Abstract
PU.1 is a member of the ets family of transcription factors and is expressed in Friend virus-induced murine erythroleukemia (MEL) cells as a consequence of proviral integration into the PU.1/Spi-1 locus. After induction of MEL cell differentiation by treatment with dimethylsulfoxide (DMSO), expression of the PU.1/Spi-1 gene decreased before induction of β-globin gene expression. Overexpression of PU.1 by using a zinc-inducible expression plasmid in MEL cells resulted in unexpected growth inhibition of the transfectants. When PU.1-overexpressing transfectants were treated with DMSO, growth inhibition became much pronounced and apoptosis was induced. Expression of the β-globin gene was not induced under this condition. Neither growth inhibition nor apoptosis was induced in MEL cells after expression of mutant PU.1 proteins with a deletion of the activation domain or the DNA-binding Ets domain irrespective of the presence of DMSO. Interestingly, β-globin gene expression was not induced in the transfectants expressing the former mutant, whereas it was induced in those expressing the latter one in the presence of DMSO. These results indicate that overexpression of PU.1 in MEL cells results in growth and differentiation inhibition and, in conjunction with DMSO treatment, apoptotic cell death. These results also suggest that the activation domain and the Ets domain of PU.1 contribute differently to induction of these effects.
MALIGNANT TRANSFORMATION is a consequence of disturbance of cell growth and differentiation that are tightly controlled by tissue-specific gene expression in normal cells. It is widely accepted that tissue-specific gene expression is regulated by transcription factors that are expressed in a cell-type– and developmental-stage–specific fashion. Therefore, inappropriate expression of such transcription factors could lead to malignant transformation of the cells.
PU.1 is a member of the ets family of transcription factors and is originally identified as a protein that binds to a purine-rich sequence (PU box) in the promoter of the mouse major histocompatibility complex (MHC) class II I-Aβ gene.1 PU.1 is expressed in hematopoietic cells, predominantly in the B-cell and macrophage lineages.1 Recent evidence has been accumulating to suggest that PU.1 is involved in the regulation of expression of many hematopoietic cell-specific genes. For example, PU.1 has been shown to bind to the enhancer region of the Ig heavy chain gene2 and light chain genes3-5 in B cells and the promoter region of the CD11 gene,6 CD18 gene,7 granulocyte-macrophage colony-stimulating factor receptor gene,8 and macrophage scavenger receptor gene9 10 in monocytic cells.
PU.1 is identical to the product of the Spi-1 proto-oncogene.11,12 In most cases of Friend virus-induced murine erythroleukemia (MEL), the provirus of spleen focus-forming virus (SFFV) integrates at the Spi-1 locus.13-16 The first stage of the MEL genesis, polyclonal expansion of erythroblasts with the capability of a limited self-renewal and terminal differentiation into erythrocyte-like cells, is observed. This initial polyclonal expansion is the result of constitutive stimulation of erythropoietin (Epo) receptor by binding of the gp55 glycoprotein encoded by the env gene of SFFV.17,18 After several weeks, more malignant leukemic cells arise.19,20 In the leukemic cells of this stage, both activation of the PU.1/Spi-1 gene by proviral integration at the locus and inactivation of the p53 tumor suppressor gene by gene mutation, gene deletion, and/or SFFV proviral integration at the locus occur.21-23 It is conceivable that these two genetic alterations are implicated in malignant transformation of MEL cells. If this is the case, PU.1 might contribute to growth enhancement and differentiation inhibition of the leukemic cells. Schuetze et al24 reported that recommitment of erythroid differentiation and loss of immortality of MEL cells coincides with a decrease in the amount of PU.1.
In this study, to further elucidate the functional role of PU.1 in MEL genesis, we introduced a zinc-inducible expression plasmid of the mouse PU.1/Spi-1 gene in MEL cells and investigated the resulting effects of overexpression of PU.1 on growth and differentiation of MEL cells. Furthermore, we made deletion mutants of PU.1 and expressed them in MEL cells to determine the region(s) required for induction of the effects.
MATERIALS AND METHODS
Plasmid construction.The full-length cDNA of the mouse PU.1/Spi-1 gene12 and a zinc-inducible expression vector containing the human metallothionein promoter, pSVneoHMT-Ter, were kindly donated by Drs D. Kabat (Department of Biochemistry and Molecular Biology, School of Medicine, Oregon Health Sciences University, Portland, OR) and M. Obinata (Department of Cell Biology, Institute of Development, Aging and Cancer, Tohuku University, Sendai, Japan), respectively. An expression plasmid of the PU.1/Spi-1 gene, pPU.1-sense, was constructed by subcloning the full-length PU.1/Spi-1 cDNA in the sense orientation under the human metallothionein promoter in pSVneoHMT-Ter. A control plasmid, pPU.1-antisense, was constructed as described above except that the cDNA was in the antisense orientation.
A mutant PU.1/Spi-1 gene encoding the PU.1 with a deletion of amino acids 166 through 255 corresponding to the Ets domain25 was generated by polymerase chain reaction (PCR) using Pfu DNA polymerase (Stratagene, La Jolla, CA). The primers used were as follows: no. 1, 5′-TGACCCACGACCGTCCAGTC; no. 2, 5′-CACAGAGCTGATGCACGTCCTCGAT; no. 3, 5′-GGACGTGCATCAGCTCTGTGAAGTG; and no. 4, 5′-GTCTCTGCGGGCGATCAGTG. The first reaction to generate the DNA region encoding amino acids 1 through 165 was performed with primers no. 1 and 2 using the full-length PU.1/Spi-1 cDNA as a template. The second reaction to generate the DNA region encoding amino acids 256 through 260 was performed with primers no. 3 and 4 using the same template. The product of the first PCR was complementary in its 3′ end to the 5′ end of the product of the second PCR, because primers no. 2 and 3 were complementary at their 5′ ends. To generate the final product, the third reaction was performed with primers no. 1 and 4 using the products of the first and second reactions as templates. DNA sequencing confirmed that no additional mutations were introduced during the PCR process. The final PCR product was subcloned in the sense orientation in pSVneoHMT-Ter to construct the expression plasmid, pPU.1-Δ E-sense, or in the antisense orientation to construct the control plasmid, pPU.1-Δ E-antisense.
Another mutant PU.1/Spi-1 gene encoding the PU.1 with a deletion of amino acids 74 through 122 that includes the glutamine-rich subdomain (amino acids 74 through 100) of the activation domain26 27 was generated by removing the corresponding nucleotides from the full-length PU.1/Spi-1 cDNA. The deleted cDNA was subcloned in the sense orientation in pSVneoHMT-Ter to construct the expression plasmid, pPU.1-Δ A-sense or in the antisense orientation to construct the control plasmid, pPU.1-Δ A-antisense.
Cell culture.A Friend virus-induced mouse erythroleukemia cell line, MEL-B8/3, was maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. For transfection, 107 cells were suspended in 1 mL of phosphate-buffered saline with 10 μg of plasmid DNAs and then transfected by electroporation at 450 V and 960 μF using a Gene Pulser (Bio-Rad, Richmond, CA). The cells were then seeded into 10 mL of culture medium, and then transfectants were selected in medium containing 500 μg/mL of G418 (Geneticin; Wako, Osaka, Japan). Clones of the transfectants were obtained using the limiting dilution method.
Southern and Northern blot analyses.Ten micrograms of high molecular weight DNA samples was digested with 50 U of appropriate restriction endonucleases, separated in a 0.7% agarose gel, and then transferred onto a nylon membrane. Twenty micrograms of total RNA samples was denatured with formamide, separated in a 1.0% agarose gel containing formaldehyde, and then transferred onto a nylon membrane.
A 0.43-kb EcoRI-Nco I fragment of mouse PU.1/Spi-1 cDNA (mentioned above), a 0.81-kb BamHI fragment of human β-globin cDNA, a 2.07-kb BamHI fragment of mouse β-actin cDNA, and a 0.76-kb Pvu II fragment of bacterial neomycin phosphotransferase DNA were used as probes. The probes were labeled with 32P-dCTP by using a random primer DNA labeling kit (Boehringer Mannheim, Mannheim, Germany). Hybridization and wash were performed as described previously.28
Western blot analysis.Fifty micrograms of total protein samples was subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a nylon membrane. The membranes were blocked in TBS-T (20 mmol/L Tris-HCl, pH 7.5, 137 mmol/L NaCl, 0.1% Tween-20) containing 5% nonfat dried milk and then probed with an antimouse PU.1 rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) appropriately diluted in TBS-T. After wash with TBS-T, bound antibody was detected by using the ECL system (Amersham, Amersham, UK).
Cell growth analysis.Cells (2 × 106) were placed in 10 mL of culture medium and the cell number was determined every 24 hours for 5 days.
DNA fragmentation analysis.DNA samples were prepared according to the method of Smith et al.29 Twenty microliters of samples was mixed with 10 μL of loading buffer (10 mmol/L EDTA, 0.25% bromophenol blue, 1% low melting point agarose, and 40% sucrose) and electrophoresed in a 1.5% agarose gel containing ethidium bromide.
Electron microscopy.The cells were fixed with 3% glutaraldehyde in phosphate buffer at 4°C for 1 hour, postfixed with osmium tetroxide at 4°C for 1 hour, and then processed and embedded in epon resin according to the routine methods. Ultrathin sections were contrasted with uranyl-acetate and lead citrate and then examined with a transmission electron microscopy (1200EX; JEOL, Tokyo, Japan) at 60 kV.
RESULTS
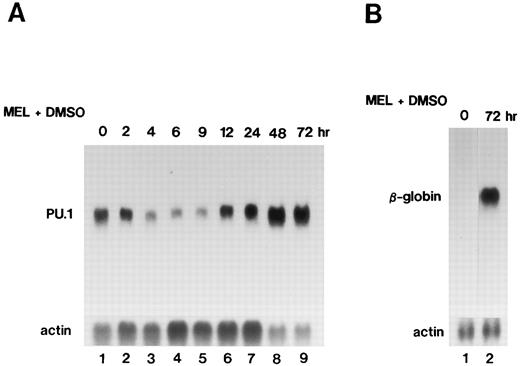
Expression of the PU.1/Spi-1 and β-globin genes during differentiation of MEL cells.When MEL-B8/3 cells were treated with 1.5% dimethylsulfoxide (DMSO) to induce differentiation into erythrocyte-like cells, expression of the PU.1/Spi-1 gene was decreased during the first several hours, with the lowest level at 6 hours, and expression of the β-globin gene was remarkably increased by 72 hours after the differentiation induction (Fig 1). Decreased expression of the PU.1/Spi-1 gene was also observed in another Friend virus-induced MEL cell line, MEL-D1, after differentiation induction by DMSO treatment (data not shown). These observations were consistent with earlier reports showing a decline of PU.1/Spi-1 mRNA in MEL cells after differentiation induction.24 25 On the basis of these observations, we hypothesized that enforced expression of the PU.1/Spi-1 gene may affect the status of growth and differentiation of MEL cells. To examine this possibility, we introduced the zinc-inducible expression plasmid of the PU.1/Spi-1 gene, pPU.1-sense, into MEL-B8/3 cells.
Northern blot analysis of expression of the PU.1/Spi-1 gene (A) and the β-globin gene (B) in MEL-B8/3 cells after the differentiation induction. Total RNA was extracted from the cells at the indicated time after the addition of 1.5% DMSO to the culture, separated in a 1.0% agarose gel, transferred to a nylon membrane, and then hybridized with 32P-labeled probes.
Northern blot analysis of expression of the PU.1/Spi-1 gene (A) and the β-globin gene (B) in MEL-B8/3 cells after the differentiation induction. Total RNA was extracted from the cells at the indicated time after the addition of 1.5% DMSO to the culture, separated in a 1.0% agarose gel, transferred to a nylon membrane, and then hybridized with 32P-labeled probes.
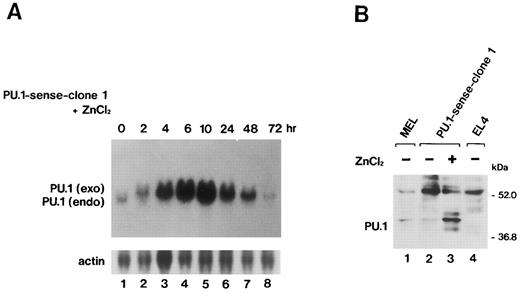
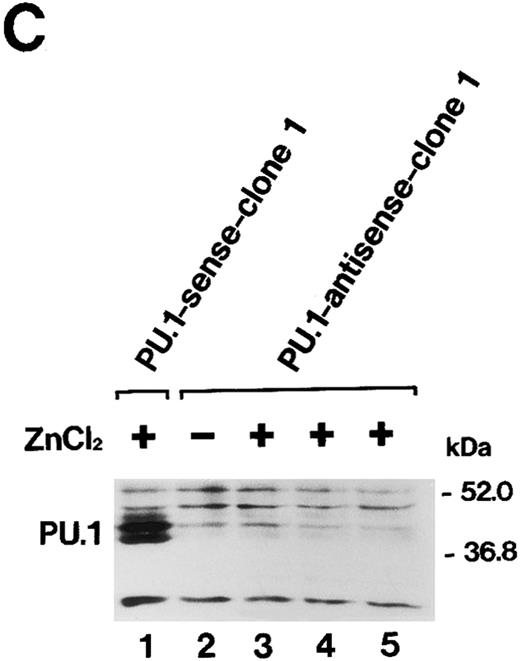
Overexpression of PU.1 in MEL cells.After the electroporation of MEL-B8/3 cells with the expression plasmid pPU.1-sense, containing the mouse PU.1/Spi-1 gene under the human metallothionein promoter, and subsequent G418 selection, several clones of transfectants, designated PU.1-sense clones, were obtained. Among these clones, PU.1-sense clone 1 and clone 2 were mainly used in subsequent analyses. Four clones of control transfectants, PU.1-antisense clone 1 and clone 2 and PU.1-mock clone 1 and clone 2, were also obtained by electroporation of MEL-B8/3 cells with control plasmids, pPU.1-antisense, and pSVneoHMT-Ter, respectively. Southern blot analysis using bacterial neomycin phosphotransferase DNA as a probe showed that all of clones retained transferred plasmid DNA (data not shown). Northern blot analysis showed transiently and significantly expressed exogenous PU.1/Spi-1 mRNA after the addition of 100 μmol/L ZnCl2 to the culture besides the 1.4-kb endogenous PU.1/Spi-1 mRNA in the PU.1-sense clone 1 cells (Fig 2A). Western blot analysis confirmed that the level of PU.1 was remarkably increased in the cells in the presence of ZnCl2 (Fig 2B). Induction of overexpression of the PU.1/Spi-1 gene and PU.1 protein by ZnCl2 was also observed in PU.1-sense clone 2 and in all of the other clones of PU.1-sense transfectants examined (data not shown). In the PU.1-antisense clone 1 cells, expression of PU.1 was decreased, but not completely suppressed, at least during the first 48 hours after the addition of ZnCl2 (Fig 2C). The similar results were observed in the PU.1-antisense clone 2 cells (data not shown).
(A) Northern blot analysis of induction of PU.1/Spi-1 gene expression in the PU.1-sense transfectants. Total RNA was extracted from the PU.1-sense clone 1 cells at the indicated time after the addition of 100 μmol/L ZnCl2 to the culture and hybridized with 32P-labeled probes. (B) Western blot analysis of overexpression of the PU.1 protein in the PU.1-sense transfectants. Total protein was extracted from the parental MEL-B8/3 cells (lane 1), PU.1-sense clone 1 cells (lane 2), and PU.1-sense clone 1 cells cultured for 8 hours with 100 μmol/L ZnCl2 (lane 3) and was probed with antimouse PU.1 antibody. Total protein of PU.1-negative T-cell lymphoma EL4 was also loaded as a negative control (lane 4). (C) Western blot analysis of suppression of expression of the PU.1 protein in the PU.1-antisense transfectants. Total protein was extracted from the PU.1-sense clone 1 cells cultured for 8 hours with 100 μmol/L ZnCl2 (lane 1), PU.1-antisense clone 1 cells (lane 2), and PU.1-antisense clone 1 cells cultured for 10 hours (lane 3), 24 hours (lane 4), and 48 hours (lane 5) with 100 μmol/L ZnCl2 and was probed with antimouse PU.1 antibody.
(A) Northern blot analysis of induction of PU.1/Spi-1 gene expression in the PU.1-sense transfectants. Total RNA was extracted from the PU.1-sense clone 1 cells at the indicated time after the addition of 100 μmol/L ZnCl2 to the culture and hybridized with 32P-labeled probes. (B) Western blot analysis of overexpression of the PU.1 protein in the PU.1-sense transfectants. Total protein was extracted from the parental MEL-B8/3 cells (lane 1), PU.1-sense clone 1 cells (lane 2), and PU.1-sense clone 1 cells cultured for 8 hours with 100 μmol/L ZnCl2 (lane 3) and was probed with antimouse PU.1 antibody. Total protein of PU.1-negative T-cell lymphoma EL4 was also loaded as a negative control (lane 4). (C) Western blot analysis of suppression of expression of the PU.1 protein in the PU.1-antisense transfectants. Total protein was extracted from the PU.1-sense clone 1 cells cultured for 8 hours with 100 μmol/L ZnCl2 (lane 1), PU.1-antisense clone 1 cells (lane 2), and PU.1-antisense clone 1 cells cultured for 10 hours (lane 3), 24 hours (lane 4), and 48 hours (lane 5) with 100 μmol/L ZnCl2 and was probed with antimouse PU.1 antibody.
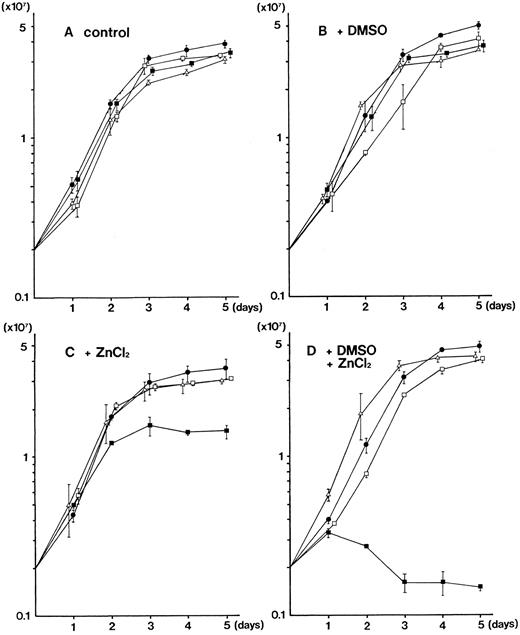
Growth inhibition of MEL cells by overexpression of PU.1.We noticed that the growth of PU.1-sense clones were slower than those of parental MEL-B8/3 cells and four clones of control transfectants in the presence of 100 μmol/L ZnCl2 . We analyzed the growth of these cells under a variety of conditions. In the ordinary culture, no significant difference was observed in the growth kinetics of parental MEL-B8/3 cells and three clones of transfectants, PU.1-sense clone 1, PU.1-antisense clone 1, and PU.1-mock clone 1 (Fig 3A). When cultured with 1.5% DMSO, no remarkable difference was observed in the growth kinetics of all cells (Fig 3B). In contrast, when 100 μmol/L ZnCl2 was added to the culture, the growth of PU.1-sense clone 1 was markedly reduced (Fig 3C). Moreover, when 1.5% DMSO was added along with 100 μmol/L ZnCl2 , the growth reduction of PU.1-sense clone 1 became much pronounced (Fig 3D). Flow cytometric analysis showed that the number of cells in G1 phase increased when PU.1-sense clone 1 was treated with ZnCl2 alone or DMSO and ZnCl2 (Kihara et al, unpublished data). The similar results were obtained when the second clone of each transfectant (PU.1-sense clone 2, PU.1-antisense clone 2, and PU.1-mock clone 2) was used (data not shown). These results indicate that overexpression of PU.1 inhibits the growth of MEL-B8/3 cells.
Growth kinetics of the parental MEL-B8/3 cells and transfectants under various culture conditions. Parental MEL-B8/3 cells (•), PU.1-sense clone 1 cells (▪), PU.1-antisense clone 1 cells (□), and PU.1-mock clone 1 cells (▵) were cultured in the ordinary medium (A), in the medium containing 1.5% DMSO (B), in the medium containing 100 μmol/L ZnCl2 (C), or in the medium containing 1.5% DMSO and 100 μmol/L ZnCl2 (D). Cells (2 × 106) were placed in 10 mL of the culture medium and the cell number was determined every 24 hours. Mean values and standard deviations of three independent experiments are shown.
Growth kinetics of the parental MEL-B8/3 cells and transfectants under various culture conditions. Parental MEL-B8/3 cells (•), PU.1-sense clone 1 cells (▪), PU.1-antisense clone 1 cells (□), and PU.1-mock clone 1 cells (▵) were cultured in the ordinary medium (A), in the medium containing 1.5% DMSO (B), in the medium containing 100 μmol/L ZnCl2 (C), or in the medium containing 1.5% DMSO and 100 μmol/L ZnCl2 (D). Cells (2 × 106) were placed in 10 mL of the culture medium and the cell number was determined every 24 hours. Mean values and standard deviations of three independent experiments are shown.
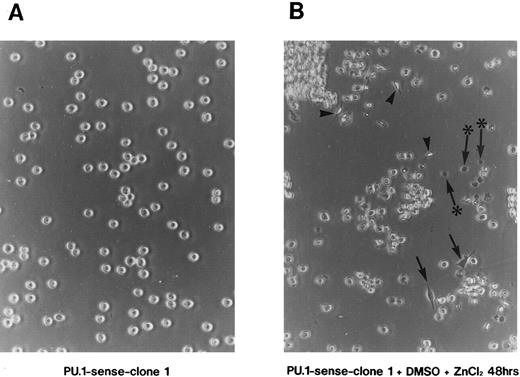
Induction of apoptosis by overexpression of PU.1 in MEL cells.When PU.1-sense clone 1 was cultured with 1.5% DMSO and 100 μmol/L ZnCl2 , morphologic changes of the cells were observed in addition to growth reduction. PU.1-sense cells, like parental MEL-B8/3 cells, were spherical and grew in suspension (Fig 4A). By 48 hours of culture under the conditions described above, some of PU.1-sense cells changed their shape to flat and attached to the bottom of culture dishes (Fig 4B, arrow) or changed to spindle-like shape (Fig 4B, arrowhead). At this time, a considerable number of dead cells were observed (Fig 4B, arrow with an asterisk). The trypan blue exclusion assay showed that 30% to 40% of cells were dead after 48 hours of culture with DMSO and ZnCl2 (data not shown). Such morphologic changes and death of the cells were also observed in PU.1-sense clone 2 and in all of the other clones of PU.1-sense cells when cultured with DMSO and ZnCl2 (data not shown). No morphologic changes and death of the cells were observed in parental MEL-B8/3 cells, PU.1-antisense clones, and PU.1-mock clones under the same culture conditions (data not shown).
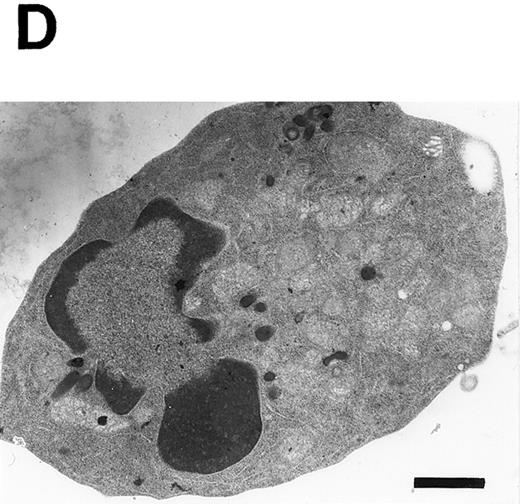
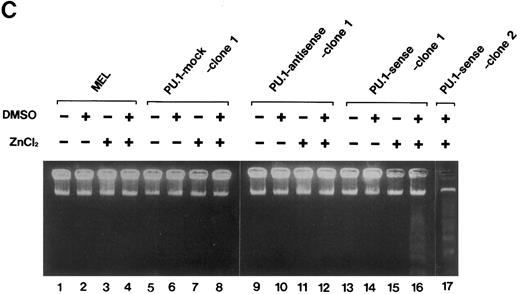
Induction of apoptosis in PU.1-overexpressing cells. (A) Morphology of PU.1-sense clone 1 cells cultured in the ordinary medium. (B) Morphology of the same cells cultured with 1.5% DMSO and 100 μmol/L ZnCl2 for 48 hours. Arrows point to the flat cells attaching to the bottom of culture dishes, arrowheads to the spindle-like cells, and arrows with an asterisk to the dead cells. (C) DNA fragmentation analysis of the parental MEL-B8/3 cells and transfectants cultured under various conditions. DNA was extracted from the cells cultured for 48 hours in the ordinary medium or in the medium containing 1.5% DMSO and/or 100 μmol/L ZnCl2 and was electrophoresed in a 1.5% agarose gel. (D) The ultrastructural appearance of a PU.1-sense clone 1 cell showing characteristic features of apoptosis after 48 hours of culture with 1.5% DMSO and 100 μmol/L ZnCl2 . Note the dense chromatin aggregation under the nuclear membrane and no remarkable changes of the mitochondria and other organelles. Scale bar, 1 μm.
Induction of apoptosis in PU.1-overexpressing cells. (A) Morphology of PU.1-sense clone 1 cells cultured in the ordinary medium. (B) Morphology of the same cells cultured with 1.5% DMSO and 100 μmol/L ZnCl2 for 48 hours. Arrows point to the flat cells attaching to the bottom of culture dishes, arrowheads to the spindle-like cells, and arrows with an asterisk to the dead cells. (C) DNA fragmentation analysis of the parental MEL-B8/3 cells and transfectants cultured under various conditions. DNA was extracted from the cells cultured for 48 hours in the ordinary medium or in the medium containing 1.5% DMSO and/or 100 μmol/L ZnCl2 and was electrophoresed in a 1.5% agarose gel. (D) The ultrastructural appearance of a PU.1-sense clone 1 cell showing characteristic features of apoptosis after 48 hours of culture with 1.5% DMSO and 100 μmol/L ZnCl2 . Note the dense chromatin aggregation under the nuclear membrane and no remarkable changes of the mitochondria and other organelles. Scale bar, 1 μm.
To determine whether the cell death observed in PU.1-sense cells is apoptosis or necrosis, we examined cleavage of DNA and fine structure of the cells. The characteristic fragmentation, a hallmark of apoptosis, was observed in DNA from the cells of PU.1-sense clone 1 and clone 2 cultured with DMSO and ZnCl2 for 48 hours (Fig 4C, lanes 16 and 17). When cultured with DMSO alone or ZnCl2 alone, DNA fragmentation was not observed in PU.1-sense clone 1 (Fig 4C, lanes 14 and 15) and clone 2 (data not shown). No fragmentation of DNA was induced under any culture conditions in parental MEL-B8/3 cells (Fig 4C, lanes 1 through 4), in PU.1-mock clone 1 (Fig 4C, lanes 5 through 8) and clone 2 (data not shown), and in PU.1-antisense clone 1 (Fig 4C, lanes 9 through 12) and clone 2 (data not shown). Electron microscopic studies of PU.1-sense cells of this stage showed the dense chromatin aggregated under the nuclear membrane with intact mitochondria and other organelles in the cytoplasm, which are characteristic features of apoptosis (Fig 4D). These results indicate that cell death induced by overexpression of PU.1 in MEL-B8/3 cells occurred by apoptosis.
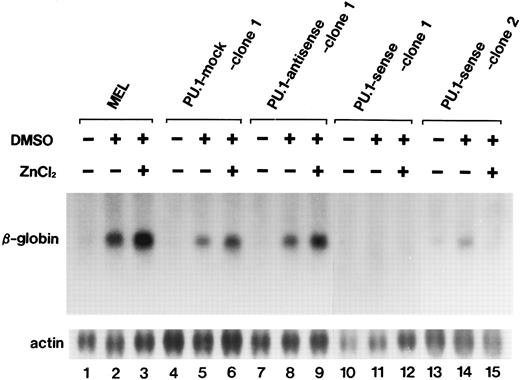
Differentiation inhibition of MEL cells by overexpression of PU.1.One of the possible functional roles of PU.1 in MEL genesis is differentiation inhibition of the cells. We next examined the effects of PU.1 overexpression on differentiation of MEL-B8/3 cells. As shown in Fig 5, expression of the β-globin gene was increased after 72 hours of DMSO treatment irrespective of the presence of 100 μmol/L ZnCl2 in parental MEL B8/3 cells (lanes 2 and 3) in PU 1-mock clone 1 (lanes 5 and 6) and clone 2 (data not shown) and in PU.1-antisense clone 1 (lanes 8 and 9) and clone 2 (data not shown). In contrast, in PU.1-sense clones, expression of the β-globin gene was hardly increased in clone 1 (lane 11) and slightly increased in clone 2 (lane 14) after DMSO treatment and was hardly increased in both clones after treatment with DMSO and ZnCl2 (lanes 12 and 15). These results suggest that overexpression of PU.1 interferes with differentiation of MEL-B8/3 cells induced by DMSO treatment. The slight increase in β-globin gene expression in PU.1-sense clones after treatment with DMSO alone might be due to interference by the increased levels of PU.1 as a result of leak of the PU.1/Spi-1 gene expression from the metallothionein promoter. In the Northern blot analysis, barely detectable levels of expression of the messages from this promoter are seen in the PU.1-sense cells treated with ZnCl2 for 0 hours (Fig 2A, lane 1). To examine whether increased expression of the β-globin gene is observed in other clones of PU.1-sense cells, we treated six other clones with 1.5% DMSO and found slightly increased expression of the gene in one clone and little increased expression in five clones. We also treated these clones with DMSO and ZnCl2 and found little increased expression in all of them (data not shown).
Northern blot analysis of induction of β-globin gene expression in the parental MEL-B8/3 cells and transfectants cultured under various conditions. Total RNA was extracted from the cells cultured for 72 hours in the ordinary medium or in the medium containing 1.5% DMSO alone or 1.5% DMSO and 100 μmol/L ZnCl2 and was hybridized with 23P-labeled probes.
Northern blot analysis of induction of β-globin gene expression in the parental MEL-B8/3 cells and transfectants cultured under various conditions. Total RNA was extracted from the cells cultured for 72 hours in the ordinary medium or in the medium containing 1.5% DMSO alone or 1.5% DMSO and 100 μmol/L ZnCl2 and was hybridized with 23P-labeled probes.
It should be taken into account that treatment of PU.1-sense cells with DMSO and ZnCl2 results in the onset of apoptosis. This raises the possibility that the cells that responded to the differentiation stimulus selectively went into apoptosis process and, as a consequence, increased expression of the β-globin gene was not observed in the Northern blot analysis. Because we had found that apoptosis can be avoided when PU.1-sense cells receive DMSO and ZnCl2 in the medium containing 30% serum (data not shown), we determined the expression of the β-globin gene in PU.1-sense clone 1 under these conditions. Expression of the gene was not increased, although the cells were viable (data not shown). This observation supports the notion that overexpression of PU.1 interferes with the differentiation of MEL-B8/3 cells.
Our results indicate that overexpression of PU.1 in MEL-B8/3 cells inhibits cell growth and differentiation and, in conjunction with DMSO treatment, leads to apoptotic cell death. To examine the reproducibility of the experiments, we established two additional clones of PU.1-sense, PU.1-antisense, and PU.1-mock cells after the second transfection of MEL-B8/3 cells with the corresponding plasmids. Overexpression of PU.1 resulted in inhibition of cell growth and differentiation and, in the presence of DMSO, induction of apoptosis in the clones of PU.1-sense cells (data not shown). These phenomena are not specific for MEL-B8/3 cells, because we observed similar effects of PU.1 overexpression in two clones of transfectants established by introduction of pPU.1-sense plasmid in another MEL cell line, MEL-D1 (data not shown).
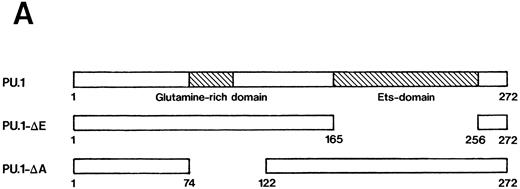
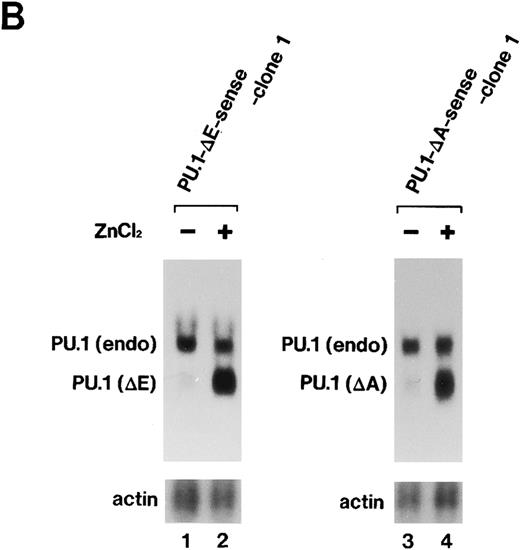
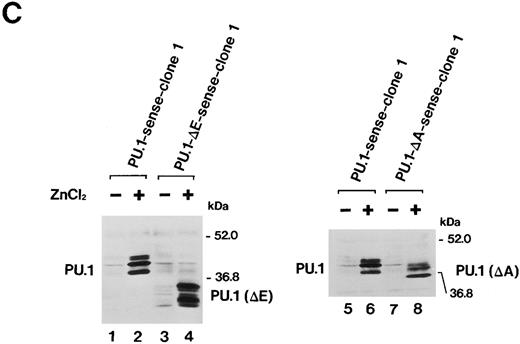
Importance of the Ets domain and the activation domain of PU.1 for induction of the effects.We next sought to determine the region(s) of PU.1 required for induction of the effects described above in MEL-B8/3 cells. We constructed two kinds of expression plasmids, pPU.1-Δ E-sense and pPU.1-Δ A-sense, as described in the Materials and Methods. The former contains a synthetic mutant PU.1/Spi-1 gene encoding PU.1 with a deletion of the region corresponding to the Ets domain and the latter contains another synthetic mutant gene encoding PU.1 with a deletion of the region including the glutamine-rich subdomain of the activation domain (Fig 6A). After the introduction of these expression plasmids into MEL-B8/3 cells, four clones of transfectants, designated PU.1-Δ E-sense clone 1 and clone 2 and PU.1-Δ A-sense clone 1 and clone 2 were obtained. Northern blot analysis showed induction of expression of the truncated messages in PU.1-Δ E-sense clone 1 and PU.1-Δ A-sense clone 1 after ZnCl2 treatment (Fig 6B, lanes 2 and 4). Western blot analysis showed that the levels of induced products of the truncated genes in these clones were comparable with that of wild-type PU.1 in PU.1-sense clone 1 (Fig 6C). Similar results were obtained in PU.1-Δ E-sense clone 2 and PU.1-Δ A-sense clone 2 after ZnCl2 treatment (data not shown). Four clones of control transfectants, PU.1-Δ E-antisense clone 1 and clone 2 and PU.1-Δ A-antisense clone 1 and clone 2, were also obtained by introducing the control plasmids into MEL-B8/3 cells.
(A) Schematic structure of wild-type PU.1 and two kinds of deletion mutants, PU.1-Δ E and PU.1-Δ A. The Ets domain and glutamine-rich domain are indicated by the hatched box. (B) Northern blot analysis of induction of expression of the mutant PU.1/Spi-1 genes, PU.1-Δ E and PU.1-Δ A, in the transfectants. Total RNA was extracted from PU.1-Δ E-sense clone 1 cells and PU.1-Δ A-sense clone 1 cells cultured for 6 hours with or without 100 μmol/L ZnCl2 and was hybridized with 32P-labeled probes. (C) Western blot analysis of induction of expression of the mutant PU.1 proteins in the transfectants. Total protein was extracted from PU.1-Δ E-sense clone 1 cells and PU.1-Δ A-sense clone 1 cells cultured for 8 hours with or without 100 μmol/L ZnCl2 and was probed with antimouse PU.1 antibody.
(A) Schematic structure of wild-type PU.1 and two kinds of deletion mutants, PU.1-Δ E and PU.1-Δ A. The Ets domain and glutamine-rich domain are indicated by the hatched box. (B) Northern blot analysis of induction of expression of the mutant PU.1/Spi-1 genes, PU.1-Δ E and PU.1-Δ A, in the transfectants. Total RNA was extracted from PU.1-Δ E-sense clone 1 cells and PU.1-Δ A-sense clone 1 cells cultured for 6 hours with or without 100 μmol/L ZnCl2 and was hybridized with 32P-labeled probes. (C) Western blot analysis of induction of expression of the mutant PU.1 proteins in the transfectants. Total protein was extracted from PU.1-Δ E-sense clone 1 cells and PU.1-Δ A-sense clone 1 cells cultured for 8 hours with or without 100 μmol/L ZnCl2 and was probed with antimouse PU.1 antibody.
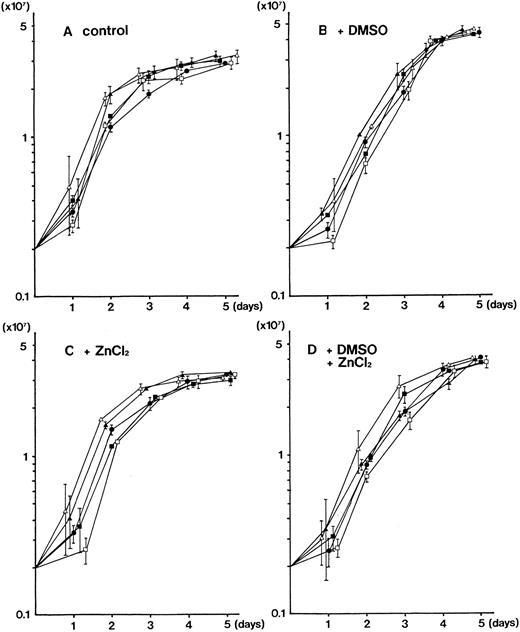
No obvious difference was observed in the growth kinetics of parental MEL-B8/3 cells and four clones of the transfectants (PU.1-Δ E-sense clone 1, PU.1-Δ E-antisense clone 1, PU.1-Δ A-sense clone 1, and PU.1-Δ A-antisense clone 1) in the ordinary culture and in the cultures with DMSO and/or ZnCl2 (Fig 7). The similar results were obtained when the second clones of each transfectant were used (data not shown).
Growth kinetics of the parental MEL-B8/3 cells and transfectants of the mutant PU.1/Spi-1 genes under various culture conditions. Parental MEL-B8/3 cells (•), PU.1-Δ E-sense clone 1 cells (▪), PU.1-Δ E-antisense clone 1 cells (□), PU.1-Δ A-sense clone 1 cells (▴), and PU.1-Δ A-antisense clone 1 cells (▵) were cultured in the ordinary medium (A), in the medium containing 1.5% DMSO (B), in the medium containing 100 μmol/L ZnCl2 (C), or in the medium containing 1.5% DMSO and 100 μmol/L ZnCl2 (D). Cells (2 × 106) were placed in 10 mL of the culture medium and the cell number was determined every 24 hours. Mean values and standard deviations of three independent experiments are shown.
Growth kinetics of the parental MEL-B8/3 cells and transfectants of the mutant PU.1/Spi-1 genes under various culture conditions. Parental MEL-B8/3 cells (•), PU.1-Δ E-sense clone 1 cells (▪), PU.1-Δ E-antisense clone 1 cells (□), PU.1-Δ A-sense clone 1 cells (▴), and PU.1-Δ A-antisense clone 1 cells (▵) were cultured in the ordinary medium (A), in the medium containing 1.5% DMSO (B), in the medium containing 100 μmol/L ZnCl2 (C), or in the medium containing 1.5% DMSO and 100 μmol/L ZnCl2 (D). Cells (2 × 106) were placed in 10 mL of the culture medium and the cell number was determined every 24 hours. Mean values and standard deviations of three independent experiments are shown.
The morphologic changes and death of cells, observed in PU.1-sense cells by 48 hours of culture with DMSO and ZnCl2 , were not induced in any of the clones of PU.1-Δ E-sense and pPU.1-Δ E-antisense cells and PU.1-Δ A-sense and pPU.1-Δ A-antisense cells under any culture conditions (data not shown). DNA fragmentation was not observed in any of the clones of PU.1-Δ E-sense and PU.1-Δ A-sense cells after 48 hours of culture with DMSO and ZnCl2 (data not shown).
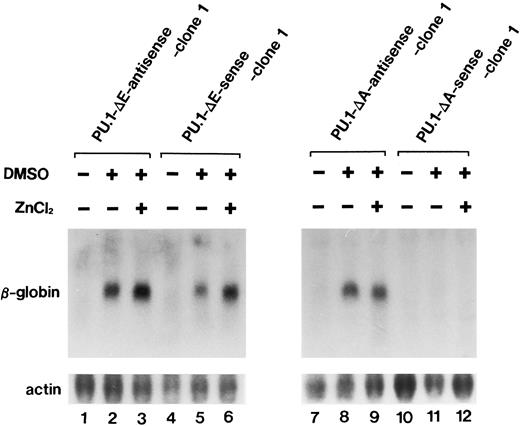
Expression of the β-globin gene was increased after 72 hours of DMSO treatment irrespective of the presence of ZnCl2 in PU.1-Δ E-sense clone 1 (Fig 8, lanes 5 and 6) and clone 2 (data not shown) and in PU.1-Δ E-antisense clone 1 (Fig 8, lanes 2 and 3) and clone 2 (data not shown). In contrast to PU.1-Δ E-sense cells, the expression of the gene was hardly detected after 72 hours of culture with DMSO alone or DMSO and ZnCl2 in PU.1-Δ A-sense clone 1 (Fig 8, lanes 11 and 12) and clone 2 (data not shown). Low levels of expression of the gene in PU.1-Δ A-sense cells cultured with DMSO alone could be due to leak of the mutant PU.1/Spi-1 gene expression from the metallothionein promoter (Fig 6B, lane 3). Increased β-globin gene expression was observed after 72 hours of DMSO treatment irrespective of the presence of ZnCl2 in PU.1-Δ A-antisense clone 1 (Fig 8, lanes 8 and 9) and clone 2 (data not shown).
Northern blot analysis of expression induction of the β-globin gene in PU.1-Δ E-sense clone 1 cells and PU.1-Δ A-sense clone 1 cells cultured under various conditions. Total RNA was extracted from the cells cultured for 72 hours in the ordinary medium or in the medium containing 1.5% DMSO alone or 1.5% DMSO and 100 μmol/L ZnCl2 and was hybridized with 32P-labeled probes.
Northern blot analysis of expression induction of the β-globin gene in PU.1-Δ E-sense clone 1 cells and PU.1-Δ A-sense clone 1 cells cultured under various conditions. Total RNA was extracted from the cells cultured for 72 hours in the ordinary medium or in the medium containing 1.5% DMSO alone or 1.5% DMSO and 100 μmol/L ZnCl2 and was hybridized with 32P-labeled probes.
These results suggest that growth inhibition and apoptosis induction may require both the region corresponding to the Ets domain and the region including the glutamine-rich domain, whereas differentiation inhibition may require the former one but not the latter one.
DISCUSSION
In this study, we showed that overexpression of PU.1 in MEL-B8/3 cells results in inhibition of growth and differentiation, and apoptotic death of the cells. These phenomena are not peculiar to MEL-B8/3 cells, because we observed similar effects of PU.1 overexpression in another MEL cell line, MEL-D1 (data not shown).
Growth inhibition of MEL cells after overexpression of PU.1 appears to be inconsistent with the notion that PU.1 activation is one of the causal reasons of malignant transformation of MEL cells. MEL cells used in this study might have different characters from those of MEL cells used by others, although the rearrangement of the PU.1/Spi-1 locus was detected in MEL-B8/3 cells in the Southern blot analysis (data not shown). Alternatively, there may be suitable levels of PU.1 expression in MEL cells for enhancement of cell growth. Much higher levels of expression of PU.1 may be disadvantageous to cell growth. Further analyses using MEL cell lines expressing PU.1 at various levels will provide further insight into mechanisms of growth inhibition of the cells by PU.1 overexpression. Delgado et al30 reported that growth and cloning efficiency of two lines of MEL cells were inhibited by treatment of the cells with anti–PU.1/Spi-1 antisense oligodeoxynucleotides. In the present study, the growth of the PU.1-antisense cells was not decreased after the addition of ZnCl2 . The discrepancy between the results may be due to the residual expression of PU.1 in our cells (Fig 3C).
In the presence of 1.5% DMSO, PU.1 overexpression in MEL cells led to prominent growth inhibition and apoptotic death of the cells. This suggests that PU.1 is involved in both of the opposite cellular fates of malignant transformation and apoptosis in MEL cells. A well-known example of the protein participating in both processes is c-Myc oncoprotein. In certain types of cells, overexpression of c-Myc causes growth enhancement under normal culture conditions but apoptosis under serum-deprived conditions.31 Like c-Myc, PU.1 might show opposite functional role under different culture conditions.
To determine the precise timing of onset of growth inhibition and apoptosis, PU.1-sense clone 1 was cultured with 1.5% DMSO and 100 μmol/L ZnCl2 for various times and then cultured in normal medium such that the total culture time becomes 48 hours. After that, the number of total cells and dead cells was determined. Growth inhibition was not observed when the culture time with DMSO and ZnCl2 was less than 12 hours and apoptosis was not observed when the culture time was less than 24 hours, indicating that commitment occurred to growth inhibition and apoptosis after 12 hours and after 24 hours, respectively (data not shown). This suggests that prominent growth inhibition observed in the culture with DMSO and ZnCl2 is not a result of apoptotic cell death.
Expression of the exogenous PU.1/Spi-1 gene was induced by 2 hours and the peak of the expression was 10 hours after the addition of ZnCl2 in the PU.1-sense cells (Fig 2A). On the other hand, as described above, the onset of growth inhibition was after 12 hours and that of apoptosis was after 24 hours of culture with DMSO and ZnCl2 . This lag might be due to the requirement of time for expression of a gene(s) activated by PU.1 and/or DMSO and involved in induction of these effects. We also observed prominent growth inhibition and apoptosis in PU.1-sense cells when treated with 100 μmol/L ZnCl2 and 5 mmol/L N,N′-hexamethylene-bis-acetamide (HMBA), which is also known as an inducer of MEL cell differentiation, supporting the notion that a gene(s) activated in the course of differentiation may be involved in the onset of these processes (data not shown). However, we cannot rule out the possibility that cytotoxicity of the differentiation inducers contributes to the induction of these effects, although DMSO or HMBA alone was not toxic to MEL cells.
Flow cytometric analysis showed that the proportion of the cells in G1 phase was increased by 18 hours and that in sub-G1 fraction was remarkably increased by 36 hours after the addition of DMSO and ZnCl2 , suggesting that cell cycle arrests at G1 phase before the onset of apoptosis (Kihara et al, unpublished data). We are currently investigating expression of the genes, which participate in cell cycle regulation and induction or inhibition of apoptosis, in PU.1-sense cells treated with DMSO and/or ZnCl2 . So far, we have obtained the preliminary results showing that expression of the c-myc, bcl-2, and p53 genes were decreased in the process of apoptosis (data not shown).
It has been shown that insulin-like growth factor (IGF )-1 interferes with c-myc–mediated apoptosis in fibroblasts.32 We found that 30% serum in the culture medium prevents apoptosis induced by the treatment with DMSO and ZnCl2 in PU.1-sense cells (data not shown). This suggests that growth factors in serum may interfere with or override apoptotic cell death, although 30% serum in the culture medium did not enhance the growth rates of MEL-B8/3 cells and PU.1-sense cells in comparison with 10% serum (data not shown).
Morphologic changes were also observed in PU.1-sense cells treated with DMSO and ZnCl2 . It is not clear if the changes precede apoptosis. The molecular basis for this phenomenon is also currently obscure. Cytoskeletal structure might be changed in some cells under this condition.
Expression of the β-globin gene was hardly detected in PU.1-sense cells after 72 hours of culture with DMSO and ZnCl2 , indicating that overexpression of PU.1 interferes with MEL cell differentiation. The similar effect was observed on differentiation induced by HMBA treatment (data not shown). These observations, along with the result showing decreased levels of PU.1/Spi-1 gene expression after differentiation induction of MEL-B8/3 cells (Fig 1A), suggests that PU.1 plays a fundamental role in the malignant transformation of MEL cells by blocking differentiation of the cells. In the same line of investigations, Quang et al33 showed that enforced expression of the PU.1/Spi-1 gene in avian primary erythroblasts inhibits differentiation normally induced by Epo treatment.
From the experiments using the deletion mutants of PU.1, we showed that the region corresponding to the Ets domain and that including the glutamine-rich domain contribute differently to the induction of the effects. It appears that the former is required for growth inhibition, apoptosis, and differentiation inhibition, whereas the latter is required for growth inhibition and apoptosis. PU.1 binds to its consensus DNA sequence through the Ets domain and activates transcription of the target genes through the activation domain. The glutamine-rich domain is a subdomain of the activation domain.26,27 It has been shown that deletion of the glutamine-rich domain results in significant loss of transcriptional activity of PU.1.27 Considering the results obtained using deletion mutants of PU.1 together with functional role of each domain, it is suggested that new activation of the gene(s) by PU.1 may be required for growth inhibition and apoptosis and that binding of PU.1 to its target sequence may be sufficient for differentiation inhibition. As discussed above, lagged onset of growth inhibition and apoptosis also supports the notion that new activation of the gene(s) is required for their induction. Overexpressed PU.1, irrespective of having the activation domain, might inhibit MEL cell differentiation by binding to the target sequence and interfering with the binding of the other transcription factor(s) that normally binds to the sequence in the course of differentiation.
By using gene targeting methods, PU.1 has been shown to be required for the development of hematopoietic cell lineages, including B and T lymphocytes, monocytes, and granulocytes.34 It is conceivable that PU.1 is expressed in many cell lineages in early times of hematopoiesis to regulate development of the cells and that PU.1 is then suppressed in cell lineages other than B-cell and macrophage lineages, in which relatively high levels of PU.1 expression are observed.1 It is tempting to speculate that the inappropriate expression of PU.1 in the cell lineages other than B cells and macrophages may trigger malignant transformation of the cells. MEL genesis accompanied by expression of PU.1 may be just such an example. Immortalization of erythroblasts after introduction of the PU.1/Spi-1 gene into cultured mouse bone marrow cells by using a retroviral vector35 also supports this hypothesis. Apoptosis observed in this study might be mechanisms to eliminate erythroid cells expressing PU.1 inappropriately. Expression levels of PU.1 and developmental stage of the cells in which PU.1 is expressed could be important for decision towards growth inhibition and apoptosis, because they were observed, at least in MEL cells, only after overexpression of PU.1 and, in particular, because the latter was induced in the presence of DMSO.
Further investigation of functional role of PU.1 in growth, differentiation, malignant transformation, and death of cells will provide further knowledge for understanding not only the molecular basis of these phenomena but also how they correlate with each other.
ACKNOWLEDGMENT
We are indebted to Dr Y. Hashimoto, the director of Sasaki Institute, and Dr C.W. Boone (National Institutes of Health, Bethesda, MD) for their critical reading of the manuscript. We thank Drs M. Obinata, D. Kabat, R. Maki, and F. Moreau-Gachelin for their kind donation of MEL-B8/3 cells, a zinc-inducible expression vector, pSVneoHMT-Ter, and the plasmids for the mouse PU.1/Spi-1 cDNA. We are grateful for financial support from Dr J. Akiyama (OB-GYN Akiyama Memorial Hospital, Hakodate, Japan) and Dr Y. Yamada (Taiho Pharmaceutical Co Ltd, Saitama, Japan).
Supported by Grants-in-Aid of Scientific Research from the Ministry of Education, Science and Culture of Japan.
Address reprint requests to Tsuneyuki Oikawa, MD, PhD, Department of Cell Genetics, Sasaki Institute, 2-2 Kanda-Surugadai, Chiyoda-ku, Tokyo 101, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal