Abstract
Two cAMP analogs, 8- and 2- [(4-bromo-2,3-dioxobutyl) thio]adenosine 3′,5′-cyclic monophosphate (8- and 2-BDB-TcAMP) have been used in probing the catalytic site of recombinant monocyte cAMP-specific phosphodiesterase (PDE4a). 2-BDB-TcAMP is a reversible and competitive inhibitor (Ki = 5.5 μmol/L) of cAMP hydrolysis by PDE4a. 8-BDB-TcAMP irreversibly inactivates the enzyme in a time- and concentration-dependent manner with a second order rate constant of 0.022 mmol/L−1min−1. The rate of inactivation of PDE4a is reduced by the presence of the substrate cAMP and specific inhibitors, rolipram and denbufylline, but not by cGMP or AMP. Reduction of the enzyme-inhibitor complex with sodium [3H]borohydride shows that 1.2 mol of the affinity label/mol of enzyme was incorporated. The radiolabeled peptide is composed of 10 amino acid residues (697 to 706) located near the carboxyl end of the proposed catalytic domain. The peptide (GPGHPPLPDK) has seven nonpolar and aliphatic residues, of which four are proline, giving the peptide a highly structured conformation. This peptide is the first to be identified in the putative catalytic domain involved in substrate recognition.
CYCLIC nucleotide phosphodiesterases (PDEs) catalyze the hydrolysis of cAMP and cGMP to AMP and GMP, respectively, and, therefore, are critical in controlling the intracellular levels of these second messengers. These enzymes are widely distributed in tissues. In human platelets, an increase in the intracellular concentration of cAMP is associated with inhibition of platelet responses to agonists including such responses as shape change, aggregation, adhesion, and release of granule contents, and in leukocytes, activation by agonists is also downregulated. Seven PDE gene families have been identified: calcium/calmodulin-dependent PDEs (PDE1); cGMP-stimulated PDE (PDE2); cGMP-inhibited PDE (PDE3); cAMP-specific PDE (PDE4); cGMP-specific PDE (PDE5); photoreceptor PDE (PDE6); and high affinity cAMP-specific PDE (PDE7).1-3 In addition, within each gene family, isotypes due to splice variants or deletions exist, for example, PDE4a-d.4
All mammalian PDE families share a conserved carboxyl terminal domain of approximately 250 amino acids with a number of highly conserved amino acid residues. The amino acid sequence in the conserved domain exhibits approximately 35% identity and similarity between PDE families and >70% homology within each family. This domain from different PDE families has been expressed and shown to possess catalytic activity in the absence of the amino terminal domain.5-8 The monocyte PDE4a exists as a 96-kD protein, but the recombinant enzyme is expressed without the N-terminal 64 amino acids and thus is an 88-kD protein that displays catalytic activity similar to the native protein.9 Therefore the conserved domain is believed to harbor the active site and has been referred to as the “catalytic domain.” We define the active site as containing the substrate binding region, the amino acids involved in the catalytic reaction and a metal binding site.10 11
Although the putative active site has been proposed to be within the conserved domain, the exact nature or location of the catalytic amino acid residue(s), substrate recognition site, or metal binding sequence(s) that makeup the active site is not known. This information is important in defining a mechanism for cyclic nucleotide hydrolysis by this group of enzymes. In addition, more exact knowledge would facilitate the rational design of inhibitors for PDE4a, which has lately been the target for several potential therapeutic agents.12,13 Based on the pH-rate profile and group-specific chemical modification, histidines, and a cysteine have been postulated to play a critical role in the hydrolysis of cGMP by CAM-PDE by Ahn et al14 and cGMP-inhibited PDE in our laboratory.15 The location of these critical amino acids in the primary structure has not yet been defined, since peptide sequences containing modified amino acids have not been identified.
Affinity labeling using a substrate analog with a reactive functional group has the potential to modify a specific amino acid residue located at or near the active site.16,17 Several cAMP analogs have been used as affinity or photoaffinity labels for cAMP-dependent kinases,18,19 phosphodiesterases,20-22 and cAMP binding proteins.23,24 In our laboratory, two new cAMP analogs, 8-[(4-bromo-2,3-dioxobutyl)thio]cAMP (8-BDB-TcAMP) and 2-(4-bromo-2,3-dioxobutyl)thio]cAMP (2-BDB-TcAMP) were synthesized; the former irreversibly inactivated PDE3a isolated from platelets, whereas the latter functioned as a reversible and competitive inhibitor of that enzyme.25
We postulated that 8-BDB-TcAMP would act as an affinity label of PDEs, other than PDE3a, which hydrolyze cAMP. We have used 8-BDB-TcAMP and 2-BDB-TcAMP to identify, for the first time, a substrate recognition site located in the active site of a recombinant monocyte cAMP-specific PDE (r-PDE4a). We report here that, while 2-BDB-TcAMP behaves as a reversible inhibitor, 8-BDB-TcAMP irreversibly inactivates r-PDE4a concomitant with incorporation of the reagent into the enzyme. The results indicate that a peptide composed of 10 amino acids (GPGHPPLPDK) located at the C-terminal end of the putative catalytic domain is the target of 8-BDB-TcAMP and that this peptide is involved in substrate recognition. A preliminary version of a portion of this work has been presented.26
MATERIALS AND METHODS
Materials.[2,8-H3]cAMP (28.4 Ci/mmol) and sodium [3H]borohydride (13.6 Ci/mmol) were purchased from DuPont NEN (Boston, MA). Dithiothreitol, cAMP, cGMP, HEPES, TrisHCl, and other buffers were purchased from Sigma (St Louis, MO). 5′-[14C]AMP (57 mCi/mmol) was from ICN (Costa Mesa, CA). The boronate gel (Affi-Gel 601 resin) used in the separation of 5′-AMP from cAMP was purchased from BioRad (Richmond, VA). ISS protein gold reagent was purchased from Enprotech (Boston, MA). All organic chemicals were of high purity grade purchased from Aldrich Chemical Co (Milwaukee, WI) or Sigma Chemical Co.
Enzyme activity assay.The enzymatic activity was assayed in 100 μL final reaction volume containing 50 mmol/L TrisHCl, pH 7.8, 5 mmol/L magnesium chloride and 0.5 mg/mL bovine serum albumin (BSA). The reaction was initiated by adding 25 μL of concentrated substrate solution containing 4 μmol/L [3H]cAMP (20,000 CPM), 200 μmol/L [14C]AMP (3,000 CPM) and 20 mmol/L MgCl2 in 50 mmol/L TrisHCl buffer, pH 7.5, and incubated for 30 minutes at 30°C. After incubation, the reaction was terminated by boiling the tubes for 2 minutes. The samples were diluted with 3 mL of 100 mmol/L HEPES buffer, pH 8.5 containing 100 mmol/L NaCl, then loaded onto boronate columns (5 mL) equilibrated with the same buffer. After washing the columns with the HEPES/NaCl buffer (20 mL), [3H]AMP was eluted with 10 mL 0.25 mol/L acetic acid. Enzymatic assays were conducted in the linear range of the reaction where less than 30% of the initial substrate was hydrolyzed. The 5′-[14C]AMP was used to calculate recovery. To measure inhibition as a function of concentration of 2-BDB-TcAMP or 8-BDB-TcAMP, varying amounts of reagent were added to the activity assay solution and the reaction was started by the addition of the enzyme. All assays were performed in triplicate and kinetic experiments were repeated at least three times. The data are expressed as means ± SEM. Coefficients of variation were less than 15%.
Enzyme purification.The recombinant human monocyte PDE4a (r-PDE4a) used in this study lacks the first 64 amino acids from the N-terminal end and was expressed in and purified from PDE-deficient yeast expression system.27 GL61 cells were harvested by centrifugation at 10,000g for 15 minutes at 4°C and resuspended in 50 mmol/L Tris HCl, pH 7.5 buffer containing 5 mmol/L MgCl2 , 20 mmol/L dithiothreitol, 0.1% thiodiglycol, 0.3% Tween 80, and 1.0 mmol/L phenylmethanesulfonyl fluoride (storage buffer). All subsequent purification steps were performed at 4°C or on ice bath in storage buffer containing in addition 1 μg/mL leupeptin, 1 μg/mL pepstatin, and 50 μmol/L tosyl phenylalanyl chloromethyl ketone (TPCK) (purification buffer). The preceding protease inhibitors were dissolved at 1,000 × the desired final concentration in methanol and added in at appropriate volumes (not exceeding 1% methanol). Cells lysed using an APV Rannie apparatus (Rainin Inst. Co, Woburn, MA) set at 12,000 psi were centrifuged at 16,000g for 60 minutes and supernatant filtered through 4 layers of cheesecloth. The filtrate was applied to a Q-Sepharose fast flow column (5 × 60 cm, from Pharmacia) previously equilibrated with purification buffer. When all unbound protein was eluted, r-PDE4a was recovered with a linear NaCl gradient (0.1 to 0.7 mol/L over 10 column volumes). The active fractions were pooled, desalted and loaded onto a heparin Sepharose CL-6B column (5 × 15 cm, from Pharmacia, Piscataway, NJ) previously equilibrated with purification buffer. The enzyme was eluted with a linear NaCl gradient (0 to 0.4 mol/L over 6 column volumes) and the pooled fractions desalted, then applied to a Q Sepharose high performance column (3.5 × 10 cm, from Pharmacia). After extensive washing with purification buffer, the PDE4a was recovered by applying a linear gradient of NaCl (0.15 to 0.3 mol/L over 15 column volumes). The purified enzyme was concentrated and stored at −70°C. On sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis, the protein displayed a Mr = 88 kD and accounted for more than 90% of the protein present.
Synthesis of 8-[(4-bromo-2,3-dioxobutyl)thio]cAMP (8-BDB-TcAMP).This compound was synthesized by the procedure of Grant et al25 except that the 8-Br-cAMP (1.0 g) was converted to 8-SH-cAMP by adding to it a suspension of 0.51 g thiourea in 6 mL 1N NaOH and, after allowing 8-Br-cAMP to dissolve completely, the solvent was removed by rotary evaporation. The 2-BDB-TcAMP was synthesized as previously described by Grant et al.25 Both 2-BDB-TcAMP and 8-BDB-TcAMP were stored with desiccant at −70°C as dry powder. Solutions were made for single use.
Inactivation of r-PDE4a by 8-BDB-TcAMP and 2-BDB-TcAMP.The r-cAS-PDE was dialyzed against 50 mmol/L HEPES, pH 7.5 buffer containing 5 mmol/L MgCl2 and 20% glycerol. 8-BDB-TcAMP and 2-BDB-TcAMP were dissolved in 50 mmol/L HEPES pH 7.5 and the reagents were incubated at 25°C with enzyme (1.0 mg/mL). At timed intervals aliquots were removed, diluted at least 1,000-fold in assay buffer and 5 mL of the diluents were assayed for activity. Control samples were treated identically but without the reagents. To test for reversibility of inactivation, the enzyme was inactivated to 35% of control activity then passed through spin columns packed with Sephadex G-50 to remove excess reagent.28 The eluent was diluted and assayed for activity. The effect of dithiothreitol on the partially inactivated r-PDE4a was tested by incubating the eluents with 20 mmol/L dithiothreitol for 1 hour at 2°C before determining the catalytic activity.
The effects of the substrate (cAMP), product (AMP), cGMP, and specific inhibitors of PDE4a on the rate of inactivation of the enzyme were evaluated by incubating the enzyme with 8-BDB-TcAMP in the presence of these compounds. The enzyme was preincubated with the compounds for 2 minutes before the inactivator was added. Timed aliquots were removed, diluted, and assayed.
Incorporation of 8-BDB-TcAMP.The enzyme (0.1 mg) was inactivated with 4 mmol/L 8-BDB-TcAMP and, when it had 10% residual enzymatic activity, the enzyme-inactivator mixture was passed through an initial Sephadex G-50 spin column equilibrated with 7 mol/L guanidine HCl in 50 mmol/L HEPES buffer, pH 7.5 to remove excess reagent. The modified enzyme was reduced by incubation for 30 minutes at 4°C with 3 mmol/L [3H]NaBH4. The mixture was passed through a second Sephadex G-50 spin column that had been equilibrated with 5 mol/L guanidine HCl in 50 mmol/L HEPES buffer, pH 7.5. The filtrate was then diafiltered to remove excess sodium [3H]borohydride. The progress of diafiltration was monitored by measuring radioactive counts of the filtrate. The protein content of the reduced complex was determined by using the ISS protein gold reagent (Enprotec, Boston, MA) with BSA as the standard.29 The radioactive (3H) content was determined by liquid scintillation counting. Control samples were similarly treated except that inactivation with 8-BDB-TcAMP was omitted.
To determine reversibility of incorporation, radiolabeled protein was divided into two equal aliquots. To one portion, dithiothreitol was added to yield 20 mmol/L, and to the other, the control, 50 mmol/L HEPES, pH 7.5 was added. Both samples were filtered using a diafiltration system (Centricon) from Amicon (Danvers, MA), and the filters were rinsed twice with buffer. The filtrate and membranes were counted for radioactivity.
Determination of Ki for 2-BDB-TcAMP inhibition of cAS-PDE. (A) The initial rate of hydrolysis of cAMP was determined in triplicate and in the presence of varying concentrations of the substrate and inhibitor. The concentrations of 2-BDB-TcAMP used were 0 μmol/L (•), 5 μmol/L (▪), 10 μmol/L (▴), and 20 μmol/L (▾). The data was fit to the equation: 1/v = 1/V + Km/V[S](1 + [I]/Ki ). (B) The slope of each line was plotted against 2-BDB-TcAMP concentrations according to the equation: slope = Km/V(1 + [I]/Ki) to give a Ki value of 5.5 ± 0.4 μmol/L (SEM).
Determination of Ki for 2-BDB-TcAMP inhibition of cAS-PDE. (A) The initial rate of hydrolysis of cAMP was determined in triplicate and in the presence of varying concentrations of the substrate and inhibitor. The concentrations of 2-BDB-TcAMP used were 0 μmol/L (•), 5 μmol/L (▪), 10 μmol/L (▴), and 20 μmol/L (▾). The data was fit to the equation: 1/v = 1/V + Km/V[S](1 + [I]/Ki ). (B) The slope of each line was plotted against 2-BDB-TcAMP concentrations according to the equation: slope = Km/V(1 + [I]/Ki) to give a Ki value of 5.5 ± 0.4 μmol/L (SEM).
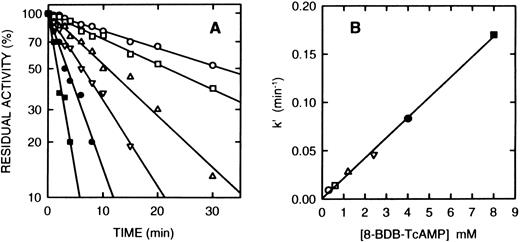
Time course of the inactivation of cAS-PDE by 8-BDB-TcAMP. (A) The enzyme was incubated at 25°C with the 8-BDB-TcAMP (○, 0.3; □, 0.6; ▵, 1.2; ▿, 2.5; •, 4; and ▪, 8 mmol/L) in 50 mmol/L HEPES buffer, pH 7.5, 20% glycerol and 5 mmol/L MgCl2 . Aliquots (2 μL) were removed, diluted 1,000-fold, and the diluted solution was assayed in triplicate. The results were corrected for the control which retained >95% of starting activity over the course of the experiment. (B) The pseudo-first-order constants k′ were plotted against the concentration of 8-BDB-TcAMP to obtain the second-order rate constant (0.022 min−1mmol/L−1).
Time course of the inactivation of cAS-PDE by 8-BDB-TcAMP. (A) The enzyme was incubated at 25°C with the 8-BDB-TcAMP (○, 0.3; □, 0.6; ▵, 1.2; ▿, 2.5; •, 4; and ▪, 8 mmol/L) in 50 mmol/L HEPES buffer, pH 7.5, 20% glycerol and 5 mmol/L MgCl2 . Aliquots (2 μL) were removed, diluted 1,000-fold, and the diluted solution was assayed in triplicate. The results were corrected for the control which retained >95% of starting activity over the course of the experiment. (B) The pseudo-first-order constants k′ were plotted against the concentration of 8-BDB-TcAMP to obtain the second-order rate constant (0.022 min−1mmol/L−1).
Modification of PDE4a.Homogeneous PDE4a, 0.85 mg in 1.5 mL (of 50 mmol/L HEPES, pH 7.2 buffer) was incubated with 4 mmol/L 8-BDB-TcAMP at 25°C and activity monitored until ∼10% remained. The protein was then loaded to a single 5 mL spin column packed with Sephadex G-50 resin and previously equilibrated with 50 mmol/L HEPES, pH 7.2 buffer containing 5 mol/L guanidine-HCl to minimize protein loss. One minute after sample application, the column was centrifuged at 7,000g for 5 minutes. This procedure removed excess 8-BDB-TcAMP. The eluent (1.7 mL) was then dialyzed overnight in 1 L, 20 mmol/L ammonium bicarbonate, pH 8.2 buffer, using Slydea Lyzer dialysis cassette (Pierce, Rockford, IL) at 4°C to minimize the loss that might take place on a second spin column. The protein was then subjected to reduction by two additions at 4°C (40 minutes apart) of 17 μL (4 mmol/L final concentration) from 200 mmol/L stock solution of [3H]NaBH4. The specific radioactivity of the stock solution was 2 × 1013 cpm/mol. The mixture was then subjected to carboxymethylation by the addition of iodoacetate to 50 mmol/L and allowed to incubate for 20 minutes at 25°C. At the end of the incubation, the solution was made 5 mol/L with guanidine-HCl, and the entire reaction mixture (2.1 mL) was then dialyzed against 2 L, 20 mmol/L ammonium bicarbonate pH 8.1 buffer, for 2 days at 4°C, with four buffer changes.
At the end of the above procedure, the modified and denatured protein (0.27 mg) was subjected to proteolytic digestion. This was accomplished by adding 13.5 μg TPCK-treated bovine pancreas trypsin to the 270 μg modified protein (5% wt/wt) and incubating at 37°C for 1 hour. This procedure was repeated once before the sample was frozen in a dry ice/acetone mixture and lyophilized to reduce the volume to 0.7 mL. The sample was then centrifuged at 13,000g using a microcentrifuge, and the labeled peptide was purified.
Purification of modified peptide.Separation of tryptic digests was performed on a Spectra-Physics high performance liquid chromatography (HPLC) with a Vydac C18 reverse phase column (1 × 25 cm) with a flow rate maintained at 1.0 mL/min. Two HPLC runs using four solvent systems were performed to isolate radioactive peptide. The tryptic mixture was first purified using 0.1% trifluoroacetic acid (TFA) in water (solvent A) and 0.1% TFA in 80% acetonitrile (solvent B). Linear gradients were started after 10 minutes and made 5% B at 20 minutes, 25% B at 120 minutes, 65% B at 260 minutes, and 100% B at 320 minutes. Aliquots of 100 μL were removed, and tritium was determined by liquid scintillation counting in 3 mL Ecolite counting fluid (ICN Pharmaceuticals Inc, Plainview, NY). The radioactive peaks were pooled and dried on a rotary-evaporator. The dry, crude peptide was dissolved in H2O and purified further on the same C18 column but using 20 mmol/L ammonium acetate, pH 6.1 (solvent C) and 20 mmol/L ammonium acetate in 50% acetonitrile (solvent D). The solvent gradient was started at the end of a 10 minutes wash with solvent C and made 35% D at 70 minutes, 60% D at 190 minutes, and 100% D at 240 minutes. Two hundred microliters of each fraction was removed and assayed for radioactivity and the fraction containing radioactivity, which also corresponds to an absorption peak at A210 nm, was selected for peptide analysis.
Peptide analysis.The amino acid sequences of isolated peptide were determined using an automated gas-phase protein/peptide sequence analyzer from Applied Biosystems (Foster City, CA), Model 470A, equipped with an on-line PTH analyzer, Model 120 and Computer Model 900A.
RESULTS
Competitive inhibition of PDE4a by 2-BDB-TcAMP.When the enzyme was incubated at 25°C in Tris.HCl, pH 7.5 buffer and mixed with 2-BDB-TcAMP (0.5 and 5 mmol/L) in the same buffer, immediate inhibition was observed and no time-dependent behavior was detected up to 1 hour. The inhibited enzyme was reactivated to 75% of starting activity when subjected to spin column filtration or diafiltration. When varying concentrations of 2-BDB-TcAMP (0.5 to 50 μmol/L) were added to PDE4a and the enzyme activity was assayed at various concentrations of the substrate (0.05 to 25 μmol/L), a kinetic profile consistent with competitive inhibition (Fig 1A) was observed with Ki of approximately 5.5 ± 0.4 μmol/L (Fig 1B). This result is similar to that observed for platelet PDE3a.25
Effect of cAMP and cGMP on inactivation of cAS-PDE by 8-BDB-TcAMP. The enzyme was preincubated with 20 mmol/L cAMP (□) or cGMP (20 mmol/L) (▵) or with buffer (○) for 2 minutes, then 5 mmol/L 8-BDB-TcAMP was added and timed aliquots were taken, and diluted 1,000-fold in 50 mmol/L HEPES buffer pH 7.5, 5 mmol/L MgCl2 . The diluted mixture (5 μL) was assayed for activity in triplicate. The data is presented as percent of control activity (buffer).
Effect of cAMP and cGMP on inactivation of cAS-PDE by 8-BDB-TcAMP. The enzyme was preincubated with 20 mmol/L cAMP (□) or cGMP (20 mmol/L) (▵) or with buffer (○) for 2 minutes, then 5 mmol/L 8-BDB-TcAMP was added and timed aliquots were taken, and diluted 1,000-fold in 50 mmol/L HEPES buffer pH 7.5, 5 mmol/L MgCl2 . The diluted mixture (5 μL) was assayed for activity in triplicate. The data is presented as percent of control activity (buffer).
Inactivation of PDE4a by 8-BDB-TcAMP.Incubation of the purified recombinant monocyte cAS-PDE with the affinity label (2 mmol/L) in the same buffer resulted in time-dependent loss of enzymatic activity. The pseudo-first order rate constant k′ is 0.0385 min−1 when 8-BDB-TcAMP is 2 mmol/L (t1/2 = 18 minutes). Samples that were passed through spin columns of Sephadex G-50 after inactivation revealed that 8-BDB-TcAMP irreversibly inactivated the enzyme, since no reactivation was observed upon gel filtration or dialysis. The enzyme is inactivated in a time- and concentration-dependent manner (Fig 2A). The reaction rate obeyed pseudo-first-order kinetics, and a plot of k′ versus concentration of 8-BDB-TcAMP gave a second-order rate of 0.022 mmol/L−1min−1 (Fig 2B). To test whether 8-BDB-TcAMP affects the Km or Vmax , the Km value for cAMP hydrolysis of partially inactivated (30% active) PDE4a was determined and found to be 4.1 μmol/L, close to the Km of the control enzyme (Km = 3.5 μmol/L). Thus, no important change of Km was observed when the kcat was reduced more than 3 times.
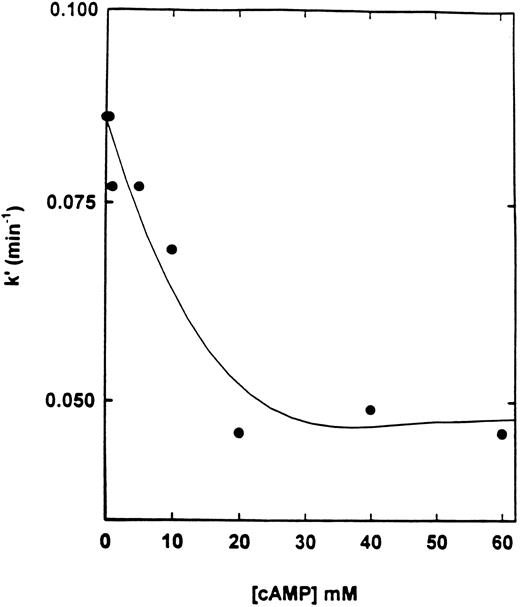
Effect of cAMP and specific inhibitors on the rate of inactivation of cAS-PDE by 8-BDB-TcAMP.The rate of inactivation of the enzyme by 8-BDB-TcAMP was markedly decreased by cAMP (20 mmol/L) but not by cGMP (Fig 3) and AMP (not shown). The protective effect of cAMP was dependent on its concentration. Figure 4 shows that 20 mmol/L cAMP is required for maximum protection; concentrations higher than 20 mmol/L did not improve the protection, and 10 mmol/L cAMP gave 50% of the maximum protection.
Effect of cAMP on the pseudo-first-order rate constant of inactivation of cAS-PDE. The enzyme was preincubated with various concentrations (1 to 60 mmol/L) of cAMP for 2 minutes then 8-BDB-TcAMP (5 mmol/L) was added and assays performed as in Fig 3.
Effect of cAMP on the pseudo-first-order rate constant of inactivation of cAS-PDE. The enzyme was preincubated with various concentrations (1 to 60 mmol/L) of cAMP for 2 minutes then 8-BDB-TcAMP (5 mmol/L) was added and assays performed as in Fig 3.
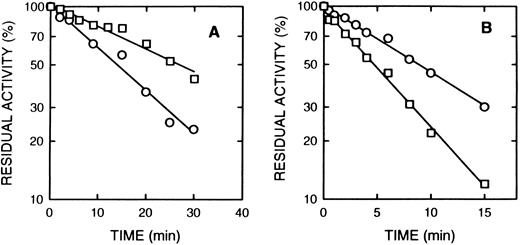
Rolipram and denbufylline are both specific inhibitors of cAS-PDE with IC50 of < 1 μmol/L.27 30 Both rolipram (0.3 μmol/L) (Fig 5A) and denbufylline (0.5 μmol/L) (Fig 5B) significantly reduced the rate of inactivation by 8-BDB-TcAMP. In control experiments at the high dilution used in the assay, neither rolipram nor denbufylline was inhibitory in the assay. The rate of inactivation of the control sample in Fig 5B is greater than that in Fig 5A because of the higher concentration of the affinity label.
The effect of rolipram and denbufylline. (A) Rolipram (0.3 μmol/L) was preincubated with cAS-PDE samples for 2 minutes and then 8-BDB-TcAMP (5 mmol/L) was added (□). The data (○) represents samples treated only with 8-BDB-TcAMP (5 μmol/L). (B) Denbufylline (0.5 μmol/L) was preincubated for 2 minutes and then 8-BDB-TcAMP (8 mmol/L) was added (○). The data (□) represent inactivation of cAS-PDE by 8 mmol/L 8-BDB-TcAMP. The timed aliquots were diluted 20,000-fold and activity determined in triplicate.
The effect of rolipram and denbufylline. (A) Rolipram (0.3 μmol/L) was preincubated with cAS-PDE samples for 2 minutes and then 8-BDB-TcAMP (5 mmol/L) was added (□). The data (○) represents samples treated only with 8-BDB-TcAMP (5 μmol/L). (B) Denbufylline (0.5 μmol/L) was preincubated for 2 minutes and then 8-BDB-TcAMP (8 mmol/L) was added (○). The data (□) represent inactivation of cAS-PDE by 8 mmol/L 8-BDB-TcAMP. The timed aliquots were diluted 20,000-fold and activity determined in triplicate.
Reagent incorporation.To quantify the amount of affinity label incorporated into the PDE4a molecule, [3H] was introduced into the modified protein by reduction with sodium [3H]borohydride (see Materials and Methods). Reduction of 8-BDB-TcAMP by sodium borohydride is expected to introduce 2 atoms of hydrogen into the two carbons of the diketo group, thus forming a diol moiety. The incorporation into modified protein consistently showed 2.6 atoms [3H]/mol protein when the enzyme had 10% residual activity, whereas the corresponding unmodified control had 0.2 atoms [3H]/mol protein. The reduced sample not incubated with 8-BDB-TcAMP maintained ∼90% of catalytic activity as compared to an identical sample not subjected to sodium [3H]borohydride. Therefore, observed net stoichiometry of labeling was 2.4 atoms [3H] per mole of enzyme. This ratio corresponds to 1.2 mol of 8-BDB-TcAMP incorporated per mole of PDE4a. The radiolabeled protein complex migrated in the same way as unmodified enzyme as determined by SDS-PAGE gel electrophoresis (data not shown).
Reversal of inactivation and incorporation by dithiothreitol.Partially inactivated enzyme was reactivated by treatment with dithiothreitol. When the enzyme, inactivated to 35% to 40% of initial activity, was incubated with 20 mmol/L dithiothreitol for 1 hour, the catalytic activity increased to 68% to 70%. The regenerated activity was retained after overnight dialysis in 50 mmol/L TrisHCl buffer, pH 7.5 containing 5 mmol/L Mg+2 and 5 mmol/L dithiothreitol. To determine if the incorporation was reversible, the same sample of modified, inactive PDE4a-8-BDB-TcAMP mixture was reduced with tritiated sodium borohydride to convert the diketone moiety to the corresponding dialcohol and to incorporate tritium into the complex. When this radiolabeled complex was incubated with 20 mmol/L dithiothreitol, 60% of radioactivity was released from the complex, presumably due to the dissociation of radioactive reagent.
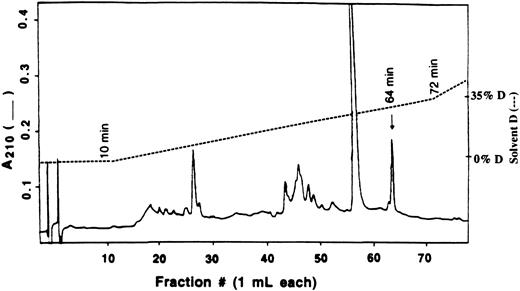
Isolation and identification of modified peptide by HPLC.The modified peptide was isolated from an enzyme sample containing 0.8 molecules of 8-BDB-TcAMP incorporated per molecule of PDE4a. When digested with trypsin and fractionated on a C18 column using the trifluoroacetic acid (TFA) solvent system, the radioactivity eluted as a peak at 129 minutes (27% solvent B). This peak was pooled, lyophilized, and repurified on the same C18 reverse phase column but equilibrated this time with 20 mmol/L ammonium acetate, pH 6.1, and eluted with a gradient to 20 mmol/L ammonium acetate containing 50% acetonitrile (solvent D). The main peak of radioactivity eluted at ∼30% solvent D and corresponded with the peptide peak marked with an arrow in Fig 6. Several other peptide peaks were observed during this procedure (Fig 6), but these did not contain significant radioactivity.
Separation of tryptic peptides by HPLC. The crude radioactive peptide from 0.1% TFA/Acetonitrile solvent system was reapplied to Vydac C18 HPLC column equilibrated with 20 mmol/L ammonium acetate, pH 6.1. The peptide peaks were eluted at 1 mL/min with a linear gradient of 20 mmol/L ammonium acetate in 50% acetonitrile after a 10 minutes wash. The only radioactive peak, indicated by an arrow, was pooled and sequenced.
Separation of tryptic peptides by HPLC. The crude radioactive peptide from 0.1% TFA/Acetonitrile solvent system was reapplied to Vydac C18 HPLC column equilibrated with 20 mmol/L ammonium acetate, pH 6.1. The peptide peaks were eluted at 1 mL/min with a linear gradient of 20 mmol/L ammonium acetate in 50% acetonitrile after a 10 minutes wash. The only radioactive peak, indicated by an arrow, was pooled and sequenced.
Table 1 reports the amino acid sequence data obtained from an aliquot of the peptide eluting at 129 minutes. The PTH-amino acids correspond to the sequence Gly697-Pro-Gly-His-Pro-Pro-Leu-Pro-Asp-Lys706, identified by reference to the amino acid sequence deduced from the cDNA sequence.27 This peptide results from cleavage by trypsin after Arg696 and Lys706, consistent with the specificity of trypsin recognition sites.31 32 Peptide 697-706 is located within the conserved catalytic domain of PDE4a.
Modified Peptide Sequence Determined by N-Terminal Sequencing
| Cycle . | PTH-Amino Acid (pmol)* . | Amino Acid Sequence . |
|---|---|---|
| . | . | Deduced From cDNA† . |
| 1 | Gly (28) | Gly697 |
| 2 | Pro (18) | Pro698 |
| 3 | Gly (53) | Gly699 |
| 4 | His (11) | His700 |
| 5 | Pro (55) | Pro701 |
| 6 | Pro (12) | Pro702 |
| 7 | ‡ | Leu703 |
| 8 | Pro (6) | Pro704 |
| 9 | Asp (11) | Asp705 |
| 10 | ‡ | Lys706 |
| Cycle . | PTH-Amino Acid (pmol)* . | Amino Acid Sequence . |
|---|---|---|
| . | . | Deduced From cDNA† . |
| 1 | Gly (28) | Gly697 |
| 2 | Pro (18) | Pro698 |
| 3 | Gly (53) | Gly699 |
| 4 | His (11) | His700 |
| 5 | Pro (55) | Pro701 |
| 6 | Pro (12) | Pro702 |
| 7 | ‡ | Leu703 |
| 8 | Pro (6) | Pro704 |
| 9 | Asp (11) | Asp705 |
| 10 | ‡ | Lys706 |
From the measured radioactivity, 71 pmol of radioactive pepide was applied to the sequencer.
The amino acid sequence of the isolated peptide determined by N-terminal sequencing is compared with the cDNA deduced protein sequence from Livi et al.27 A second peptide, Ile-Ser-Gly-Try-Gly-Asn, was also present and was identified as derived from bovine pancreatic trypsin.30 31
No PTH derivative was detected at ≥5 pmol.
DISCUSSION
The cAMP analogs, 8-BDB-TcAMP and 2-BDB-TcAMP, both can be considered potential active site-directed inhibitors of monocyte PDE4a. However, 2-BDB-TcAMP, although it effectively interacts with the enzyme, does not form a covalent complex with PDE4a but instead is a reversible competitive inhibitor. This result is similar to an earlier observation made by Grant et al25 with platelet PDE3a. The inability of 2-BDB-TcAMP to covalently modify these enzymes suggests that there is no appropriate enzyme nucleophile in proximity to the bromoketo group of 2-BDB-TcAMP when it is bound to the substrate site on phosphodiesterase. In contrast, 8-BDB-TcAMP irreversibly inactivates r-PDE4a by the formation of a covalent complex. The protection against loss of catalytic activity offered by the substrate suggests that the inactivation is a consequence of reaction at the substrate binding site. The rate of inactivation of PDE3a by 8-BDB-TcAMP was also reduced by cAMP.25 The similar mode of inactivation of r-PDE4a and PDE3a with these two substrate analogs suggests that the substrate recognition moiety in the two enzymes is similar, as might be predicted from the high degree of sequence homology in the putative catalytic domain.
Although the r-PDE4a and platelet PDE3a both interact with 8-BDB-TcAMP and 2-BDB-TcAMP, the catalytic sites of the two enzymes exhibit distinct differences. PDE3a is known to be competitively inhibited by cGMP with a Ki = 0.1 μmol/L.33 Recent evidence 15 suggests that there are two nonidentical but overlapping sites for cAMP and cGMP. In contrast, PDE4a is unaffected by cGMP and no evidence exists for a binding site for that nucleotide. The data in this paper showing that monocyte PDE4a inactivation by 8-BDB-TcAMP was not affected by cGMP is consistent with the previous conclusions. The fact that partial inactivation of r-PDE4a by 8-BDB-TcAMP does not cause a shift in the Km value for cAMP hydrolysis can be interpreted to indicate the presence of a binding site for the substrate analog. The observed stoichiometry of incorporation of 1.2 mol of 8-BDB-TcAMP per mole of protein suggests modification of one site.
It has previously been proposed that rolipram, a selective and competitive inhibitor of PDE4a, exerts its anti-inflammatory action by inhibiting the catalytic activity of phosphodiesterase.13 34 However, the hypothesis that rolipram (and denbufylline) inhibits the catalytic activity of PDE4a by binding in the putative catalytic domain has not been shown directly. The protection data presented here provides the first direct evidence that these drugs inhibit the catalytic activity of the enzyme by binding at or near the substrate binding site. However, the data do not rule out the presence of other nondetected binding sites for these inhibitors. For example, these agents could also be binding at the putative regulatory domain, which may not have a direct influence on the catalytic activity, but may exert allosteric conformational changes.
Several enzyme systems have been investigated by the affinity labeling approach utilizing bromoketo substituents of nucleotides. Such a functional group has the potential to react covalently with various amino acid side chains in proteins. Lysine,35 tyrosine,36 cysteine and histidine 37,38 have been modified by a bromoketo substituent of a nucleotide analogue. 8-[(4-Bromo-2,3-dioxobutyl)thio]-adenosine 5′-triphosphate (8-BDB-TA-5′-TP) was found to be an affinity label for pyruvate kinase, reacting with a cysteine residue at the substrate binding site.39 NAD-+specific isocitrate dehydrogenase was irreversibly inactivated and covalently modified at an aspartate residue by 2-(4-bromo-2,3-dioxobutylthio)-adenosine 5′-diphosphate.40,41 In glutathione S-transferase, tyrosine in the catalytic site was modified by S-(4-bromo-2,3-dioxobutyl)glutathione concomitant with inactivation.42
PDE4a is inactivated by a covalent reaction between an amino acid residue of the enzyme and the bromoketo moiety of 8-BDB-TcAMP, then a thiol-containing compound is likely to reverse the reaction, thereby restoring free enzyme. The reversal of activity in partially inactivated enzyme and the displacement of the affinity label from modified protein complex by dithiothreitol indicate that the reagent is capable of reacting covalently with the complex, plausibly at the protein-analog linkage bond. Such a reaction is predicted to regenerate an enzyme capable of catalyzing cAMP hydrolysis.
The interaction of PDE4a with rolipram or denbufylline shows distinct differences from that observed with the 8-BDB-TcAMP. Although a low concentration (< 1 μmol/L) of rolipram or denbufylline affords protection against inactivation, the amount of cAMP required to protect the enzyme to a similar extent is significantly higher (10 to 20 mmol/L). The high cAMP requirement may be because the fact that cAMP is a substrate and, thus, is continuously being metabolized. It is also possible (although we have no evidence to support this hypothesis) that there are two binding sites for this analogue, of which cAMP binds to only one site, thus offering only partial protection.
Following the covalent modification of PDE4a by 8-BDB-TcAMP, reduction with (3H)NaBH4 allowed the incorporation of radioactivity at about 1 mol/mol enzyme. Tryptic digestion and HPLC fractionation allowed the identification and isolation of a single radioactive peptide. N-terminal sequencing established the sequence as Gly-Pro-Gly-His-Pro-Pro-X-Pro-Asp-X. The cDNA construct used in this study has been completely sequenced,27 and comparison of the peptide sequence with the cDNA-derived sequence revealed that the affinity compound labeled this enzyme near the C-terminal portion of the catalytic domain. The peptide sequence corresponds to amino acids 697-706: Gly-Pro-Gly-His-Pro-Pro-Leu-Pro-Asp-Lys. There are only two PDEs with low Km for cAMP: PDE3 and PDE4. However, it is not clear whether their substrate recognition sites are the same. A search of the protein data base revealed 80% identity (8 of 10 amino acids) with rat PDE4 but not with other PDEs. This peptide is therefore consistent with a unique substrate recognition site in PDE4.
It would have been desirable to identify directly the amino acid in r-PDE4a, modified by 8-BDB-TcAMP; however, radioactivity released after each cycle of the Edman degradation was too low to compare quantitatively. If Lys706 were modified by 8-BDB-TcAMP, the cleavage by trypsin should not have occurred, and, thus, this derivatization is unlikely. We cannot distinguish between modification of His700 and Asp705 because both are known to react with this class of affinity reagents and both regenerate the unmodified amino acid during the sequencing reactions. Modification of histidine can produce an unstable peptide.38 Histidines are known to play a role in other PDEs as either a catalytic residue14 or metal binding ligand.10 11 Since the reaction of 8-BDB-TcAMP forms an ester with aspartate residues, such a compound could hydrolyze, especially when subjected to low pH, as in TFA (pH = 2). Aspartic acid could also be important as a metal binding ligand or participate as part of the substrate binding site. The protection by cAMP makes it highly likely that this peptide is close to or part of the active site.
We have shown that 8-BDB-TcAMP is effective as an affinity label of the catalytic site of PDEs. These analogs can also be used to probe cAMP binding sites of other proteins. The data support the notion that specific inhibitors confer their effect by binding at the catalytic site and that the catalytic site of PDE4a is likely to be located within the conserved domain.
ACKNOWLEDGMENT
We thank Rita Stewart for meticulous manuscript preparation.
Supported in part by a research grant from the National Institutes of Health (NIH) HL46341 (RWC), Walter P. Lomax, Jr Fellowship Award from the Southeastern Pennsylvania Chapter of the American Heart Association and NIH HL03375 (GAO), and from NSF MCB-94-23108 (RFC).
Address correspondence to Robert W. Colman, MD, Sol Sherry Thrombosis Research Center, Temple University School of Medicine, 3400 North Broad St, Philadelphia, PA 19140.

. (B) The slope of each line was plotted against 2-BDB-TcAMP concentrations according to the equation: slope = Km/V(1 + [I]/Ki) to give a Ki value of 5.5 ± 0.4 μmol/L (SEM).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/3/10.1182_blood.v89.3.1019/8/m_bl_0019f1.jpeg?Expires=1767765343&Signature=I0~ZLLMNuaKVxAF7Kz-cAIJNH-CKQ-5F4oKHOgDu-LPtmlWl1-dH1zHWEFc0lh2uVuh8fJfs~Oojx80qTzqQ1CAaP1xT33RKWJ54ecgpHijt7G-Cp4fmLO6HBOUyNx7uaOmAviSkX8vL5hGX99rFIFmIsmghV73wEi6L-043bqPcj-O1IFl5-vQh3iArCfsNy2m9r757Tqw99mcUH6oZ9hj50IGDGA1CN-Ozoi2-QqaMi4rY2AAVmcAinMKL02n4Tm1E3xWMuk~9hrwb2QDTIBIgVSqX7N7pXDtEo5POUqUp~KsZA7F7mO5R49dxWBgVSfyr9h5IPaZmy7HYgDZ65w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal