arg486-Trp/Asn361-Asp | Czech | 10/10 | 14/14 | C/C | C/C |
| 8 | 5-7 | 25 | C1456T/splicing 101-1G-A | Arg486-Trp/splice-site mutation | Czech | 10/10 | 14/15 | C/C | C/C |
| 9 | 30-90 | 25 | G994A/C1594T | Gly332-Ser/Arg532-Trp | Czech | 10/10 | 17/17 | C/C | C/C |
| 10 | 9 | 28 | C1456T/1060delAAG | Arg486-Trp/del Lys354 | Slovak | 10/10 | 14/15 | C/C | C/C |
| 11 | 15 | 42 | G1174A/? | Ala392-Thr/? | Slovak | 10/10 | 15/16 | C/C | C/C |
| 12 | 20-30 | ND | G1529A/G1529A | Arg510-Gln/Arg510-Gln | English | 10/10 | 14/14 | C/C | C/C |
| 13 | 34 | 12 | G1529A/A1081G | Arg510-Gln/Asn361-Asp | German | 10/10 | 14/16 | C/C | C/C |
| 14 | 35 | 43 | C1456T/G1010A | Arg486-Trp/Arg337-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 15 | 5 | 20 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 16 | 56 | 200* | G1529A/G1281T | Arg510-Gln/Glu427-Asp | German | 10/10 | 14/17 | C/C | C/C |
| 17 | 8 | 18 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | Slovak | 10/10 | 14/14 | C/C | C/C |
| 18 | 5 | 30 | G787A/? | Gly263-Arg/? | German | 10/10 | 14/16 | C/C | C/C |
| 19 | 18 | 14 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 20 | 19 | 70 | 1060delAAG/? | del Lys 354/? | Slovak | 10/10 | 15/15 | C/C | C/C |
| 21 | ND | ND | 628delGT/? | Frameshift/? | English | 10/10 | 14/15 | C/C | C/C |
| 22 | ND | ND | G1529A/G994A | Arg510-Gln/Gly332-Ser | English | 10/10 | 14/17 | C/C | C/C |
| 23 | ND | ND | C1456T/G1127T | Arg486-Trp/Ser376-Ile | English | 10/10 | 14/15 | C/C | C/C |
| 24 | ND | 25 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | English | 10/10 | 14/14 | C/C | C/C |
| 25 | ND | ND | G1529A/C1456T | Arg510-Gln/Arg486-Trp | English | 10/10 | 14/14 | C/C | C/C |
| 26 | 14 | 45 | G1529A/C1454T | Arg510-Gln/Ser485-Phe | German | 10/19 | 14/12 | C/A | C/T |
| 27 | 13 | 17 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 28 | 8 | 17 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 29 | 66 | 10 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| Control | 0-1 | 100 |
| Patient No. . | Reticulocytes (%) . | Residual PK Activity (%) . | Mutations . | Amino Acid Substitutions . | Nationality . | (T)n in Intron I . | (ATT)n in Intron J . | C/A 1705 . | C/T 1992 . |
|---|---|---|---|---|---|---|---|---|---|
| . | . | . | (alleles 1 and 2) . | . | . | . | . | . | . |
| 1 | 60-80 | 6 | G1529A/1203insAGC | Arg510-Gln/ins Ser after Cys401 | German | 10/19 | 14/12 | C/A | C/T |
| 2 | 25 | 10 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 3 | 20-40 | 14 | C1594T/G994A | Arg532-Trp/Gly332-Ser | Slovak | 10/10 | 17/17 | C/C | C/C |
| 4 | 53-80 | 21 | Splicing 101-1G-A/G1493A | Splice-site mutation/Arg498-His | Czech | 10/19 | 15/13 | C/A | C/T |
| 5 | 4-5 | 87 | G1006T/? | Ala336-Ser/? | Czech | 10/10 | 14/14 | C/C | C/C |
| 6 | 60-90 | 12 | C1594T/1060delAAG | Arg532-Trp/del Lys354 | Czech | 10/10 | 17/15 | C/C | C/C |
| 7 | 8-10 | 24 | C1456T/A1081G | Arg486-Trp/Asn361-Asp | Czech | 10/10 | 14/14 | C/C | C/C |
| 8 | 5-7 | 25 | C1456T/splicing 101-1G-A | Arg486-Trp/splice-site mutation | Czech | 10/10 | 14/15 | C/C | C/C |
| 9 | 30-90 | 25 | G994A/C1594T | Gly332-Ser/Arg532-Trp | Czech | 10/10 | 17/17 | C/C | C/C |
| 10 | 9 | 28 | C1456T/1060delAAG | Arg486-Trp/del Lys354 | Slovak | 10/10 | 14/15 | C/C | C/C |
| 11 | 15 | 42 | G1174A/? | Ala392-Thr/? | Slovak | 10/10 | 15/16 | C/C | C/C |
| 12 | 20-30 | ND | G1529A/G1529A | Arg510-Gln/Arg510-Gln | English | 10/10 | 14/14 | C/C | C/C |
| 13 | 34 | 12 | G1529A/A1081G | Arg510-Gln/Asn361-Asp | German | 10/10 | 14/16 | C/C | C/C |
| 14 | 35 | 43 | C1456T/G1010A | Arg486-Trp/Arg337-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 15 | 5 | 20 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 16 | 56 | 200* | G1529A/G1281T | Arg510-Gln/Glu427-Asp | German | 10/10 | 14/17 | C/C | C/C |
| 17 | 8 | 18 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | Slovak | 10/10 | 14/14 | C/C | C/C |
| 18 | 5 | 30 | G787A/? | Gly263-Arg/? | German | 10/10 | 14/16 | C/C | C/C |
| 19 | 18 | 14 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 20 | 19 | 70 | 1060delAAG/? | del Lys 354/? | Slovak | 10/10 | 15/15 | C/C | C/C |
| 21 | ND | ND | 628delGT/? | Frameshift/? | English | 10/10 | 14/15 | C/C | C/C |
| 22 | ND | ND | G1529A/G994A | Arg510-Gln/Gly332-Ser | English | 10/10 | 14/17 | C/C | C/C |
| 23 | ND | ND | C1456T/G1127T | Arg486-Trp/Ser376-Ile | English | 10/10 | 14/15 | C/C | C/C |
| 24 | ND | 25 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | English | 10/10 | 14/14 | C/C | C/C |
| 25 | ND | ND | G1529A/C1456T | Arg510-Gln/Arg486-Trp | English | 10/10 | 14/14 | C/C | C/C |
| 26 | 14 | 45 | G1529A/C1454T | Arg510-Gln/Ser485-Phe | German | 10/19 | 14/12 | C/A | C/T |
| 27 | 13 | 17 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 28 | 8 | 17 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| 29 | 66 | 10 | G1529A/G1529A | Arg510-Gln/Arg510-Gln | German | 10/10 | 14/14 | C/C | C/C |
| Control | 0-1 | 100 |
Patients no. 1 to 12 have been previously reported.24
Abbreviation: ND, not determined.
See Materials and Methods.
Hemoglobin concentration, red blood cell (RBC) count, PK enzyme activity, mean cellular volume, and enzyme kinetic parameters were determined by standard hematologic methods and according to the recommendations of the International Committee for Standardization in Haematology.25 The persistence of the PK-M2 isoenzyme in RBCs of patients no. 1, 2, 3, 6, 9, 16, and 29 was determined by immunotitration with a human monospecific PK-M2 antibody.26
DNA preparation.Genomic DNA was isolated from frozen total blood with a Nucleon-DNA-extraction kit from Scotlab Bioscience (Wiesloch, Germany).
Mutation analysis.All mutations were analyzed by direct sequencing of PCR-amplified fragments from genomic DNA. The sequencing included all exons, the entire 5′- and 3′-untranslated regions, and a further 240 nucleotides (nts) of the promoter of the PK-L/R gene.12 Mutation analysis was performed according to a previously described protocol24 based on a set of intron-derived PCR primers. The sequences of the primers used to amplify the 3′-untranslated region were 5′-AAG CTC CGT GGC TTC CTC-3′ and 5′-GAC ACA CTC ATT TTT GGC-3′.
All mutations were confirmed by independent DNA analysis techniques such as allele-specific oligonucleotide hybridization24 or PCR analysis combined with restriction enzyme analysis, when a restriction site was affected, or by direct length comparison of PCR fragments covering the deletion or insertion (with patients no. 6, 10, 20, and 21). Table 2 lists an overview of restriction enzymes used for detection of all but one of the mutations found.
Restriction Enzymes Used to Detect Mutations in the PK-L/R Gene
| Mutation . | Exon . | Recognition Sequence . | Restriction Enzyme for Normal Site . | Restriction Enzyme for Mutated Site . |
|---|---|---|---|---|
| 101-1G-A | n: gGGCCA | Sau96I: GGNCC | Hae I: WGGCCW | |
| m: aGGCCA | ||||
| 628delGT | 6 | n: CCGGGTCG | ScrFI: CCNGG | |
| m: CCGGCG | Nci I; Bcn I: CCSGG | |||
| G787A | 7 | n: CCCGGG | Sma I; Xma I: CCCGGG | EcoRII: CCWGG |
| m: CCCAGG | Msp I; HpaII: CCGG | |||
| G994A | 8 | n: GACGGC | No restriction site found | No restriction site found |
| m: GACAGC | ||||
| G1006T | 8 | n: TGGCAC | MaeIII: GTNAC | |
| m: TGTCAC | ||||
| G1010A | 8 | n: CACGGG | BscGI: CCCGT | |
| m: CACAGG | (− strand) | |||
| 1060delAAG | 8 | n: AGAAGA | MboII: GAAGA | |
| m: AGATGA | ||||
| A1081G | 8 | n: TGCAAC | CviRI: TGCA | |
| m: TGCGAC | ||||
| G1127T | 9 | n: GAGCATGA | BspHI: TCATGA | |
| m: GATCATGA | Mbo I; Dpn I: Sau3AI: GATC | |||
| G1174A | 9 | n: GTCGCC | MaeIII: GTNAC | |
| m: GTCACC | ||||
| 1203insAGC | 9 | n: CTGCAT | Pst I: CTGCAG | |
| m: CTGCAGCA | Bby I: GCAGC | |||
| G1281T | 10 | n: GGAGGC | Mnl I: CCTC | Fok I: GGATG |
| m: GGATGC | (− strand) | |||
| C1454T | 11 | n: GTCTCG | BsmAI: GTCTC | |
| m: GTTTCG | ||||
| C1456T | 11 | n: GTCTCG | BsmAI: GTCTC | |
| m: GTCTTG | ||||
| G1493A | 11 | n: CACCCGCTC | AccBSI: GAGCGG | TaqII: |
| m: CACCCACTC | (− strand); Aci I: CCGC | CACCCA(n)11 | ||
| G1529A | 11 | n: CCGAGG | Sty I: CCWWGG | |
| m: CCAAGG | ||||
| C1594T | 11 | n: GCCGGG | Msp I; HpaII: CCGG | |
| m: GCTGGG | ScrFI: CCNGG |
| Mutation . | Exon . | Recognition Sequence . | Restriction Enzyme for Normal Site . | Restriction Enzyme for Mutated Site . |
|---|---|---|---|---|
| 101-1G-A | n: gGGCCA | Sau96I: GGNCC | Hae I: WGGCCW | |
| m: aGGCCA | ||||
| 628delGT | 6 | n: CCGGGTCG | ScrFI: CCNGG | |
| m: CCGGCG | Nci I; Bcn I: CCSGG | |||
| G787A | 7 | n: CCCGGG | Sma I; Xma I: CCCGGG | EcoRII: CCWGG |
| m: CCCAGG | Msp I; HpaII: CCGG | |||
| G994A | 8 | n: GACGGC | No restriction site found | No restriction site found |
| m: GACAGC | ||||
| G1006T | 8 | n: TGGCAC | MaeIII: GTNAC | |
| m: TGTCAC | ||||
| G1010A | 8 | n: CACGGG | BscGI: CCCGT | |
| m: CACAGG | (− strand) | |||
| 1060delAAG | 8 | n: AGAAGA | MboII: GAAGA | |
| m: AGATGA | ||||
| A1081G | 8 | n: TGCAAC | CviRI: TGCA | |
| m: TGCGAC | ||||
| G1127T | 9 | n: GAGCATGA | BspHI: TCATGA | |
| m: GATCATGA | Mbo I; Dpn I: Sau3AI: GATC | |||
| G1174A | 9 | n: GTCGCC | MaeIII: GTNAC | |
| m: GTCACC | ||||
| 1203insAGC | 9 | n: CTGCAT | Pst I: CTGCAG | |
| m: CTGCAGCA | Bby I: GCAGC | |||
| G1281T | 10 | n: GGAGGC | Mnl I: CCTC | Fok I: GGATG |
| m: GGATGC | (− strand) | |||
| C1454T | 11 | n: GTCTCG | BsmAI: GTCTC | |
| m: GTTTCG | ||||
| C1456T | 11 | n: GTCTCG | BsmAI: GTCTC | |
| m: GTCTTG | ||||
| G1493A | 11 | n: CACCCGCTC | AccBSI: GAGCGG | TaqII: |
| m: CACCCACTC | (− strand); Aci I: CCGC | CACCCA(n)11 | ||
| G1529A | 11 | n: CCGAGG | Sty I: CCWWGG | |
| m: CCAAGG | ||||
| C1594T | 11 | n: GCCGGG | Msp I; HpaII: CCGG | |
| m: GCTGGG | ScrFI: CCNGG |
Boldface letters designate mutated nts.
Abbreviations: n, normal; m, mutated.
Nomenclature.The nt numbers are designated according to the PK/R-cDNA sequence, with the A of the initiation codon ATG being assigned number 1.13 Amino acids are counted according to the deduced PK/R-isoenzyme amino acid sequence.6 7
The designation of exons by numbers and introns by letters follows the system we used previously.24
Mutations were designated according to the recommendations of the Cystic Fibrosis Genetic Analysis Consortium.27
Analysis of four polymorphic sites.The polymorphic site C/A at nt 170514 was examined via amplification of exon 12 and restriction enzyme analysis with RcaI (GIBCO BRL, Eggenstein, Germany). The (ATT)n microsatellite in intron J was examined as previously described.23 Furthermore, we analyzed and used the two polymorphic markers (T)n , with n = 10 or 19, in intron I and C/T at nt 1992, which are described elsewhere.28
Isoelectric focusing.Isoelectric focusing and immunologic visualization of normal and mutant RBC PK were performed as described previously.5
RESULTS
Description of mutations in the PK-L/R gene.Using a solid-phase PCR sequencing strategy, we analyzed 29 unrelated patients and their families for mutations in the PK-L/R gene. All patients were caucasians of Central European origin.
The mutations found are summarized in Table 1. Additionally, the reticulocyte counts and residual PK activities of the patients and their haplotypes comprising the polymorphic sites (ATT)n in intron J,23 (oligo T)n in intron I,28 C/A at nt 1705,9 24 and C/T at nt 199228 are listed.
In 58 alleles at risk, we detected the mutations in all but five. Among the 17 different mutations, 13 were missense mutations causing single–amino acid changes (G787A, G994A, G1006T, G1010A, A1081G, G1127T, G1174A, G1281T, C1454T, C1456T, G1493A, G1529A, and C1594T). Two others were an in-frame triplet deletion and insertion leading to a one–amino acid loss or gain, respectively (1060delAAG and 1203insAGC). Furthermore, one frameshift deletion (628delGT) and one splice-site mutation (splicing 101-1G-A) were found.
Like Baronciani et al,12 we also found some minor differences in the promoter sequence as compared with the original report by Kanno et al29: insertion of one adenine between nts 41 and 42 and of two adenines between nts 55 and 56.
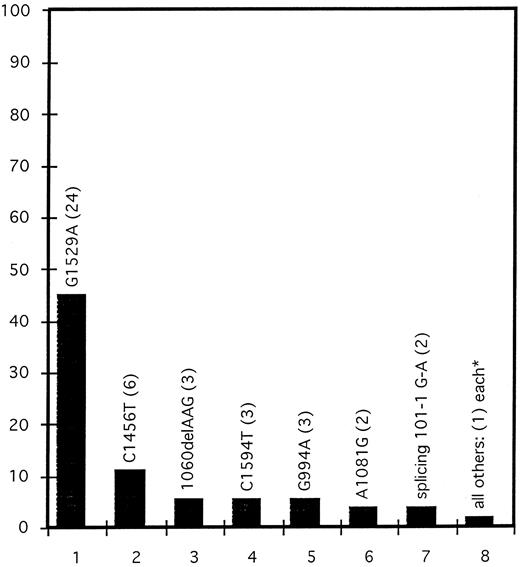
Frequency distribution of PK mutations.The frequency distribution of all our 53 mutations is shown in Fig 1. Clearly, the most frequent mutation is the G1529A missense mutation that leads to an Arg510-Gln exchange occurring in 24 alleles, ie, in 45.3% of all mutated chromosomes. Nine patients carry it homozygously, and six have it in combination with a second mutation in a compound heterozygous status. The G1529A mutation is much more frequent among the patients of German and English origin (14 of 18 patients are affected homozygously or heterozygously) than among patients from Czechia/Slovakia (1 homozygote among 11 PKD patients). On the other hand, the missense mutation C1594T (3 of 11 patients), the deletion 1060delAAG (3 of 11), and the splice-site mutation splicing 101-1G-A (2 of 11) have been found exclusively in the Czechia/Slovakia group and never among German and English people.
Frequency distribution of PK mutations. Twenty-nine PKD patients were analyzed for mutations in the PK-L/R gene by DNA sequencing. Of 53 mutations found, 17 were different. Frequency distribution of all individual mutations is shown. *Ten mutations that have been found only once.
Frequency distribution of PK mutations. Twenty-nine PKD patients were analyzed for mutations in the PK-L/R gene by DNA sequencing. Of 53 mutations found, 17 were different. Frequency distribution of all individual mutations is shown. *Ten mutations that have been found only once.
The second most frequent mutation, the missense mutation C1456T (six affected alleles), is equally distributed in the German/English group and in the Czechia/Slovakia group.
Polymorphisms.To obtain information on the genetic background of patients carrying the G1529A mutation, we compared the distribution of polymorphic sites in the PK-L/R gene: the (ATT)n microsatellite of intron J,23 a polymorphic oligo (T)n stretch in intron I, which occurs in the two allelic forms (T)10 and (T)19 ,28 and the two polymorphic sites at nt 1705 (A/C)14 and at nt 1992 (C/T)28 (Table 1). In all four polymorphic markers, they were completely concordant, showing the following marker alleles: (ATT)14 , (T)10 , nt 1705-C, and nt 1992-C.
Clinical status of patients homozygous for the mutation G1529A.PKD patients homozygous for the same mutation could be expected to show similar hematologic dysfunctions and clinical symptoms. However, this is not the case (Table 3). We compared the available hematologic and clinical data from a group of nine PKD patients homozygous for the mutation G1529A. The data for three homozygous siblings of patients no. 2 and 17 are also presented in Table 3. These were not included in the analysis of frequency distribution.
Hematologic Data From PKD Patients Homozygous for the Mutation G1529A
| Patient No. . | Sex . | Hemoglobin (g/dL) . | Residual PK Activity (%) . | RBCs (×1012/L) . | MCV (fL) . | Reticulocytes (%) . | RBC t1/2 (d)3-150 . | Clinical Manifestation3-151 . |
|---|---|---|---|---|---|---|---|---|
| 293-152 | F | 5.8 | 10 | 1.82 | 128 | 66 | 2.4 | Severe |
| 2a | M | 10.0 | 10 | 2.34 | 122 | 25 | 4.0 | Severe |
| 2b3-152 | M | 9.0 | 13 | 2.52 | 121 | 29 | 4.0 | Severe |
| 12 | M | 8.3 | ND | ND | ND | 25 | 3.7 | Severe |
| 19 | M | 7.2 | 14 | 2.32 | 107 | 18 | ND | Less severe |
| 27 | M | 7.6 | 17 | 2.70 | 106 | 13 | 7.0 | Less severe |
| 28 | M | 12.1 | 17 | 3.59 | 107 | 8 | 11.3 | Moderate |
| 17a | M | 12.2 | 18 | 3.74 | 95 | 8 | 13.2 | Moderate |
| 17b | F | 9.2 | 22 | 2.56 | 94 | 6 | 14.1 | Moderate |
| 17c | F | 8.1 | 13 | 2.92 | 94 | 8 | ND | Moderate |
| 15 | M | 10.9 | 20 | 3.74 | 91 | 5 | 15.3 | Moderate |
| 24 | F | 9.0 | 25 | 3.24 | 92 | ND | 13.7 | Moderate |
| Normal | M 16.7 | 100 | 4.0-6.5 | 80-95 | <1 | 25-30 | ||
| F 13.89 |
| Patient No. . | Sex . | Hemoglobin (g/dL) . | Residual PK Activity (%) . | RBCs (×1012/L) . | MCV (fL) . | Reticulocytes (%) . | RBC t1/2 (d)3-150 . | Clinical Manifestation3-151 . |
|---|---|---|---|---|---|---|---|---|
| 293-152 | F | 5.8 | 10 | 1.82 | 128 | 66 | 2.4 | Severe |
| 2a | M | 10.0 | 10 | 2.34 | 122 | 25 | 4.0 | Severe |
| 2b3-152 | M | 9.0 | 13 | 2.52 | 121 | 29 | 4.0 | Severe |
| 12 | M | 8.3 | ND | ND | ND | 25 | 3.7 | Severe |
| 19 | M | 7.2 | 14 | 2.32 | 107 | 18 | ND | Less severe |
| 27 | M | 7.6 | 17 | 2.70 | 106 | 13 | 7.0 | Less severe |
| 28 | M | 12.1 | 17 | 3.59 | 107 | 8 | 11.3 | Moderate |
| 17a | M | 12.2 | 18 | 3.74 | 95 | 8 | 13.2 | Moderate |
| 17b | F | 9.2 | 22 | 2.56 | 94 | 6 | 14.1 | Moderate |
| 17c | F | 8.1 | 13 | 2.92 | 94 | 8 | ND | Moderate |
| 15 | M | 10.9 | 20 | 3.74 | 91 | 5 | 15.3 | Moderate |
| 24 | F | 9.0 | 25 | 3.24 | 92 | ND | 13.7 | Moderate |
| Normal | M 16.7 | 100 | 4.0-6.5 | 80-95 | <1 | 25-30 | ||
| F 13.89 |
Determined by 51Cr labeling.35
Severity of the clinical manifestation of the disease has been classified using the following criteria: number, t1/2, and MCV of RBCs, degree of reticulocytosis, susceptibility to infections, physical capacity (generally good in category “moderate”), development of a secondary hemochromatosis, and life expectancy of not more than ∼40 years for category “severe.”
Died recently at the age of 41 years (no. 29) and 35 years (no. 2b).
It is evident that the patients are affected differently. Clinical symptoms range from a mild compensated hemolysis (patients no. 15, 17a, 17b, 17c, 24, and 28) to intermediate anemias (patients no. 19 and 27) and severe anemias (patients no. 2a, 2b, 12, and 29). All show low residual enzyme activities between approximately 10% and 25% of normal, of which the more severe cases have lower PK activities than the milder forms. Reticulocyte counts vary between approximately 5% and 8% in the group of slightly affected patients to 25% to 66% in the cases seriously affected. The severely affected patients no. 2a, 2b, and 29 are characterized by a compensatory persistence of the M2-type enzyme in their RBCs, which accounts for about half of the residual PK activity shown in Table 3.
DISCUSSION
We analyzed a group of 29 patients with PKD of RBCs for mutations at the level of the PK-L/R gene. Seventeen different mutations could be distinguished. According to the latest reports on mutations in the PK-L/R gene,7,8,12-20,22,24 30-32 six mutations are novel and have not been described previously.
A previous report from our group included six patients in whom the mutation in the second allele had not been found, giving rise to the assumption that these could be located in regulatory regions of the PK-L/R gene.24 After reexamination of these patients, the second mutation was detected in four (patients no. 7, 8, 9, and 10). Patients no. 5 and 11, who are suspected to be true heterozygotes due to a high residual PK activity, have been confirmed to be by repeated sequencing of their DNA and that of their available relatives. All six patients are included in this study under the same numbers.
Concerning the putative functional consequence of the exchanged amino acid, one would expect that only nonconservative exchanges should alter the enzymatic and protein-chemical properties considerably. All mutations were classic nonconservative exchanges altering the properties of the affected amino acids in a way like Arg/Gln (charged/uncharged) or Ser/Phe (hydrophilic/hydrophobic) and would explain the observed changes of the mutated enzyme (Table 1). A further argument for the functional relevance of the observed mutations is the fact that only amino acids are affected that are highly conserved between the PK isoenzymes and PKs of different species, as also shown by Baronciani and Beutler13 for other PK mutations.
The overall number of different mutations reported to be associated with PKD is now 637-24,30-32 (and this study; Table 1). They are scattered throughout the whole coding region with no preference for the active site, as determined by crystallographic analysis of the cat muscle PK.33 However, no mutation has been detected until now in exon 1 that is restricted to the erythrocyte-specific isoenzyme PK-R. Furthermore, no regulatory mutation has been found associated with PKD. The only mutations that do not affect the coding regions directly are the four splice-site mutations detected by Kanno et al,17,19 Bianchi et al,21 and our group.24
Most mutations have only been found once. But there is a clear accumulation of some mutations with a strong ethnic and regional background. Among 53 altered chromosomes, we detected 24 G1529A and six C1456T mutations. Interestingly, the G1529A mutation is much more frequent among our German and English patients (14 of 18, or 77.8%) than among patients from Czechia and Slovakia (1 of 11, or 9.1%). Together with the second most frequent mutation, C1456T, nearly every PKD patient of our English/German group carried at least one of these two mutations. This high frequency allows an easy screening by genomic PCR/restriction enzyme analysis (Table 2) for suspected PKD cases in these countries.
A Western/Northern European origin of the G1529A mutation is also confirmed by the data from Baronciani and Beutler.12 13 Seventy-two percent of a group of 25 white North American PKD patients carried it. But as our study shows, it is much less frequent in a population of predominantly Slav origin.
The nine patients homozygous for G1529A have identical haplotypes [(ATT)14/(T)10/C1705/C1992] defined by all four polymorphic markers investigated. The use of the two new informative polymorphic markers (normal chromosomes: (T)10/C1992, 72%, v (T)19/T1992, 28%; PKD chromosomes: (T)10/C1992, 95%, v (T)19/T1992, 5%) and the analysis of a larger group of homozygous patients further substantiate the single-origin hypothesis for this mutation.28
In a study of Italian PKD patients, Bianchi et al22 have shown that among the 16 mutated alleles found, there are only three G1529A mutations but seven C1456T mutations, which (C1456T) is the second most frequent in our group. However, the G1529A mutation has never been found among Japanese PKD patients. Here, the mutation C1468T predominates, as has been reported by Kanno et al.19
The strong ethnic and regional differences in the occurrence of certain PKD mutations even among patients of European origin indicate that most of the mutations cannot be very old, at least not as old as the separation of European and East Asian ethnic groups and probably younger than the extended European migrations in the first 1,000 years A.D.
A comparison of a group of nine PKD patients and also three siblings of patients no. 2 and 17, all homozygous for the same mutation G1529A, enabled us to ask the question as to the manifestation of the disease in different persons with an equal genetic background with respect to the cause of the mutation in the PK-L/R gene. Patients homozygous for the same mutation should show a similar appearance of the anemia. However, the severity of the disease and the well-being of the patients differs dramatically (Table 3).
PKD patients homozygous for the mutation G1529A show residual R-type PK activities between approximately 10% and 25%, and the reticulocytosis varies between 5% and 66%. In general, the severity of the disease correlates with the impairment of the hematologic parameters: the lower the residual PK activity and the higher the reticulocyte counts, the more the patients are affected.
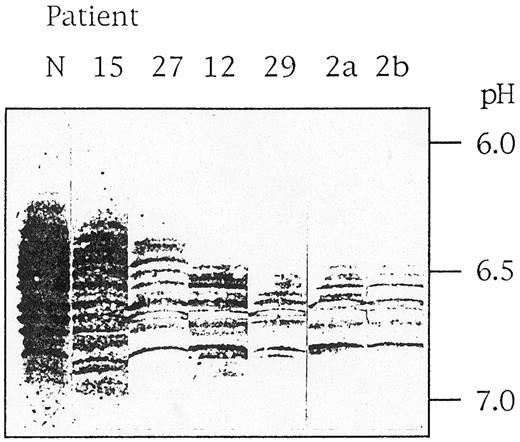
How can these differences in clinical manifestation be explained? The mutation G1529A leads to an amino acid substitution of arginine 510 by a glutamine, which destabilizes the enzyme protein. This can be demonstrated in vitro by the observation of a lower thermostability at 53°C and an increasing sensitivity to proteolytic degradation of the mutant enzyme compared with the normal enzyme (G.J., unpublished results, October 1990). In vivo, this PK variant is characterized by an accelerated intracellular proteolytic degradation. We analyzed the peptide degradation pattern by isoelectric focusing and immunoblotting using monospecific antibodies against R-type PK. Such a pattern of mutant G1529A-PK-R from one moderate case (patient no. 15), one less severe case (patient no. 27), and four severe cases (patients no. 12, 29, 2a, and 2b) is compared with normal PK-R in Fig 2. It is evident that the more severe cases correlate with a significant loss of undegraded enzyme (running in the more acidic parts of the gel) and are associated with a less complex degradation pattern due to faster elimination of unstable peptides. Therefore, the actual PK activity is the result of at least two factors. The constant factor is the basic mutation, resulting in an enzyme with reduced activity but also with altered stability. This interferes with the second, variable factor, the genetically determined, individually different intracellular proteolytic activity. However, it cannot be ruled out that still other individual genetically determined differences in RBC metabolism modulate the basic effect of the mutation. A mainly genetic and nonenvironmental explanation for the differences in clinical manifestation is also probable from the analysis of close relatives, as can be seen with the two severe cases of the brothers 2a and 2b and with the three moderate cases of the sisters 17a, 17b, and 17c (Table 3). The most severe cases (patients no. 2a, 2b, and 29) are furthermore characterized by a compensatory persistence of the M2-type enzyme, which is about half of the overall residual activity listed in Table 3.
Pattern of PK-R immunoreactive polypeptides in RBCs of PKD patients. Stroma-free hemolysates of a normal individual (N) and six PKD patients (no. 15, 27, 12, 29, 2a, and 2b) were subjected to isoelectric focusing in ultrathin polyacrylamide gels, and immunoreactive bands were visualized using a monospecific antibody against PK-R.5
Pattern of PK-R immunoreactive polypeptides in RBCs of PKD patients. Stroma-free hemolysates of a normal individual (N) and six PKD patients (no. 15, 27, 12, 29, 2a, and 2b) were subjected to isoelectric focusing in ultrathin polyacrylamide gels, and immunoreactive bands were visualized using a monospecific antibody against PK-R.5
In general, the situation in PKD seems to be similar to other genetic disorders like cystic fibrosis.34 It has become clear that genetic and environmental factors apart from PK genotype also play an important role in the overall severity of the disease.
ACKNOWLEDGMENT
We are most grateful to Dr D.W. Gorst of the Lancaster Acute Hospital, NHS Trust (Lancaster, UK), for help in the arrangement of British PKD patients. We want to thank the following Czech, Slovakian and British colleagues for the fruitful collaboration and the donation of blood samples: Dr Brabec, Institute of Hematology and Blood Transfusion, Praha, Czechia, Dr Sakalova, Clinics for Hematology and Blood Transfusion KRVI, Bratislava, Slovakia, Dr Baglin, Addenbrooke's NHS Trust, Cambridge; Dr Bhavnani, Wigan and Leigh Health Service NHS Trust, Wigan; Dr Summerfield and Dr Chandler, General Hospital, Middlesbrough; Dr Bolton-Maggs, Alder Hey Children's Hospital, Liverpool, Dr Parker, Torrington Speedwell Practice, London; and Dr Marsh, St. George's Hospital Medical School, London.
Supported by a grant from the Deutsche Forschungsgemeinschaft (Th 459/2-2).
Address reprint requests to Bernd-Joachim Thiele, PhD, Institute of Biochemistry, Faculty of Medicine, Humboldt-University Berlin (Charité), Hessische Str. 3-4, 10115 Berlin, Germany.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal