Abstract
Epstein-Barr virus (EBV) has been shown to establish latency in resting B lymphocytes of the peripheral blood. This creates a virus reservoir in contrast to lytic virus replication, which is thought to be restricted to differentiated epithelial cells in vivo. So far, the route of transmission between B cells and the production of progeny virus in the epithelial tissue has remained unclear. Reverse transcriptase–polymerase chain reaction (RT-PCR) and immunohistochemistry analysis of 16 patients with acute infectious mononucleosis (IM) and 25 healthy seropositive donors was performed to detect lytic replication gene products in B lymphocytes of the peripheral blood. Transcriptional activity was found in peripheral blood B lymphocytes (PBLs) for BZLF1 in 88%, BALF2 in 50%, and BcLF1 in 25% of the tested IM patients. All positive results were further confirmed in enriched B-cell populations by antigen determination using immunostaining with the APAAP technique. Furthermore, we detected transcripts for BZLF1 in 72% and for BALF2 in 16% of peripheral B lymphocytes of healthy seropositive donors. In contrast to patients with IM, no signals for BcLF1 were ever found in healthy seropositive donors. In these individuals, lytic replication of EBV is probably restricted by immunologic and gene regulatory mechanisms, whereas in the absence of immunologic control, reflected here by IM patients, the production of infectious virus becomes visible in PBLs.
EPSTEIN-BARR VIRUS (EBV) is a ubiquitous lymphotrophic γ-herpesvirus that persists lifelong in the human body. Primary infection with EBV is asymptomatic during early childhood. However, in young adults and in older adolescents, nearly half of the cases of primary infection with the virus result in infectious mononucleosis (IM).1 IM is a lymphoproliferative disorder associated with fever, pharyngeal inflammation, and cervical lymphadenopathy. Primary infection is serologically characterized by the appearence of anti–virus capsid antigen (VCA) IgM and anti–early antigen (EA) IgM and transient development of anti–EA IgG in approximately 80% of infected individuals. Anti–VCA IgG, which persists throughout the life of normal individuals, occurs almost concurrently and continues to increase during the course of IM. Anti–EBV nuclear antigen (EBNA) IgG is absent during acute infection.2 Cell-mediated immunity to IM includes a cytotoxic T lymphocyte (CTL) response against lytic and latent EBV gene products,3 increased natural killer cell activity,4 and secretion of cytokines, including interleukin-1 (IL-1),5 IL-2,6 IL-4,7 IL-6,8 and interferon alpha and gamma.9 Normally, IM is a self-limiting disease that is controlled after initial infection by memory CTLs, which are thought to be of major importance in providing continuous immunosurveillance. In rare cases, EBV-infected individuals develop severe diseases associated with either lytic virus replication (oral hairy leukoplakia of the tongue,10 chronic fatigue syndrome,11 and chronic active EBV infection12 ) or latent virus replication (Burkitt's lymphoma [BL],13 undifferentiated nasopharyngeal carcinoma,14 and Hodgkin's disease15 ). The main portal for EBV to enter the human body is the oropharynx, and it is likely that both terminally differentiated epithelium16-18 and B cells19 are infected. In vitro infection of peripheral blood lymphocytes (PBLs) or umbilical cord lymphocytes induces their transformation into proliferating lymphoblasts (LCLs). This process is called immortalization, leading to the expression of a limited set of viral genes. These latent genes are EBV nuclear antigens (EBNA), 1, 2, 3A, 3B, 3C, and EBNA-LP, three membrane proteins named latent membrane protein 1 (LMP1), LMP2A, and LMP2B (also known as TP1 and TP2), and two highly expressed RNA species, “EBV-encoded RNAs” (EBER1 and EBER2).20,21 After immortalization, LCLs spontaneously switch to lytic replication in vitro and are easily inducible by chemical treatment,22 demonstrating that B cells are permissive for lytic replication.
In vivo EBV infection in the peripheral blood has been shown to be restricted to small resting B cells23,24 expressing only LMP2A or B cells that are driven by physiologic signals19 representing the “EBNA1-only programme.”24 The function of LMP2A is to block the signaling pathway that leads to proliferation of the B cell and probably triggers virus lytic replication.25 Although LMP2A has been shown to be a target for CTL response, it cannot be eliminated, because resting B cells lack the costimulatory molecule B7.23,26 The EBNA1 protein is essential for replication of the EBV genome in proliferating cells. All latent viral proteins except EBNA1 have been shown to be targets for CTLs.22,27-29 The prevention of EBNA1 processing and presentation on MHC1 molecules has been demonstrated to depend on a glycine-alanine repeat sequence in the coding region of EBNA1.30
Less is known about EBV reactivation and regulation of viral persistence. It is possible that EBV-infected B cells responding to physiologic signals such as cytokines, steroid hormones,31 tumor growth factor,32 and T cells expressing the CD40 ligand33 switch to lytic EBV replication similar to proliferating T cells infected by human T-cell lymphotrophic virus type I and human immunodeficiency virus type I.34 To study the route of EBV transmission from circulating peripheral B cells to epithelial tissue of the oropharynx, we analyzed transcripts of EBV lytic genes in B cells from 16 IM patients and five seronegative and 25 seropositive donors. We used a set of primers and probes for reverse transcriptase–polymerase chain reaction (RT-PCR) and nested PCR detecting the BZLF1 and BRLF1 reading frame encoding the immediate early Zta and Rta transactivator proteins, respectively, the BALF2 reading frame encoding the early DNA-binding protein EA, and the BcLF1 reading frame encoding the late VCA. We further confirmed our data by an immunohistochemical determination method for the immediate-early (BZLF1), early (EA), and late (VCA) proteins.
MATERIALS AND METHODS
Cell lines.The following cell lines were used to establish RT-PCR. BJAB, an EBV-negative cell line derived from a sporadic BL,35 served as a negative control. P3HR1/16,36 Akata,37 and Raji,38 EBV-positive BL lines, were used as positive controls after chemical treatment with phorbol 12-myristate 13-acetate (PMA) and butyrate.22 To test the sensitivity of primer-probe combinations, the lymphoblastoid cell line B95-8 (ATCC RL CRL 1612), established from marmoset B cells by immortalization with EBV in throat washings of a person with IM, was used for titration assays with an increasing number of cells of the EBV-negative cell line Hela S3 (ATCC CCL 2.2). B95-8 immortalized cells are known to contain approximately 5% cells in lytic cycle,39 and therefore were not induced by chemical treatment. All cells were maintained in RPMI 1640 medium supplemented with 1 mmol/L glutamine, 10% fetal serum, and 10 μg/mL gentamycin (GIBCO, Eggenstein, Germany).
Blood donors.Heparinized blood was taken from 16 patients with IM at the Medical School of the University of Lübeck and at the military hospital of German Bundeswehr Hamburg. The diagnosis of IM was based on clinical symptoms such as pharyngitis, cervical lymphadenopathy, fever, and atypical lymphocytes in blood smear, and was further confirmed by indirect immunofluorescence (Freca-Fluor; Fresenius, Bad Homburg, Germany) according to the following criteria: anti–EA IgG greater than 1:20, anti–VCA IgM greater than 1:20, anti–VCA IgG greater than 1:80, and anti–EBNA antibodies less than 1:10. Clinical symptoms existed for 3 to 28 days. In all cases, PBLs were purified by Ficoll (Biochrom, Berlin, Germany) density centrifugation. B lymphocytes were further enriched by negative selection using monoclonal antibodies and complement, as well as density centrifugation according to the manufacturer's protocol (One Lambda, Los Angeles, CA). The purity and titer of B lymphocytes was further controlled by FACS analysis (Coulter, Krefeld, Germany) and immunostaining with anti–CD19 antibody (Dako, Hamburg, Germany). The procedure resulted in a population of 85% to 90% pure B lymphocytes. For further studies, blood samples from 25 seropositive donors and five seronegative donors from the staff of the Institut für Medizinische Mikrobiologie und Hygiene, Regensburg, were prepared as described earlier. Serology was performed by a standard assay (Biotest, Frankfurt, Germany).40 In cases of seronegative donors, the absence of anti–VCA IgG, anti–VCA IgM, and anti–EBNA IgG antibodies was confirmed by immunofluorescence analysis.
RNA preparation and RT-PCR amplification.RNA was isolated from snap-frozen or freshly prepared cell pellets from enriched B lymphocytes by extraction in 1 mL RNAzol (WAK-Chemie, Bad Homburg, Germany) per 1 × 107 cells. Cells were homogenized by pipetting, 1 vol chloroform was added, and the suspension was mixed vigorously. The homogenate was centrifuged in a benchtop centrifuge for 30 minutes at 15,000 rpm and 4°C, and RNA in the upper aqueous phase was precipitated with 1 vol prechilled isopropanol. Then, the precipitate was pelleted by centrifugation for 30 minutes at 15,000 rpm and 4°C, washed once with prechilled 70% ethanol, and resuspended in diethyl-pyrocarbonate–treated water with a final concentration of 40 U RNAsin and 5 mmol/L dithiothreitol (Promega, Heidelberg, Germany).
RT-PCR assays were performed with aliquots of RNA representing 2 × 106 cells per reaction. RNA samples were initially heated for 5 minutes to 65°C, removing secondary structures, followed by incubation at 37°C for 10 minutes to allow 3′ primer annealing for reverse transcription. RT-PCR was performed in a one-tube reaction with MMLV-RT (Perkin Elmer Cetus, Branchburg, NJ) or with Tth-polymerase (Boehringer, Mannheim, Germany) for the BZLF1 and BRLF1 RT-PCRs according to the protocol of the manufacturer. For BALF2 and BcLF1 RT-PCR, samples were pretreated with 10 U DNAse (Ambion, Austin, TX) for 15 minutes at 37°C followed by denaturing of the enzyme for 5 minutes at 99°C, to avoid amplification of DNA contamination. PCR products from 35 rounds of amplification were separated on 2% agarose gels and then Southern-transferred onto positively charged nylon membranes (Boehringer) and hybridized with oligonucleotides specific for the amplified DNA sequences end-labeled with Digoxigenin (Dig) (Boehringer). The labeled probes were detected with alkaline phosphatase (AP) anti-Dig conjugated antibodies (Boehringer). CSPD* (Tropix; Sigma, Deisenhofen, Germany) was used as a substrate for the AP. Membranes were exposed for 5 minutes, and in some cases up to 1 hour. Primer-probe combinations are listed in Table 1, which presents nucleotide sequence and exon derivation (where appropriate) according to the coordinates of the B95-8 EBV genome. For quality control of RNA samples, we used histone 3.3 RT-PCR according to the method of Futscher et al.41
Oligonucleotide Primers and Probes Used in RT-PCR and Nested PCR Analysis
| Transcript . | Primer or Probe Designation . | Genome Coordinates . | Oligonucleotide Sequence . |
|---|---|---|---|
| BZLF1 (Zta) RNA | 5′ RT-PCR primer | 103194-103180 | 5′ ATTGCACCTTGCCGCCACCTTTG 3′ |
| 3′ RT-PCR primer | 102486-102463 | 5′ CGGCATTTTCTGGAAGCCACCCGA 3′ | |
| 5′ nested PCR primer | 103077-103057 | 5′ GACCAAGCTACCAGAGTCTAT 3′ | |
| 3′ nested PCR primer | 102530-102512/102657-102655 | 3′ CAGAATCGCATTCCTCCAGCGA 3′ | |
| Probe | 102772-102795 | 5′ CACTGCTGCTGCTGTTTGAACAGT 3′ | |
| BRLF1 (Rta) RNA | 5′ RT-PCR primer | 106161-106137 | 5′ ACAGGACACAACACCTCACTACAC 3′ |
| 3′ RT-PCR primer | 104399-104422 | 5′ ACTGCCTTGGACTTGGTTGACAGC 3′ | |
| 5′ nested PCR primer | 106136-106126/105183-105177 | 5′ CAAACAGACGCAGATGAGGC 3′ | |
| 3′ nested PCR primer | 104766-104787 | 5′ GCGGTGCCTATGGTGGCAGG 3′ | |
| Probe | 104868-104850 | 5′ AGTGATGCGCTACGTGTTA 3′ | |
| BALF2 (EA) RNA | 5′ RT-PCR primer | 163094-163072 | 5′ GTCAAGATGTTCAAGGACGTGG 3′ |
| 3′ RT-PCR primer | 162856-162878 | 5′ CTCATAGCACATACAGATGGGC 3′ | |
| 5′ nested PCR primer | 162998-162978 | 5′ GGTCAAGAGCTGCTACCACG 3′ | |
| 3′ nested PCR primer | 162881-162901 | 5′ GGAGATGTCCTGCAGGATGG 3′ | |
| Probe | 162911-162930 | 5′ GCGGTAAAACAGCTGGGTGA 3′ | |
| BcLF1 (VCA) RNA | 5′ RT-PCR primer | 136231-136210 | 5′ TATGCCCAATCCCAAGTACACG 3′ |
| 3′ RT-PCR primer | 135867-135888 | 3′ TGGACGGGTGGAGGAAGTCTTC 3′ | |
| 5′ nested PCR primer | 136137-136176 | 5′ ACACAGCAGCTACCGGTGGA 3′ | |
| 3′ nested PCR primer | 135995-136014 | 5′ TGTTGCAGGGAGTAGGTCTC 3′ | |
| Probe | 136119-136138 | 5′ ACGCGAGGAGAAGGTTATTC 3′ | |
| Histone 3.3* | 5′ RT-PCR primer | — | 5′ CCACTGAACTTCTGATTCGC 3′ |
| 3′ RT-PCR primer | 3′ GCGTGCTAGCTGGATGTCTT 3′ |
| Transcript . | Primer or Probe Designation . | Genome Coordinates . | Oligonucleotide Sequence . |
|---|---|---|---|
| BZLF1 (Zta) RNA | 5′ RT-PCR primer | 103194-103180 | 5′ ATTGCACCTTGCCGCCACCTTTG 3′ |
| 3′ RT-PCR primer | 102486-102463 | 5′ CGGCATTTTCTGGAAGCCACCCGA 3′ | |
| 5′ nested PCR primer | 103077-103057 | 5′ GACCAAGCTACCAGAGTCTAT 3′ | |
| 3′ nested PCR primer | 102530-102512/102657-102655 | 3′ CAGAATCGCATTCCTCCAGCGA 3′ | |
| Probe | 102772-102795 | 5′ CACTGCTGCTGCTGTTTGAACAGT 3′ | |
| BRLF1 (Rta) RNA | 5′ RT-PCR primer | 106161-106137 | 5′ ACAGGACACAACACCTCACTACAC 3′ |
| 3′ RT-PCR primer | 104399-104422 | 5′ ACTGCCTTGGACTTGGTTGACAGC 3′ | |
| 5′ nested PCR primer | 106136-106126/105183-105177 | 5′ CAAACAGACGCAGATGAGGC 3′ | |
| 3′ nested PCR primer | 104766-104787 | 5′ GCGGTGCCTATGGTGGCAGG 3′ | |
| Probe | 104868-104850 | 5′ AGTGATGCGCTACGTGTTA 3′ | |
| BALF2 (EA) RNA | 5′ RT-PCR primer | 163094-163072 | 5′ GTCAAGATGTTCAAGGACGTGG 3′ |
| 3′ RT-PCR primer | 162856-162878 | 5′ CTCATAGCACATACAGATGGGC 3′ | |
| 5′ nested PCR primer | 162998-162978 | 5′ GGTCAAGAGCTGCTACCACG 3′ | |
| 3′ nested PCR primer | 162881-162901 | 5′ GGAGATGTCCTGCAGGATGG 3′ | |
| Probe | 162911-162930 | 5′ GCGGTAAAACAGCTGGGTGA 3′ | |
| BcLF1 (VCA) RNA | 5′ RT-PCR primer | 136231-136210 | 5′ TATGCCCAATCCCAAGTACACG 3′ |
| 3′ RT-PCR primer | 135867-135888 | 3′ TGGACGGGTGGAGGAAGTCTTC 3′ | |
| 5′ nested PCR primer | 136137-136176 | 5′ ACACAGCAGCTACCGGTGGA 3′ | |
| 3′ nested PCR primer | 135995-136014 | 5′ TGTTGCAGGGAGTAGGTCTC 3′ | |
| Probe | 136119-136138 | 5′ ACGCGAGGAGAAGGTTATTC 3′ | |
| Histone 3.3* | 5′ RT-PCR primer | — | 5′ CCACTGAACTTCTGATTCGC 3′ |
| 3′ RT-PCR primer | 3′ GCGTGCTAGCTGGATGTCTT 3′ |
According to Futscher et al.41
Antigen determination by immunostaining.B lymphocytes were fixed on cover slips by 4 minutes' incubation in a mixture containing methanol:acetone (1:1) at −20°C. Unspecific staining was reduced by preincubation with preimmune rabbit serum (Dako, Hamburg, Germany). Immunostaining was performed using the following monoclonal antibodies: anti-BZLF1 (Dako), anti-EA, and anti-VCA (Chemikon, Tenekula). For detection, we used the AP (APAAP) technique (Dako). Slides were counterstained with hematoxylin and mounted in Immunoamount (Shandon, Astmoor, UK). B95-8 served as a positive control and BJAB cells as a negative control, respectively.
RESULTS
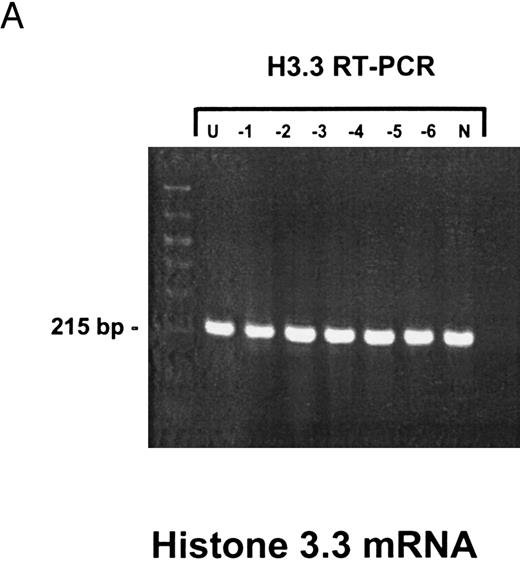
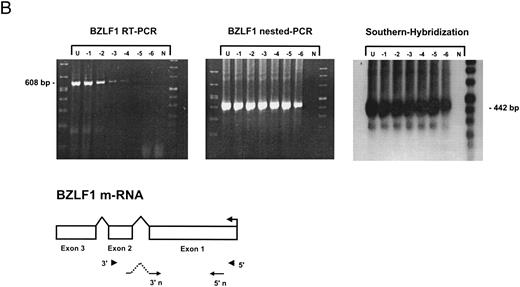
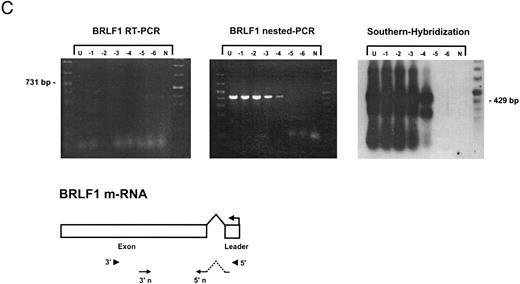
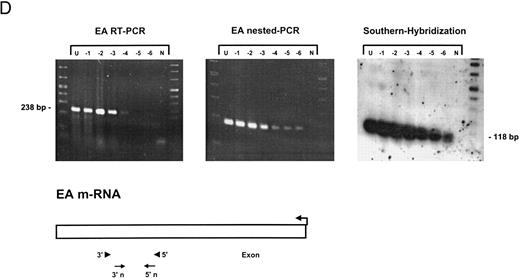
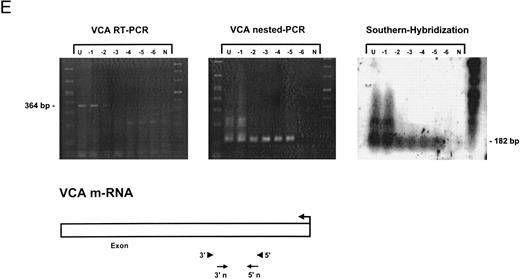
Sensitivity of RT-PCR for lytic EBV transcripts was enhanced by nested PCR and hybridization with Dig-labeled probes.The sensitivity of the RT-PCRs and nested PCRs was tested in series of 10-fold dilutions with P3HR1/16, Akata, and Raji cells treated for 36 hours with PMA and with untreated B95-8 cells in a background of EBV-negative Hela S3 cells. Before RNA extraction, cells were counted and mixed to generate samples containing 2 × 106 cells with a decreasing number of EBV-positive cells (dilutions from 10−1 to 10−6). Ten percent of the RT-PCR amplification product was used as a substrate for nested PCR. Typical results of RT-PCR, nested PCR, and hybridization with a Dig-labeled probe are shown in Fig 1B to E. Histone 3.3 RT-PCR was used to control the quality of the RNA preparations before EBV-specific RT-PCR and nested PCR. Results are presented in Fig 1A.
Detection of EBV lytic-cycle transcripts by RT-PCR amplification followed by nested PCR in RNA preparations from cell mixtures containing decreasing amounts of EBV-positive cells. RNA samples from a total of 2 × 106 cells were used as a template (U, undiluted). Negative controls (N) were performed with 2 × 106 EBV-negative Hela S3 cells. Dilutions are 10−1 to 10−6. (A) Quality control for B95-8 RNA preparation using histone 3.3 RT-PCR.41 Primer-probe combinations used for detection of BZLF1 (B), BRLF1 (C), BALF2 (D), and BcLF1 (E) are shown in the diagrams below the relevant transcripts. For BZLF1, BALF2, and BcLF1, RT-PCR and nested PCR were performed with B95-8 cells. For BRLF1 RNA detection where transcripts could not be obtained with the B95-8 cell line, we used PMA-treated P3HR1/16 cells. Nested PCR products yielding the predicted size (indicated at right) were further controlled by Southern hybridization with a specific probe.
Detection of EBV lytic-cycle transcripts by RT-PCR amplification followed by nested PCR in RNA preparations from cell mixtures containing decreasing amounts of EBV-positive cells. RNA samples from a total of 2 × 106 cells were used as a template (U, undiluted). Negative controls (N) were performed with 2 × 106 EBV-negative Hela S3 cells. Dilutions are 10−1 to 10−6. (A) Quality control for B95-8 RNA preparation using histone 3.3 RT-PCR.41 Primer-probe combinations used for detection of BZLF1 (B), BRLF1 (C), BALF2 (D), and BcLF1 (E) are shown in the diagrams below the relevant transcripts. For BZLF1, BALF2, and BcLF1, RT-PCR and nested PCR were performed with B95-8 cells. For BRLF1 RNA detection where transcripts could not be obtained with the B95-8 cell line, we used PMA-treated P3HR1/16 cells. Nested PCR products yielding the predicted size (indicated at right) were further controlled by Southern hybridization with a specific probe.
BZLF1 transcripts could be detected in all cell lines tested. Even in dilutions of two cells per sample (10−6), BZLF1 transcripts were found both in PMA-treated P3HR1/16, Akata, or Raji cells and in noninduced B95-8 cells. The strongest signals (Fig 1B) were obtained with induced Raji cells, which contain about 50 EBV genomes per cell and therefore express BZLF1 in high copy numbers after chemical treatment. Because we used a nested PCR and hybridization with Dig-labeled oligonucleotides, our approach is at least 100 times more sensitive than that used in a previous report by Tierney et al.42
The RT-PCR for BRLF1 was performed to detect transcripts containing the small 5′ leader exon and thus allowed detection of the bicistronic 3-kb BRLF1/BZLF1 RNA. In all cell lines tested, we could barely detect BRLF1 PCR products in the RT-PCR amplification. Even in cases of induced Raji cells, we were able to detect only a weak RT-PCR signal after hybridization of the amplified products. The nested PCR amplification clearly illustrated the efficiency of using a further primer pair. A BRLF1-specific signal was detected in dilutions up to 10−4 in PMA-treated Raji and P3HR1/16 cells (Fig 1C) and in dilutions up to 10−3 in Akata cells. No signal from spliced transcript of BRLF1 was obtained in the uninduced B95-8 cell line. The use of new primer pair combinations also did not increase the sensitivity, indicating that the bicistronic BRLF1/BZLF1 RNA is expressed in a lower copy number than the BZLF1 transcript derived from the Z promotor (Zp) in all cell lines tested.
Transcripts for EA could be detected in high copy numbers in PMA-induced P3HR1/16, Akata, and Raji cell lines and also in uninduced B95-8 (Fig 1D). For 2 × 106 and 2 × 105 PMA-treated P3HR1/16 or Raji cells, we could only obtain a smear, indicating an extremely high expression rate of EA in those induced cell lines (data not shown).
For late gene expression, we used an RT-PCR and nested PCR for the reading frame BcLF1 (VCA). We were able to detect signals in uninduced B95-8 cells up to a dilution of 10−5 for BcLF1 RNA transcripts (Fig 1E). Even the use of new RT-PCR and nested PCR primer combinations could not increase the sensitivity for detection of BcLF1 transcripts, demonstrating that the BcLF1 and BALF2 reading frames are not equally expressed in the cell lines examined. No signals were ever obtained in Raji cells, which lack the distal part of the BALF2 coding region, and the EBV-negative cell line BJAB.
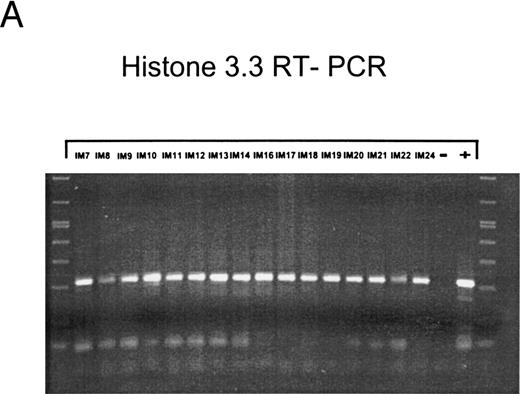
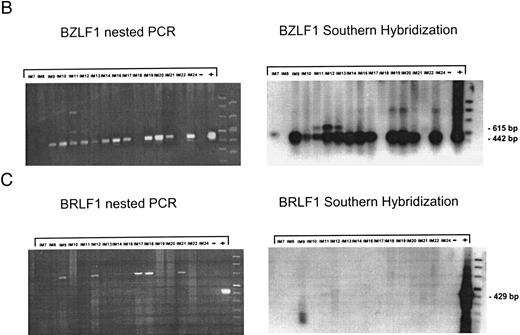
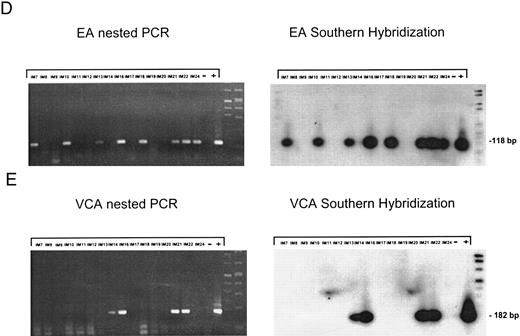
EBV lytically replicates in B cells in IM.During the course of primary infection, serologic data indicate lytic replication of EBV, and free virus DNA can be detected in the serum of patients with IM by PCR.43 Further, the tendency of B lymphocytes for lytic replication was demonstrated in vitro by induction of lytic gene expression by several methods and is obvious by the low percentage of spontaneous lytic replication in LCL.44 Therefore, we assumed that lymphoid cells in vivo play a crucial role in transmission of virus infection to epithelial tissue. To examine lytic virus replication in the peripheral blood, RNA was prepared from enriched B lymphocytes derived from 16 patients with serologically confirmed IM. For quality control of the prepared RNA samples, we used histone 3.3 RT-PCR (Fig 2A). Each sample was tested on at least two occasions of independent RT-PCR assays. Figure 2B to E presents the results of nested PCR and hybridization with Dig-labeled probes of samples from 16 IM patients assayed in parallel. Each track shows the PCR product from 2 × 106 B cells derived from IM patients, or from 2 × 106 B95-8 or P3HR1/16 cells treated with PMA as a positive control and 2 × 106 BJAB cells as a negative control.
Nested PCR screening and probe hybridization for BZLF1 (B), BRLF1 (C), BALF2 (D), and BcLF1 (E) in RNA preparations from freshly prepared and enriched B cells of IM patients. RNA from 2 × 106 cells was used as a template. Before the EBV-specific PCR, RNA quality was controlled by histone 3.3 RT-PCR (A).41 Positive controls were performed with 2 × 104 PMA-treated P3HR1/16 cells for BRLF1 and 2 × 106 untreated B95-8 cells for BZLF1, BALF2, and BcLF1 RT-PCR and nested PCR. For negative controls, we used 2 × 106 EBV-negative BJAB cells.
Nested PCR screening and probe hybridization for BZLF1 (B), BRLF1 (C), BALF2 (D), and BcLF1 (E) in RNA preparations from freshly prepared and enriched B cells of IM patients. RNA from 2 × 106 cells was used as a template. Before the EBV-specific PCR, RNA quality was controlled by histone 3.3 RT-PCR (A).41 Positive controls were performed with 2 × 104 PMA-treated P3HR1/16 cells for BRLF1 and 2 × 106 untreated B95-8 cells for BZLF1, BALF2, and BcLF1 RT-PCR and nested PCR. For negative controls, we used 2 × 106 EBV-negative BJAB cells.
BZLF1-specific mRNA products (442 bp) could be detected in 13 of 16 patients (Fig 2B). Interestingly, in some cases, we found a second (615 bp) BZLF1-specific transcript (Fig 2B: IM11, IM12, IM13, and IM22, lanes 5, 6, 7, and 15) indicating an unspliced BZLF1 RNA. Prior treatment of the RNA samples with DNAse confirmed the RNA-specific amplification of a 615-bp BZLF1 transcript. The BZLF1-specific transcript was then excised from the agarose gel and purified. Sequencing demonstrated that the PCR product resulted from a combination of 3′ RT primer with 5′ nested primer containing the 124 bp of intron 1 of BZLF1. Contamination of the nested PCR by the 3′ RT primer can be explained by the fact that 10% of the first PCR was used as a template for the nested PCR without prior purification (see Materials and Methods).
In the case of IM8, the RNA probe was probably degraded, because of the weak histone 3.3 signal (Fig 2A, lane 3). However, detection of Zta protein by immunohistochemistry also resulted in weak signals for BZLF1 in 0.04%, for EA protein in 0.02%, and for VCA protein in 0.02% of purified B cells. These results suggest the onset of immunologic control in patient no. IM8, since the decrease in RNA and protein synthesis can be detected a long time before the serologic markers (Table 2). In patient no. IM18, no BZLF1-specific PCR product could be detected, whereas immunostaining by the APAAP technique clearly showed expression of Zta protein in 0.2% of purified peripheral B lymphocytes. Therefore, we assume that the PCR failed in this case, probably due to heparin contamination, which blocks activity of the Tth-polymerase.
Summary of Serologic Data, Nested RT-PCR Analysis, and Antigen Determination in PBLs From IM Patients
| Patient No./Age (yr)/Sex . | Duration . | Serologic Data . | Nested PCR . | |||||
|---|---|---|---|---|---|---|---|---|
| . | . | EA IgG . | VCA IgM . | VCA IgG . | EBNA IgG . | BZLF1 (Zta) . | BALF2 (EA) . | BcLF1 (VCA) . |
| IM7/19/M | 21 | 1:160 | 1:20 | 1:640 | <1:10 | + | + | − |
| IM8/14/F | 21 | 1:80 | 1:640 | 1:1,280 | <1:10 | − | − | − |
| IM9/14/F | 14 | 1:320 | 1:640 | 1:1,280 | <1:10 | + | − | − |
| IM10/15/F | 12 | 1:160 | 1:640 | 1:1,280 | <1:10 | + | + | − |
| IM11/16/M | 11 | 1:10 | 1:320 | 1:1,280 | <1:10 | + | − | − |
| IM12/17/M | 12 | 1:640 | 1:640 | 1:1,280 | <1:10 | + | − | − |
| IM13/15/F | 7 | 1:80 | 1:320 | 1:640 | <1:10 | + | + | − |
| IM14/23/M | 6 | 1:80 | 1:320 | 1:640 | <1:10 | + | − | + |
| IM16/16/M | 14 | 1:160 | 1:80 | 1:640 | <1:10 | + | + | + |
| IM17/25/F | 28 | 1:160 | 1:20 | 1:640 | <1:10 | + | − | − |
| IM18/22/F | 3 | 1:10 | 1:20 | 1:640 | <1:10 | − | + | − |
| IM19/18/M | 14 | 1:1,280 | 1:640 | 1:2,560 | <1:10 | + | − | − |
| IM20/23/M | 10 | 1:160 | 1:640 | 1:2,560 | <1:10 | + | − | + |
| IM21/20/M | 16 | 1:640 | 1:320 | 1:1,280 | <1:10 | + | + | + |
| IM22/28/M | 9 | 1:160 | 1:1,280 | 1:320 | <1:10 | + | + | − |
| IM24/26/F | 14 | 1:64 | 1:32 | 1:1,024 | <1:10 | + | + | − |
| Patient No./Age (yr)/Sex . | Duration . | Serologic Data . | Nested PCR . | |||||
|---|---|---|---|---|---|---|---|---|
| . | . | EA IgG . | VCA IgM . | VCA IgG . | EBNA IgG . | BZLF1 (Zta) . | BALF2 (EA) . | BcLF1 (VCA) . |
| IM7/19/M | 21 | 1:160 | 1:20 | 1:640 | <1:10 | + | + | − |
| IM8/14/F | 21 | 1:80 | 1:640 | 1:1,280 | <1:10 | − | − | − |
| IM9/14/F | 14 | 1:320 | 1:640 | 1:1,280 | <1:10 | + | − | − |
| IM10/15/F | 12 | 1:160 | 1:640 | 1:1,280 | <1:10 | + | + | − |
| IM11/16/M | 11 | 1:10 | 1:320 | 1:1,280 | <1:10 | + | − | − |
| IM12/17/M | 12 | 1:640 | 1:640 | 1:1,280 | <1:10 | + | − | − |
| IM13/15/F | 7 | 1:80 | 1:320 | 1:640 | <1:10 | + | + | − |
| IM14/23/M | 6 | 1:80 | 1:320 | 1:640 | <1:10 | + | − | + |
| IM16/16/M | 14 | 1:160 | 1:80 | 1:640 | <1:10 | + | + | + |
| IM17/25/F | 28 | 1:160 | 1:20 | 1:640 | <1:10 | + | − | − |
| IM18/22/F | 3 | 1:10 | 1:20 | 1:640 | <1:10 | − | + | − |
| IM19/18/M | 14 | 1:1,280 | 1:640 | 1:2,560 | <1:10 | + | − | − |
| IM20/23/M | 10 | 1:160 | 1:640 | 1:2,560 | <1:10 | + | − | + |
| IM21/20/M | 16 | 1:640 | 1:320 | 1:1,280 | <1:10 | + | + | + |
| IM22/28/M | 9 | 1:160 | 1:1,280 | 1:320 | <1:10 | + | + | − |
| IM24/26/F | 14 | 1:64 | 1:32 | 1:1,024 | <1:10 | + | + | − |
Abbreviations: F, female; M, male; +, detectable; −, not detectable.
Duration of clinical symptoms (mo).
Figure 2B presents data for BRLF1-specific RNA transcripts obtained from the same 16 IM patients. None of the RNA samples showed a transcriptional signal for BRLF1. Since we tested different primer combinations for detection of the BRLF1/BZLF1 transcript, this suggests that during the course of IM, the Rp promotor is less active than the Zp promotor and that BZLF1 immediate-early transcripts are mainly derived from Zp.
Nested PCR for BALF2 mRNA showed a detectable signal in eight of 16 tested IM samples (Fig 2C). For patients no. IM9, IM12, and IM19 (lanes 3, 6, and 12), the failure to detect BALF2-specific transcript correlates with strongly elevated anti–EA IgG titers. In these cases, the immune system seems to eliminate lytically replicating B cells during primary infection. For patients no. IM14 and IM20, we could not detect a BALF2-specific PCR signal, although we detected EA protein in 0.08% of the enriched B cells.
Detection of RNA from the late gene product BcLF1, which indicates the production of progeny virus, was successful in four of 16 cases, IM14, IM16, IM20, and IM21 (Fig 2E, lanes 8, 9, 14, and 15). In cases IM16 and IM21, all levels of the lytic gene expression cascade could be shown, representing the production of progeny virus in B cells of the peripheral blood in these patients.
In summary, detection of transcriptional activity of the immediate-early gene BZLF1 and genes of the early and late stage of lytic replication indicated active replication of EBV in the peripheral blood of patients with IM. The overall results from serologic data, nested PCR analysis, and hybridization with labeled probes for 16 IM patients are presented in Table 2.
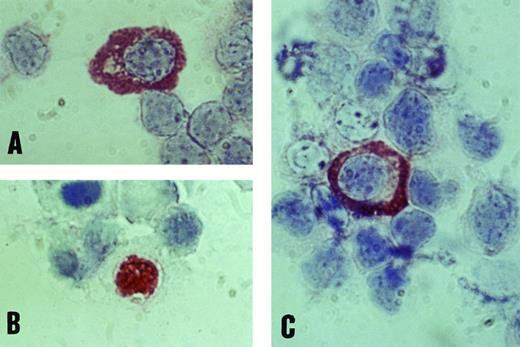
Immunochemical detection of EBV lytic-cycle proteins in B cells of the peripheral blood in IM.To confirm the data from PCR analysis, we further tested viral protein expression in enriched B-cell populations by APAAP antigen determination. Purification of peripheral blood mononuclear cells was controlled by FACS analysis and resulted in 85% to 90% pure B cells. Using monoclonal antibodies against BZLF1 (Zta), BALF2 (EA), and BcLF1 (VCA), we obtained nuclear staining for Zta and cytoplasmic staining for EA and VCA proteins in most patients with IM (Fig 3). The number of B cells expressing lytic EBV proteins fluctuated between 0.01% and 1.09%, with a tendency to decrease after the first week of acute IM. No specific protein signals could be detected in any of 25 healthy EBV-positive donors, not even in the case of donor no. 25, who represented an acute EBV reactivation state with elevated anti–EA IgM titers and a specific BALF2 PCR signal. Our data show that in contrast to the early phase of IM, EBV lytic replication is effectively controlled by the immune system in immunocompetent individuals.
Positive staining of B lymphocytes with monoclonal antibodies against EBV antigens EA (A), BZLF1 (B), and VCA (C) by immunostaining with the APAAP technique (original magnification × 1,000).
Positive staining of B lymphocytes with monoclonal antibodies against EBV antigens EA (A), BZLF1 (B), and VCA (C) by immunostaining with the APAAP technique (original magnification × 1,000).
BZLF1 is transcriptionally active in B cells derived from healthy seropositive donors.Recently, a specific CTL response against the immediate-early protein Zta has been reported in healthy seropositive donors,45 suggesting expression of this protein in the peripheral blood. Therefore, we tested the transcriptional activity for BZLF1, BRLF1, BALF2, and BcLF1 in a further set of RT-PCR and nested PCR experiments with RNA preparations from 25 healthy seropositive and five seronegative donors that had been confirmed by immunofluorescence analysis. In four of 25 seropositive donors (patients no. 1, 2, 23, and 25), anti–EA IgG antibodies, and in one case, anti–EA IgM (donor no. 25), were also detected, indicating acute EBV reactivation. Aliquots of RNA representing 2 × 106 B cells were subjected to RT-PCR and nested PCR analysis using primer-probe combinations described above. We used 2 × 106 non–PMA-treated B95-8 cells for BZLF1, BALF2, and BcLF1 RT-PCR and 2 × 106 P3HR1/16 cells treated with PMA for BRLF1 RT-PCR as a positive control. For negative controls, we tested 2 × 106 BJAB cells. Table 3 shows the overall data from serologic and nested PCR analysis for healthy positive donors.
Summary of Serologic Data and Nested RT-PCR Analysis in PBLs From Seropositive Donors
| Patient No. . | Serologic Data . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | ELISA . | IF . | Nested RT-PCR . | . | . | . | ||||||
| . | EA IgM . | EA IgG . | EBNA IgG . | VCA IgM . | VCA IgG . | EBNA IgG . | Zta . | EA . | VCA . | . | . | . |
| 1 | − | + | ++ | + | + | − | ||||||
| 2 | − | + | ++ | + | + | − | ||||||
| 3 | − | − | ++ | + | − | − | ||||||
| 4 | − | − | ++ | + | − | − | ||||||
| 5 | − | − | ++ | + | − | − | ||||||
| 6 | − | − | ++ | + | − | − | ||||||
| 7 | − | − | ++ | + | − | − | ||||||
| 8 | − | − | − | − | 1:512 | <1:10 | − | − | − | |||
| 9 | − | − | + | + | − | − | ||||||
| 10 | − | − | − | − | − | − | − | − | − | |||
| 11 | − | − | ++ | + | − | − | ||||||
| 12 | − | − | ++ | − | − | − | ||||||
| 13 | − | − | ++ | − | − | − | ||||||
| 14 | − | − | ++ | + | − | − | ||||||
| 15 | − | − | − | − | − | − | − | − | − | |||
| 16 | − | − | − | − | − | − | − | − | − | |||
| 17 | − | − | ++ | − | − | |||||||
| 18 | − | − | ++ | − | − | − | ||||||
| 19 | − | − | ++ | + | − | − | ||||||
| 20 | − | − | ++ | − | − | |||||||
| 21 | − | − | ++ | + | − | − | ||||||
| 22 | − | − | ++ | + | − | − | ||||||
| 23 | − | + | ++ | + | + | − | ||||||
| 24 | − | − | − | − | − | − | − | − | − | |||
| 25 | + | + | ++ | + | + | − | ||||||
| 26 | − | − | ++ | − | − | − | ||||||
| 27 | − | − | ++ | + | − | − | ||||||
| 28 | − | − | − | − | − | − | − | − | − | |||
| 29 | − | − | ++ | + | − | − | ||||||
| 30 | − | − | ++ | + | − | − | ||||||
| Patient No. . | Serologic Data . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | ELISA . | IF . | Nested RT-PCR . | . | . | . | ||||||
| . | EA IgM . | EA IgG . | EBNA IgG . | VCA IgM . | VCA IgG . | EBNA IgG . | Zta . | EA . | VCA . | . | . | . |
| 1 | − | + | ++ | + | + | − | ||||||
| 2 | − | + | ++ | + | + | − | ||||||
| 3 | − | − | ++ | + | − | − | ||||||
| 4 | − | − | ++ | + | − | − | ||||||
| 5 | − | − | ++ | + | − | − | ||||||
| 6 | − | − | ++ | + | − | − | ||||||
| 7 | − | − | ++ | + | − | − | ||||||
| 8 | − | − | − | − | 1:512 | <1:10 | − | − | − | |||
| 9 | − | − | + | + | − | − | ||||||
| 10 | − | − | − | − | − | − | − | − | − | |||
| 11 | − | − | ++ | + | − | − | ||||||
| 12 | − | − | ++ | − | − | − | ||||||
| 13 | − | − | ++ | − | − | − | ||||||
| 14 | − | − | ++ | + | − | − | ||||||
| 15 | − | − | − | − | − | − | − | − | − | |||
| 16 | − | − | − | − | − | − | − | − | − | |||
| 17 | − | − | ++ | − | − | |||||||
| 18 | − | − | ++ | − | − | − | ||||||
| 19 | − | − | ++ | + | − | − | ||||||
| 20 | − | − | ++ | − | − | |||||||
| 21 | − | − | ++ | + | − | − | ||||||
| 22 | − | − | ++ | + | − | − | ||||||
| 23 | − | + | ++ | + | + | − | ||||||
| 24 | − | − | − | − | − | − | − | − | − | |||
| 25 | + | + | ++ | + | + | − | ||||||
| 26 | − | − | ++ | − | − | − | ||||||
| 27 | − | − | ++ | + | − | − | ||||||
| 28 | − | − | − | − | − | − | − | − | − | |||
| 29 | − | − | ++ | + | − | − | ||||||
| 30 | − | − | ++ | + | − | − | ||||||
Abbreviations: +, detectable; −, not detectable; ++, elevated titers; IF, immunofluorescence; ELISA, enzyme-linked immunosorbent assay.
In 18 of 25 EBV-seropositive donors (72%), we were able to detect BZLF1-specific RNA (442 bp). As with IM patients, we could distinguish between unspliced and spliced RNA of BZLF1. In donors no. 2, 8, 12, 18, and 26, there was clear evidence of the unspliced BZLF1 RNA (615 bp), which was controlled by treatment of the RNA samples with DNAse before PCR analysis. Results from three of the individuals were confirmed 2 weeks later with additional blood samples (data not shown).
When the same RNA samples were screened for the 3-kbp bicistronic transcripts starting from the Rp, no specific transcripts were detected from any of these donors, despite prolonged gel exposure. Note that in the same assay, signals were obtained readily from the induced P3HR1/16 cells (data not shown). In four cases (patients no. 1, 2, 23, and 25), we detected a BALF2-specific RNA signal. These observations were consistent with the serologic data indicating EBV lytic replication in the peripheral blood, possibly caused by a secondary condition (angina), since we could show a correlation in at least one of the four cases (patient no. 25). No positive signals were ever obtained in the seronegative control group.
The same panel of healthy donors were tested for the presence of BcLF1 RNA, which indicates a complete lytic replication cycle in the peripheral blood. None of the 25 seropositive donors yielded a BcLF1-specific signal, not even patient no. 25, who showed an elevated anti–EA IgM titer. Therefore, in contrast to patients with IM, transcriptional activity of BZLF1 but no productive EBV replication could be detected in healthy seropositive donors.
DISCUSSION
There are several observations that favor a role for B cells in the lytic EBV cycle; high titers of antibodies against lytic-cycle proteins during the early phase of primary infection and high amounts of free virus DNA in the serum of IM patients,43,46 indicating productive virus replication in the peripheral blood. Furthermore, a small percentage of lymphoid cells in vitro spontaneously switch to lytic replication and have been shown to support lytic replication after treatment with PMA,22 Ca ionophor, and anti-IgG antibodies.47
In an effort to understand the role of peripheral B lymphocytes in the transmission of virus infection to epithelial tissue in the oropharynx, we used RT-PCR in combination with nested PCR to increase the sensitivity for detecting lytic EBV transcripts for the immediate-early genes BZLF1 (Zta) and BRLF1 (Rta), early gene BALF2 (EA), and late gene BcLF1 (VCA). Using this PCR-based method, we were able to detect transcription of BZLF1 and BALF2 from a two-cell input, Rp-derived bicistronic transcripts from a 200-cell input, and BcLF1-specific transcripts from a 20-cell input. Assuming that the number of infected cells in IM is one in 102 to one in 103,48,49 and during postconvalescence is one in 104 to one in 106,50 we anticipated the detection of productive EBV replication by testing 2 × 106 enriched B cells per assay.
The results from IM patients demonstrated that during the course of primary infection, the spectrum of lytic EBV transcripts reflected by BZLF1 (Zta)-, BALF2 (EA)-, and BcLF1 (VCA)-specific transcripts is expressed in vivo in circulating B cells. No bicistronic BZLF1/BRLF1 transcripts derived from the Rp promotor could be detected. Interestingly, we found a second BZLF1-specific signal containing the 124 bp of the first BZLF1 intron. Our findings were further confirmed by immunohistochemistry analysis, where we demonstrated protein expression in isolated B cells ranging from 0.01% to 1.09% for BZLF1, EA, and VCA protein, with a tendency to decrease during convalescence. To our knowledge, this is the first demonstration that lytic EBV proteins are expressed in primary infected B-lymphoid cells in vivo. In contrast, no lytic EBV proteins were ever detected in healthy seropositive donors.
Previous studies by Tierney et al42 have reported the establishment of latency in peripheral blood mononuclear cells during the course of IM and failed to detect BZLF1 transcripts in the majority of patients with IM. In contrast, we have shown BZLF1 transcripts in 13 of 18 IM patients. We believe this is due to our more sensitive nested PCR detection method. Our results are in accordance with the RT-PCR assay used in the same study, which detected both F promotor (Fp) and Q promotor (Qp) usage in peripheral B cells from most of the IM patients examined. This approach was not able to distinguish between transcripts derived from either promotor. Since the Fp-driven transcripts were only expressed in cells entering the lytic replication cycle,39 51 this is in accordance with lytically replicating cells in the peripheral blood during IM.
Interestingly, those investigators further describe Fp usage in healthy long-term virus carriers, which leads us to conclude that lytic EBV replication in the B-cell pool also takes place in healthy seropositive donors. As demonstrated in bone marrow transplantation studies, reconstitution with EBV-negative bone marrow leads to a loss of secretion of infectious EBV in the oropharynx of the recipient, whereas reconstitution with seropositive bone marrow leads to a change in the serotype of the donor.52 53 Production of progeny virus in the oropharynx must therefore occur via lytically replicating PBLs that infiltrate the epithelial tissue of the oropharynx.
To compare the situation during primary infection with the situation in healthy seropositive donors, we extended the present approach to 25 seropositive healthy donors. We demonstrated BZLF1-specific RNA signals in 72% of the tested individuals. As shown in IM patients, two different forms of BZLF1 transcripts could be detected, probably pointing to a possible posttranscriptional regulation mechanism of BZLF1.54 Differential splicing of the BZLF1 hnRNA has also been reported in NPC patients,55 which may lead to abortion of lytic EBV replication in these cells. Furthermore, as in patients with IM, we were unable to detect a BRLF1-specific signal in samples from seropositive donors. We therefore assume that in EBV-infected B cells in vivo, BZLF1 transcripts are mainly derived from the Zp and not from the Rp. Detection of EA transcripts was restricted to subjects who showed elevated IgGs (donor no. 1, 2, 23, and 25) or IgM (donor no. 25) titers against EA, indicating an acute EBV reactivation. None of the seropositive donors showed VCA-specific transcripts, demonstrating control of the lytic EBV cycle by an intact immune system.
The frequency with which transcripts of the immediate-early, early, and late viral cascade are expressed is an additional interesting feature. The viral replicative cycle is almost certainly initiated, but it appears to be aborted before expression of late gene products and genome replication. This is supported by observations by Decker et al,50 who demonstrated an absence of viral genome replication in the peripheral blood of healthy seropositive donors by an in situ lysis gel technique. The viral strategy to restrict lytic reactivation in peripheral lymphoid cells is further reflected by the development of several regulation mechanisms, which are well characterized for the immediate-early gene BZLF1. This includes transcriptional56-60 and posttranscriptional control,54 posttranslational regulation by protein-protein interaction61-63 and modification,64 as well as the CTL response depending on MHC1 alleles of the infected individual.45
In summary, our experiments clearly demonstrate that B cells of the peripheral blood are capable, at least to a certain extent, of lytic EBV replication. Previously, in accordance with our data, parallels between B-cell biology and EBV pathogenesis have provided a model of virus transmission from the circulating peripheral B cells to the epithelial cells of the oropharynx.24 This implies migration of EBV-infected B cells to the mucosal epithelium and secretion of IgA triggered by physiologic signals such as IL-10 or tumor growth factor β. Since EBV encodes an IL-10 homolog (BCRF1)65 during lytic replication, it is likely that recruitment of virus-infected B cells to the epithelium could be controlled by EBV itself. Here, production of infectious EBV might occur either by lytically replicating B cells per se or by infection of epithelial cells in the oropharynx via an IgA-mediated pathway,66 cell fusion,67 or penetration of free virus via gp220/350 binding to the cellular CD21 receptor.
ACKNOWLEDGMENT
We thank Dr Siegers (ENT Department, University of Lübeck School of Medicine) and Dr Georgi (ENT Department, military hospital of the German Bundeswehr) for providing samples from IM patients, and J. Middeldorp (Boxtel, The Netherlands) for the monoclonal EBNA1 antibody. We also thank Dr Hans Kirchner, Dr Gregor Bein, and Mark Ibberson for critical discussion. Our special thanks to Christian Gerdes and Bernhard Leschonszki for synthesizing oligonucleotides, and to Mirko Ritter and Susanne Schmaus for excellent technical assistance.
Supported by the Deutsche Forschungsgemeinschaft (Wo 227/6).
Address reprint requests to Fritz M. Schwarzmann, PhD, Institut für Medizinische Mikrobiologie und Hygiene, Franz Josef Strauβ Allee 11, D-93053 Regensburg, Germany.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal