Abstract
Treatment of freshly isolated acute promyelocytic leukemia (APL) cells and the myelogenous leukemia cell lines, NB4, HL-60, and U937, with all-trans retinoic acid (ATRA) results in a remarkable elevation in the amounts of Stat1α and Stat2 proteins. Stat1α protein levels are augmented by ATRA as a consequence of elevated amounts of the corresponding transcripts. The retinoid increases the levels of nuclear complexes that are capable of binding to interferon (IFN)-regulated consensus sequences and contain Stat1 and/or Stat2 proteins, and causes a rapid and long-lasting elevation in Stat1α tyrosine phosphorylation. Transient transfection experiments show that ATRA enhances the transactivating properties of Stat1α observed on an appropriate reporter gene, in the presence of the RARα retinoic acid receptor, but not in the presence of the PML-RAR protein. Treatment of NB4 cells with ATRA is associated with a remarkable upregulation of the two IFN-responsive genes IFN-responsive factor 1 and 2′-5′ oligoadenylate synthetase, as well as with an augmentation in the levels of IFNα secretion. Our data show that ATRA is capable of modulating the amounts and the state of activation of some of the components of the IFN intracellular signaling pathways. They also suggest that the retinoid can bypass IFN/IFN-receptor interactions and induce the expression of IFN-regulated genes.
ALL-TRANS retinoic acid (ATRA) is a strong cyto-differentiating agent1 that is extremely active in the clinical treatment of acute promyelocytic leukemia (APL; M3 according to the French-American-British classification [FAB]) patients.2 APL patients undergo complete clinical remission upon treatment with a single course of this drug.2 Disappearance of the leukemic blasts from the bone marrow (BM) of patients treated with ATRA is the consequence of an overriding of the maturation block along the granulocytic pathway triggered by the retinoid.3 In vitro challenge of freshly isolated APL blasts or the APL-derived NB4 cell line with pharmacological concentrations of ATRA and other synthetic retinoids causes maturation towards cells that morphologically and biochemically resemble neutrophilic granulocytes.4 Granulocytic maturation seems to be the consequence of an activation of the normal RARα type of nuclear retinoic acid receptors or the aberrant PML-RAR receptor, which is synthesized in APL cells as a consequence of the typical t(15:17) balanced chromosomal translocation observed in this type of leukemia.5-7 However, the molecular mechanisms and the intracellular pathways involved in the process of granulocytic maturation are still unknown.
Interferons (IFNs) are potent cytokines with antiviral, antiproliferative, and cyto-differentiating activities.8 Two classes of IFNs, ie, type I (IFNα/β) and type II (IFNγ), acting on different receptors, are known.8 IFNα is active in the treatment of chronic myelogenous and hairy cell leukemias.9,10 Many aspects of the intracellular pathways activated by the interaction between IFNs and their receptors have been elucidated. On ligand binding, both type I and type II IFN receptors cause the activation of Janus-type tyrosine kinases (Jak1 and Tyk2 in the case of IFNα/β, Jak1, and Jak2 in the case IFNγ), which results in tyrosine phosphorylation of other intracellular protein substrates.11 These substrates are known as signal transducers and activators of transcription (Stat). Stat proteins belong to an expanding family of proteins, which are tyrosine phosphorylated and activated in response to various types of cytokines.12 IFN α/β causes the tyrosine phosphorylation of Stat 1α, Stat 1β (Stat1β is a splicing variant of Stat1 lacking the last 38 amino acids at the carboxyl terminus of the protein) and Stat2, which migrate from the cytoplasm to the nucleus and along with p48/ISGF3-γ (p48) form the transcriptionally active complex known as ISGF3.11 ISGF3 binds to the IFN-stimulated responsive elements (ISRE) consensus sequence and activates the transcription of a series of genes containing this cis-acting regulatory element.13,14 However, IFNα/β, like IFNγ, is also capable of causing the tyrosine phosphorylation and homodimerization of Stat1 proteins that translocate to the nucleus and bind to consensus sequences known as GAS (γ-IFN activated site).15
Several reports have shown synergistic interactions between ATRA and IFNs; however, the mechanisms underlying the cross-talk between the intracellular pathways activated by the retinoid and the cytokines need to be defined. Very recently, Chelbi-Alix et al,16 as well as ourselves,17 showed that IFNs induce the expression of PML-RAR in the APL-derived NB4 cell line. In addition, we showed that IFNs increase the expression of certain ATRA-regulated genes in the same cellular context.17 In this report, our results show that ATRA induces the expression of Stat1α and Stat2 in NB4 cells, as well as Stat1 in freshly isolated APL blasts, HL-60, and U937 myeloid cell lines. Furthermore, the retinoid causes tyrosine phosphorylation of Stat1α and activation of nuclear complexes containing Stat1/Stat2 heterodimers and Stat1 homodimers. Finally ATRA increases the expression of two IFN-responsive genes like interferon responsive factor 1 (IRF1) and 2′-5′ oligoadenylate synthetase (OAS) and augments the secretion of IFNα in NB4 cells.
MATERIALS AND METHODS
Cell culture conditions and reagents.The NB4 APL cell line18 was a kind gift of Dr Michel Lanotte (Unite' INSERM 301, “Genetique cellulaire et moleculaire des leucemies”). HL-60 and U937 myeloid leukemic cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD). Cells were routinely seeded at 4 × 105/mL in RPMI 1640 containing 10% fetal calf serum (FCS; GIBCO-BRL, Gaithersburg, MD). HeLa S3 (HeLa) and COS-7 fibroblasts (ATCC) were cultured as monolayers in DMEM medium containing 10% FCS (GIBCO-BRL). Cultures were free from mycoplasma as assessed using the Hoechst 33258 fluorescent dye system (Farbwerke Hoechst AG, Frankfurt, Germany). ATRA was purchased from Sigma (St Louis, MO). Stock solutions of ATRA (10−2 mol/L) were prepared in dimethylsulfoxide under dimmed light and stored at -80°C, protected from light until use. Recombinant human granulocyte colony-stimulating factor (G-CSF ) (specific activity 108 U/mg protein) was from Amgen Inc (Thousand Oaks, CA). Recombinant IFNα A/D (IFNα; specific activity, 6.4 × 107 U/mg protein) was a kind gift from Dr M. Brunda (Hoffmann-La Roche, Nutley, NJ), and recombinant IFNγ (specific activity, 2 × 107 U/mg protein) was from Roussel-UCLAF (Paris, France).
Preparation of freshly isolated APL and chronic myelogenous leukemia (CML) cells.Following informed consent, BM aspirates from 5 patients with a classical form of APL (M3 according to the FAB classification) presenting the typical t(15-17) chromosomal translocation were withdrawn. Almost pure preparations of APL cells were obtained by dextran sedimentation and consisted of more than 90% APL blasts as assessed by morphology. Leukemic cells were resuspended in RPMI 1640 containing 10% FCS and cultured in the various experimental conditions described in the report.
Extract preparation, electrophoretic shift assays, immunoprecipitation, and Western blot analysis.For electrophoretic mobility shift assays (EMSA), nuclear extracts, from 1 × 107 NB4 or HeLa cells grown under various experimental conditions, were prepared exactly as described by Dignam et al.19 Nuclear extracts were kept frozen in aliquots at −80°C until use. Oligonucleotides were 32P-labeled with [γ-32P] adenosine triphosphate, using polynucleotide kinase. The nuclear extracts (5 to 10 μg) were incubated with 0.5 ng of radiolabeled DNA probe (20,000 cpm) at room temperature for 20 minutes in 10 mmol/L Tris-HCl (pH 7.5), 50 mmol/L NaCl, 1 mmol/L dithiothreitol (DTT), 1 mmol/L EDTA, 5% glycerol, and 1 μg of poly(dI-dC) to inhibit nonspecific binding of the probe to nuclear extract proteins. DNA-protein complexes were resolved by electrophoresis through 4% polyacrylamide gels containing 50 mmol/L Tris, 0.38 mol/L glycine, and 2 mmol/L EDTA. The gels were subsequently dried and autoradiographed with intensifying screens at −80°C.
The following double stranded oligonucleotides were used for EMSA: −5′CCTGATTTCCCCGAAATGATG3′ corresponding to nucleotide -130/-110 of the IRF1 gene promoter region, containing a GAS site (pIRE20 ). −5′GATCCTCGGGAAAGGGAAACCGAAACTGAAGCC3′ corresponding to nucleotide −121/−89 of the ISG15 gene promoter region, containing an ISRE site (ISRE21 ). 5′GATCGGGAGGCGTGGCCTGGGCGGGACTGGGGAGTGGCGAGATC3′ corresponding to nucleotide −80/−44 of human immunodeficiency virus-1 (HIV-1) long-terminal repeat, containing a Sp1 site.22
Gel supershift assays were performed as described above with the exception that, before incubation of oligonucleotide probes with nuclear extracts, 1 or 2 μL of TransCruz gel supershift Stat1, Stat2, or p53 antibody (Santa Cruz Biotechnolgy, Santa Cruz, CA) was added to the reaction mixture and incubated for 30 minutes at 4°C.
Immunoprecipitations were performed using rabbit polyclonal antibodies raised against a peptide common to Stat1α and Stat1β (Santa Cruz) according to the protocol of the manufacturer. Immunoprecipitates were subjected to Western blot analysis as detailed below using either a mouse monoclonal antibody raised against tyrosine phosphate (PY20, Santa Cruz) or the anti-Stat1α antibody.
For Western blot analysis, total cellular extracts from NB4, U937, HL-60, and freshly isolated APL cells (approximately 3 × 106) were prepared at 4°C in 50 μL of suspension buffer containing 0.1 mol/L NaCl, 10 mmol/L Tris/HCl (pH 7.6), 1 mmol/L EDTA, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 2.5 mg/mL soybean trypsin inhibitor, and 100 μg/mL phenylmethylsulfonylfluoride (all from Sigma). The cells were lysed by adding 50 μL of lysis buffer containing 100 mmol/L Tris/HCl (pH 6.8), 200 mmol/L dithiothreitol, 4% sodium dodecyl sulfate (SDS), 0.2% bromophenol blue, and 20% glycerol. Samples were then boiled for 10 minutes, sonicated, and centrifuged. Aliquots of the supernatants were fractionated by electrophoresis on 10% SDS acrylamide gel and electrotransferred to nitrocellulose membranes, blocked with 5% nonfat powdered milk in TBS-T (20 mmol/L Tris/HCl [pH 7.6], 137 mmol/L NaCl, 0.1% Tween-20), and then incubated for 2 hours at 37°C with rabbit polyclonal antibodies raised against peptides corresponding to Stat1α, Stat2, Stat3, Stat4, p48/ISGF3-γ, Jak1, Jak2, and Tyk2 (Santa Cruz). Following extensive washing with TBS-T, the blots were incubated for 90 minutes with peroxidase conjugated goat anti-rabbit IgG. Following washing with TBS-T, immunoreactive protein bands were visualized with a chemiluminescence-based procedure using the enhanced chemiluminescence detection kit (Amersham) according to the instructions of the manufacturer.
Isolation of Stat1α cDNA and Northern blot analysis.A cDNA containing the entire coding region of Stat1α was cloned by reverse transcriptase-polymerase chain reaction (RT-PCR). Briefly, PCR amplification of the Stat1α transcript was performed using total RNA extracted from NB4 cells treated with 10−6 mol/L ATRA for 4 days. Total RNA was prepared from NB4 or freshly isolated APL cells, as described.23 The poly(A+) RNA fraction was selected on an oligo-dT cellulose column (Pharmacia, Uppsala, Sweden) according to standard conditions.24 One microgram of poly(A+) RNA was used for reverse transcription with the gene AMP kit (Cetus Perkin Elmer, Norwalk, CT) according to the instructions of the manufacturer. The cDNA was amplified with the following couples of amplimers containing restriction sites for BamHI at their 5′ ends to allow easy subcloning: 5′GTGGATCCATGTCTCAGTGGTACGAACT3′ and 5′CAGGATCCGCTCTATACTGTGTTCATCA3′ corresponding to nucleotides 197-216 and corresponding to the complement of nucleotides 2400-2419, respectively, of the published Stat1α sequence.25 Amplification conditions of the cDNA were: 94°C, 1 minute, 55°C, 2 minutes, and 72°C, 3 minutes for 30 cycles. The primary structure of the cDNA coding for Stat1α was verified by oligonucleotide sequencing following subcloning in the BamHI site of the eukaryotic expression vector pSG5 (Pharmacia) (pSG5-Stat1).
Northern blot analysis was performed on total RNA preparations. The following probes were used for Northern blot analysis: a full-length human 2′-5′OAS cDNA26; a full-length human IRF-1 cDNA27; a cDNA fragment corresponding to the 3′ noncoding region of human β-actin28; the full-length cDNA coding for human Stat1 obtained by RT-PCR. The various probes were labeled to a specific radioactivity of (1 to 2) × 109 cpm/μg with a commercially available kit using random hexanucleotide primers and [32P] dCTP (Amersham). Hybridization and washing were performed as already described.23
Transactivation experiments.Transient transfection experiments in COS-7 cells were performed according to a standard calcium-phosphate coprecipitation procedure29 using the following plasmids: pSG5-RARα (from Dr P.G. Pelicci, Perugia, Italy) and pSG5-PML/RARα29 containing the human RARα and PML-RAR cDNAs, respectively, under the control of the SV-40 early T-antigen promoter enhancer; pIRE-TK-CAT, containing the chloramphenicol acetyl transferase (CAT) reporter gene under the control of three copies of the GAS site contained in pIRE of the IRF1 gene placed in front of the viral thymidine kinase gene; pSG5-Stat1, containing Stat1α under the control of the SV40 early T antigen; pnlsLACZ containing the bacterial β-galactosidase gene under the control of the early T antigen of the SV40 enhancer-promoter30 (from Dr A. Weisz, Napoli, Italy). The plasmid pIRE-TK-CAT was constructed by inserting three copies of the double stranded oligonucleotide 5′GCCTGATTTCCCCGAAATGACGG3′, corresponding to nucleotide −126 to −109 of the promoter of the IRF1 gene, within the BamHI and HindIII sites of pBLCAT2 plasmid.31 After leaving the DNA coprecipitate in contact with cells for 16 hours, fresh medium 10% charcoal stripped FCS, to eliminate endogenous retinoids alone or fresh medium containing ATRA 10−6 mol/L or IFNα 500 U/mLwas added and cells were further incubated for 36 hours. At the end of the treatment, cells were obtained and processed for determination of CAT and β-galactosidase enzymatic activity. CAT and β-galactosidase activities were measured on cell extracts according to standard procedures, as described.28 The results are expressed as relative CAT activity, which is the ratio of CAT activity produced by the reporter in counts per minute of acetylated chloramphenicol divided by the β-galactosidase activity produced by pnlsLACZ expressed in absorbance units at 420 nm.
Determination of IFNα immunoreactive protein.IFNα was determined in the growth medium of NB4 cells using a commercially available enzyme-linked immunosorbent assay (ELISA) kit (Cytoscreen immunoassay kit; Biosource International, Camarillo, CA) according to the instructions of the manufacturer.
RESULTS
ATRA induces Stat1α and p48, but not IFN-receptor–coupled Janus-kinases in NB4 cells.The effects of ATRA, IFNα, and IFNγ on the steady-state levels of four members of the family of Stat proteins were evaluated in NB4 cells and the results of this analysis are shown in Fig 1A. In basal conditions, NB4 cells synthesize detectable amounts of Stat1α, Stat2, and Stat3. However, to visualize Stat1α and Stat2, 10 μg of total cellular proteins were used, whereas for Stat3, 100 μg of an equivalent preparation were used. Within the limits of our assay, Stat4 is not synthesized at appreciable levels in our experimental conditions. Treatment of NB4 cells with ATRA (10−6 mol/L), IFNα (1,000 U/mL), and IFNγ (1,000 U/mL) for 4 days gives rise to a remarkable upregulation of Stat1α. Using polyclonal antibodies recognizing both Stat1α and Stat1β, we observed that the NB4 steady state levels of the latter protein are much lower than those of the former one (data not shown). However, Stat1α and Stat1β are coregulated on treatment with ATRA, IFNα, and IFNγ (data not presented). Similarly, the steady state levels of Stat2 are increased by the three stimuli although the level of induction is lower than that observed for Stat1α. ATRA, IFNα, and IFNγ have no effect on the basal levels of Stat3 protein and they do not cause the appearance of Stat4. The steady-state levels of Stat5 were not evaluated. On Western blotting analysis, a band corresponding to Stat6 is observed neither in NB4 cells in basal conditions nor following treatment with ATRA or IFNs (data not shown). Extracts obtained from HeLa cells were used as controls for these experiments. Stat1α, Stat2, and Stat4 are not synthesized at appreciable level in this cell line in basal conditions. Treatment of HeLa cells with 1,000 U/mL of IFNα for 4 days leads to the appearance of significant amounts of the three proteins.
Effects of ATRA and IFNs on the expression of Stat, p48, Jak 1, Jak 2, and Tyk 2 proteins in NB4 and HeLa cells. NB4 (4 ×105/mL) or HeLa (a confluent 25 cm2 flask) cells were treated for 4 days with medium alone (Medium) and medium containing ATRA (10−6 mol/L), IFNα (1,000 U/mL), and IFNγ (1,000 U/mL) as indicated. Cells were harvested, lysed, and subjected to Western blot analysis, using antibodies specific for Stat 1α, Stat 2, Stat 3, and Stat 4 (A), p48 (B), Jak1, Jak 2, and Tyk 2 (C). For Stat 3 and Jak 1, 100 μg of protein/lane were used, whereas for all the other samples, 10 mg of protein/lane were used. In (B), the apparent molecular weight of standard proteins (carbonic anhydrase [28 kD], ovalbumin [43 kD], bovine serum albumin [69 kD], and phosphorylase B [105 kD]) is indicated on the right. The apparent molecular weight of Stat proteins, Jak 1, Jak 2, and Tyk 2 is indicated on the right of (A) and (C). Western blots representative of at least two independent experiments are shown.
Effects of ATRA and IFNs on the expression of Stat, p48, Jak 1, Jak 2, and Tyk 2 proteins in NB4 and HeLa cells. NB4 (4 ×105/mL) or HeLa (a confluent 25 cm2 flask) cells were treated for 4 days with medium alone (Medium) and medium containing ATRA (10−6 mol/L), IFNα (1,000 U/mL), and IFNγ (1,000 U/mL) as indicated. Cells were harvested, lysed, and subjected to Western blot analysis, using antibodies specific for Stat 1α, Stat 2, Stat 3, and Stat 4 (A), p48 (B), Jak1, Jak 2, and Tyk 2 (C). For Stat 3 and Jak 1, 100 μg of protein/lane were used, whereas for all the other samples, 10 mg of protein/lane were used. In (B), the apparent molecular weight of standard proteins (carbonic anhydrase [28 kD], ovalbumin [43 kD], bovine serum albumin [69 kD], and phosphorylase B [105 kD]) is indicated on the right. The apparent molecular weight of Stat proteins, Jak 1, Jak 2, and Tyk 2 is indicated on the right of (A) and (C). Western blots representative of at least two independent experiments are shown.
Stat1α and Stat2 are part of the IFNα-dependent ISGF3 transcriptional complex along with the nuclear protein p48, which represents the DNA binding component.11 As shown in Fig 1B, detectable amounts of p48 are observed in NB4 cells incubated for 4 days in medium alone. Whereas ATRA causes a slight albeit significant induction of p48, neither IFNα nor IFNγ affects the level of expression of this protein. The lack of p48 induction by IFNα is not consistent with what is observed in HeLa cells, where the cytokine is a strong inducer of the protein.
Jak1, Jak2, and Tyk2 are tyrosine kinases associated with type I and type II IFN receptors.11 As illustrated in Fig 1C, the three proteins are detected in NB4 cells grown in basal conditions. An increase in Jak1 levels is observed on treatment of cells for 4 days with IFNα and IFNγ, whereas ATRA does not cause any effect on the expression of the protein. Jak2 and Tyk2 are not significantly modulated by the retinoid or the two cytokines.
Given the level of inducibility of Stat1α following treatment of NB4 cells with ATRA, we focused our attention on this protein. As shown by Fig 2A, in NB4 cells, induction of Stat1α by ATRA is dose-dependent, being evident at 10−7 mol/L and maximal at 10−6 mol/L. The concentrations of ATRA at which Stat1α is induced are the same as those necessary to induce morphological and biochemical maturation of NB4 promyelocytes into cells resembling normally differentiated granulocytes.18,23,29,32 Maximal granulocytic maturation of NB4 promyelocytes following challenge with 10−6 mol/L ATRA is obtained between the third and the fourth day of treatment.18 32 The lower than basal level of Stat1α protein shown in Fig 2A in the lanes corresponding to 10−9 and 10−8 mol/L ATRA is an occasional finding and was not reproduced in two other experiments.
Dose- and time-dependent induction of Stat 1α protein and Stat1 mRNA by ATRA in NB4 cells. (A) NB4 cells (4 × 105/mL) were treated for 4 days with medium alone (medium) and medium containing the indicated concentrations of ATRA. Cells were harvested, lysed and the cell extracts (10 μg of protein/lane) subjected to Western blot analysis, using antibodies specific for Stat 1α. (carbonic anhydrase [28 kD], ovalbumin [43 kD], bovine serum albumin [69 kD], phosphorylase B [105 kD], and myosin heavy chain [215 kD]). A Western blot representative of two independent experiments is shown. (B) NB4 cells (4 × 105/mL) were treated for the indicated lengths of time with 10−6 mol/L ATRA. Western blot analysis was conducted as described in (A). The apparent molecular weight of standard proteins (carbonic anhydrase [28 kD], ovalbumin [43 kD], bovine serum albumin [69 kD], phosphorylase B [105 kD], and myosin heavy chain [215 kD]) is indicated on the right. A Western blot representative of three independent experiments is shown. (C) NB4 cells (4 × 105/mL) were treated for the indicated lengths of time with 10−6 mol/L ATRA. Total RNA was isolated and subjected (20 μg/lane) to Northern blot analysis. Filters were sequentially hybridized with cDNA probes coding for Stat 1α and β-actin. A Northern blot representative of two independent experiments is shown.
Dose- and time-dependent induction of Stat 1α protein and Stat1 mRNA by ATRA in NB4 cells. (A) NB4 cells (4 × 105/mL) were treated for 4 days with medium alone (medium) and medium containing the indicated concentrations of ATRA. Cells were harvested, lysed and the cell extracts (10 μg of protein/lane) subjected to Western blot analysis, using antibodies specific for Stat 1α. (carbonic anhydrase [28 kD], ovalbumin [43 kD], bovine serum albumin [69 kD], phosphorylase B [105 kD], and myosin heavy chain [215 kD]). A Western blot representative of two independent experiments is shown. (B) NB4 cells (4 × 105/mL) were treated for the indicated lengths of time with 10−6 mol/L ATRA. Western blot analysis was conducted as described in (A). The apparent molecular weight of standard proteins (carbonic anhydrase [28 kD], ovalbumin [43 kD], bovine serum albumin [69 kD], phosphorylase B [105 kD], and myosin heavy chain [215 kD]) is indicated on the right. A Western blot representative of three independent experiments is shown. (C) NB4 cells (4 × 105/mL) were treated for the indicated lengths of time with 10−6 mol/L ATRA. Total RNA was isolated and subjected (20 μg/lane) to Northern blot analysis. Filters were sequentially hybridized with cDNA probes coding for Stat 1α and β-actin. A Northern blot representative of two independent experiments is shown.
As documented by Fig 2B, the retinoid-dependent induction of Stat1α preceeds and accompanies granulocytic maturation, being evident 48 hours from the beginning of the treatment and maximal at 96 hours. In some experiments, increased levels of Stat1α were seen at 24 hours; however, in no instance did we observe upregulation at earlier time points (data not shown). The Northern blot analysis shown in Fig 2C shows that induction of the Stat1α protein by ATRA is, at least partially, the consequence of an increase in the corresponding mRNA. In fact, undifferentiated NB4 promyelocytes express detectable levels of two transcripts of approximately 3.0 and 4.4 Kb, which hybridize with Stat1α cDNA. At present, the nature of the low molecular weight Stat1α transcript (or Stat1β, since Northern blot analysis does not distinguish between the two types of mRNA) is obscure, although it may represent an immature form of RNA or an alternatively spliced gene product. Upregulation of Stat1 transcripts by ATRA is a relatively rapid and long lasting process. A significant accumulation of Stat1 transcripts is evident at 2 hours and the levels of the mRNA remain elevated at least until 96 hours.
ATRA induces Stat1α protein and Stat1 mRNA in freshly isolated APL cells, U937 and HL-60 myeloid cell lines.To investigate whether the induction of Stat1α by ATRA is a peculiarity of NB4 cells or it is a more general phenomenon, the expression of the corresponding gene was studied at the protein and RNA levels in freshly isolated APL cells and in other myeloid cell lines. As shown in Fig 3A, in basal conditions, promyelocytes from all the APL cases are characterized by detectable amounts of Stat1α protein (in the case of APL3, the Stat1α band is visible only on longer exposure of the film). In vitro challenge of leukemic cells isolated from APL1 and APL2 patients with ATRA (10−6 mol/L) for 4 days leads to an increase in the levels of Stat1α protein. In APL1, this increase is of the same magnitude as that observed following incubation of the cells with IFNα (1,000 U/mL) for the same length of time. In two other APL cases (APL3 and APL4), treatment with ATRA alone does not induce Stat1α protein. However, in these two patients, G-CSF, another granulocyte-specific cytokine and differentiating agent is capable of upregulating the expression of the protein. This effect is further enhanced by the addition of ATRA to the growth medium. In basal conditions, the U937 and HL-60 myeloid cell lines show a significant level of expression of Stat1α protein. The two cell lines respond to ATRA treatment with an elevation in the amounts of the polypeptide. In the experimental conditions used, IFNα is a more powerful inducer of Stat1α than ATRA in HL-60 but not in U937 cells. The Northern blots shown in Fig 4B show that ATRA-, G-CSF- or (ATRA+G-CSF )-dependent induction of Stat1α in three representative APL patients is, at least in part, the result of increased accumulation of the two Stat1 transcripts.
Effects of ATRA, IFNα, G-CSF and the combination of G-CSF and ATRA on Stat 1α protein and Stat 1 mRNA in freshly isolated APL cells and the two myelogenous leukemia cell lines U937 and HL-60. (A) Leukemic cells obtained from the BM of 4 different APL patients, U937 and HL-60 cells were seeded at a concentration of 4 × 105/mL and treated for 4 days as indicated. Cells were obtained, lysed, and the cell extracts (10 μg of protein/lane) subjected to Western blot analysis, using antibodies specific for Stat 1α. (B) Freshly isolated leukemic cells (4 × 105/mL) from three APL patients were treated for 4 days, as indicated. Total RNA was isolated and subjected (20 μg/lane) to Northern blot analysis. Filters were sequentially hybridized with cDNA probes coding for Stat 1α and β-actin. For all the experiments shown, the following stimuli were used: ATRA (10−6 mol/L), G-CSF (10 ng/mL), the combination between ATRA and G-CSF, and IFNα (1,000 U/mL).
Effects of ATRA, IFNα, G-CSF and the combination of G-CSF and ATRA on Stat 1α protein and Stat 1 mRNA in freshly isolated APL cells and the two myelogenous leukemia cell lines U937 and HL-60. (A) Leukemic cells obtained from the BM of 4 different APL patients, U937 and HL-60 cells were seeded at a concentration of 4 × 105/mL and treated for 4 days as indicated. Cells were obtained, lysed, and the cell extracts (10 μg of protein/lane) subjected to Western blot analysis, using antibodies specific for Stat 1α. (B) Freshly isolated leukemic cells (4 × 105/mL) from three APL patients were treated for 4 days, as indicated. Total RNA was isolated and subjected (20 μg/lane) to Northern blot analysis. Filters were sequentially hybridized with cDNA probes coding for Stat 1α and β-actin. For all the experiments shown, the following stimuli were used: ATRA (10−6 mol/L), G-CSF (10 ng/mL), the combination between ATRA and G-CSF, and IFNα (1,000 U/mL).
EMSA with ISRE and pIRE oligonucleotides using nuclear extracts obtained from NB4 and HeLa cells treated with ATRA and IFNα. (A) Nuclear extracts obtained from NB4 or HeLa cells treated for various lengths of time with the indicated stimuli were incubated with (32P)-radiolabeled ISRE, pIRE, or Sp1 double-stranded oligonucleotides in the absence (−) or in the presence (+) of a 250-fold excess of the respective cold oligonucleotide (Sp. comp.) or the same concentration of a negative-control oligonucleotide containing mutated GAS and ISRE consensus sequences (GAS/ISRE mutant oligonucleotide, Santa Cruz) (non-Sp. comp.) and subjected to EMSA. Specific retarded complexes are indicated with arrows on the left. An EMSA representative of three independent experiments is shown. (B) Nuclear extracts obtained from NB4 cells treated for 4 days with medium alone (Medium) and medium containing ATRA or IFNα were preincubated for 30 minute at 4°C either in the absence (−) or in the presence of the indicated antibodies. Following this preincubation, the extracts were incubated with (32P)-radiolabeled ISRE and pIRE for a further 20 minute at room temperature, and subsequently subjected to EMSA. Specific and nonsupershifted retarded bands are indicated by arrows. An EMSA representative of two independent experiments is shown. In all the experiments, ATRA was used at a concentration of 10−6 mol/L and IFNα at a concentration of 1,000 U/mL.
EMSA with ISRE and pIRE oligonucleotides using nuclear extracts obtained from NB4 and HeLa cells treated with ATRA and IFNα. (A) Nuclear extracts obtained from NB4 or HeLa cells treated for various lengths of time with the indicated stimuli were incubated with (32P)-radiolabeled ISRE, pIRE, or Sp1 double-stranded oligonucleotides in the absence (−) or in the presence (+) of a 250-fold excess of the respective cold oligonucleotide (Sp. comp.) or the same concentration of a negative-control oligonucleotide containing mutated GAS and ISRE consensus sequences (GAS/ISRE mutant oligonucleotide, Santa Cruz) (non-Sp. comp.) and subjected to EMSA. Specific retarded complexes are indicated with arrows on the left. An EMSA representative of three independent experiments is shown. (B) Nuclear extracts obtained from NB4 cells treated for 4 days with medium alone (Medium) and medium containing ATRA or IFNα were preincubated for 30 minute at 4°C either in the absence (−) or in the presence of the indicated antibodies. Following this preincubation, the extracts were incubated with (32P)-radiolabeled ISRE and pIRE for a further 20 minute at room temperature, and subsequently subjected to EMSA. Specific and nonsupershifted retarded bands are indicated by arrows. An EMSA representative of two independent experiments is shown. In all the experiments, ATRA was used at a concentration of 10−6 mol/L and IFNα at a concentration of 1,000 U/mL.
ATRA increases the levels of nuclear DNA-binding complexes containing Stat1 and Stat2 in NB4 cells.To investigate whether Stat1α induction by ATRA is accompanied by an increase in the levels of DNA-binding complexes containing the protein, nuclear extracts were prepared at different times following treatment of NB4 cells with the retinoid. The nuclear extracts were used to perform EMSAs using two oligonucleotides representing the ISRE of the IFN-responsive ISG15 gene and the pIRE sequence of the IRF1 gene, respectively. The ISRE is known to bind the ISGF3 complex,11 whereas pIRE has been reported to bind Stat1α-homodimers.33 Figure 4A shows that undifferentiated NB4 promyelocytes contain low but detectable levels of nuclear proteins capable of retarding the migration of both the ISRE and the pIRE oligonucleotide. Treatment of NB4 cells with ATRA for 2 hours causes an increase in the ISRE retarded band, which is much more prominent at 24 and 96 hours. Following treatment of NB4 cells with IFNα for 4 days a significant increase in the levels of an ISRE retarded band is evident. The mobility of the ATRA-modulated ISRE retarded band is the same as that observed following treatment of both NB4 and HeLa cells with IFNα. This suggests that ATRA and IFNα activate or induce the same ISRE binding proteins. Similarly to what is observed with the ISRE oligonucleotide, ATRA increases the levels of a nuclear protein complex capable of interacting with the pIRE oligonucleotide. The kinetics of the ATRA-dependent induction of the pIRE complex are slower than those observed for the ISRE counterpart. In fact, a minimum of 24-hour treatment is necessary to observe a significant elevation of the pIRE retarded band relative to baseline. The mobility of the pIRE-retarded band is the same as that observed following treatment of NB4 and HeLa cells with IFNα. The specificity of the effects caused by ATRA and IFNα on the levels of the ISRE and pIRE complexes are substantiated by the fact that the levels of Sp1 binding proteins are not altered in any of the experimental conditions taken into consideration. In addition, ISRE, pIRE, and Sp1 retarded bands are specifically competed for by preincubation with the respective cold oligonucleotides. Figure 4B shows that the ISRE retarded band induced by ATRA contains Stat1 and Stat2 or immunologically related proteins, since clear supershifts are observed in the presence of specific antibodies directed against the two proteins. Notice that the Stat1 antibody used for supershift analysis does not distinguish between Stat1α and Stat1β. As expected, the same antibodies supershift the ISRE-binding complexes induced by IFNα in both NB4 and HeLa cells (data obtained in this last cell line are not presented). A similar experiment depicted in Fig 4B shows that Stat1, but not Stat2, is also contained in the ATRA- and IFNα-modulated pIRE complexes.
ATRA increases tyrosine phosphorylation of Stat1α in NB4 cells.The presence of DNA-binding complexes containing Stat1α in the nucleus of NB4 cells treated with ATRA suggests that the retinoid may not only induce Stat1α but also activate the protein. To investigate this point, we evaluated whether ATRA treatment of NB4 cells causes tyrosine phosphorylation of Stat1α. As shown in Fig 5, Stat1α is tyrosine phosphorylated in basal conditions. On challenge of cells for 30 minute with ATRA (10−6 mol/L), a significant increase in the level of tyrosine phosphorylation is evident. Increased tyrosine phosphorylation is maintained until at least 24 hours, a time point at which no significant induction in the levels of Stat1α protein are observed (see Fig 2B). Elevation of tyrosine phosphorylation at 30 minutes is quantitatively similar to that obtained following treatment of NB4 cells with IFNα (1,000 U/mL), which was used as a positive control for our experiments. More careful time-course experiments (data not shown) demonstrated that maximal ATRA-dependent tyrosine phosphorylation of Stat1α is observed following challenge with the retinoid (10−6 mol/L) for 1 to 2 hours. However, a major increase in this type of posttranslational modification is already detectable following 15 minutes of stimulation with ATRA, whereas a 5-minute period of treatment with the compound is ineffective. Augmented tyrosine phosphorylation of Stat1α by ATRA is not a peculiarity of the NB4 cell line and it is not related to PML-RAR expression, since the phenomenon is also observed in HL-60 myeloblasts. However, in this cell type, the increase is evident at an earlier time point (5 minutes) and it is more transient, lasting less than 24 hours (data not shown).
Effects of ATRA and IFNα on the tyrosine phosphorylation state of Stat 1α in NB4 cells. Cell lysates from NB4 cells unstimulated and stimulated with ATRA (10−6 mol/L) or IFNα (1,000 U/mL) for the indicated amount of time were immunoprecipitated with anti-Stat 1α/β antibodies. Two equivalent aliquots (30 μL) of the the various immmunoprecipitates (200 μL) were subjected to Western blot analysis in parallel and the corresponding filters were probed with antiphosphotyrosine (upper panel) and anti-Stat1α (lower panel) antibodies, respectively. The molecular weight of Stat1α is indicated on the left. Ip, immunoprecipitatory antibody; WB, Western blot antibody. A representative experiment out of three independent repeats is depicted.
Effects of ATRA and IFNα on the tyrosine phosphorylation state of Stat 1α in NB4 cells. Cell lysates from NB4 cells unstimulated and stimulated with ATRA (10−6 mol/L) or IFNα (1,000 U/mL) for the indicated amount of time were immunoprecipitated with anti-Stat 1α/β antibodies. Two equivalent aliquots (30 μL) of the the various immmunoprecipitates (200 μL) were subjected to Western blot analysis in parallel and the corresponding filters were probed with antiphosphotyrosine (upper panel) and anti-Stat1α (lower panel) antibodies, respectively. The molecular weight of Stat1α is indicated on the left. Ip, immunoprecipitatory antibody; WB, Western blot antibody. A representative experiment out of three independent repeats is depicted.
In unstimulated NB4 cells, we observe a certain degree of Stat2 tyrosine phosphorylation, as assessed by specific immunoprecipitation and subsequent blotting of the immunoprecipitate with antiphosphotyrosine antibodies. ATRA (10−6 mol/L) does not seem to affect the constitutive phosphorylation level of the protein (data not shown), at least until 48 hours of treatment.
ATRA can activate an IFN-responsive promoter through Stat1α in COS-7 cells.To further support the concept that ATRA is capable of activating Stat1α, transactivation experiments using an artificial GAS (derived from the pIRE of the IRF1 gene) containing promoter driving the expression of the CAT reporter gene (pIRE-TK-CAT) were performed in COS-7 fibroblasts. This cell line was selected as a model system because of its very low level of expression of retinoic acid receptors and because of the difficulty in transfecting NB4 cells. In this cell line, we first evaluated the ability of our Stat1α construct to confer IFNα susceptibility on the pIRE-TK-CAT reporter gene. As shown in Fig 6A, COS-7 cells transfected with the pIRE-TK-CAT plasmid alone show low levels of CAT enzymatic activity. IFNα treatment causes a significant elevation of CAT activity. This suggests that COS-7 cells contain IFNα receptors as well as all the other components of the signal transduction system. Cotransfection of 2.5 μg of Stat1α cDNA-expressing plasmid causes a significant increase in the sensitivity of the reporter gene to IFNα. Sensitivity to IFNα is partially lost following cotransfection with 5.0 μg of Stat1α, because of an increase in the cytokine-independent activation of the reporter gene. As expected,20 in the presence of optimal concentrations of Stat1α (2.5 μg), the pIRE-TK-CAT reporter gene is much more responsive to IFNγ (16-fold induction of CAT activity over baseline levels with 1,000 U/mL of the cytokine) than to IFNα (data not shown).
Effects of IFNα and ATRA on the activity of a pIRE-containing promoter in the presence of Stat1α. COS-7 cells were transiently cotransfected with pIRE-TK-CAT (1 μg) and pnlsLACZ (0.5 μg) in the absence or presence of the indicated amounts of Stat 1α, RARα, or PML-RAR, as indicated. The total quantity of DNA transfected was always maintained at 10 μg by addition of an appropriate amount of the plasmid pBluescript. Sixteen hours after transfection, medium was changed and the incubation continued for further 36 hours with the indicated concentrations of IFNα or ATRA. At the end of each treatment, cells were obtained and processed for the measurement of CAT and β-galactosidase enzymatic activities. (A) Increasing amounts of pSG5-Stat1 construct were cotransfected with pIRE-TK-CAT and pnlsLACZ. Cells were incubated in the absence (□) or in the presence (▨) of an optimal concentration of IFNα (500 U/mL). The fold-increase relative to control conditions is indicated in parenthesis. (B) Increasing amounts of pSG5-Stat1 construct in the presence of a fixed amount of RARα (50 ng) were cotransfected with pIRE-TK-CAT and pnlsLACZ. Cells were incubated in the absence (□) or in the presence (▪) of an optimal concentration of ATRA (10−6 mol/L). The fold-increase relative to control conditions is indicated in parenthesis. (C) PSG5-Stat1 (0.5 μg), RARα (50 ng) were cotransfected with pIRE-TK-CAT and pnlsLACZ. Cells were incubated with increasing concentrations of ATRA, as indicated. (D) 50 ng each of RARα or PML-RAR were cotransfected with pIRE-TK CAT and pnlsLACZ in the absence or in the presence of PSG5-Stat1 (0.5 μg), as indicated. Cells were incubated with medium alone (□) or medium containing 10−6 mol/L ATRA (▪). All the results are the mean ± SD of three replicate dishes and are expressed as relative CAT activity, which is the ratio of CAT/β-galactosidase activity. All the data shown are representative of at least three independent experiments.
Effects of IFNα and ATRA on the activity of a pIRE-containing promoter in the presence of Stat1α. COS-7 cells were transiently cotransfected with pIRE-TK-CAT (1 μg) and pnlsLACZ (0.5 μg) in the absence or presence of the indicated amounts of Stat 1α, RARα, or PML-RAR, as indicated. The total quantity of DNA transfected was always maintained at 10 μg by addition of an appropriate amount of the plasmid pBluescript. Sixteen hours after transfection, medium was changed and the incubation continued for further 36 hours with the indicated concentrations of IFNα or ATRA. At the end of each treatment, cells were obtained and processed for the measurement of CAT and β-galactosidase enzymatic activities. (A) Increasing amounts of pSG5-Stat1 construct were cotransfected with pIRE-TK-CAT and pnlsLACZ. Cells were incubated in the absence (□) or in the presence (▨) of an optimal concentration of IFNα (500 U/mL). The fold-increase relative to control conditions is indicated in parenthesis. (B) Increasing amounts of pSG5-Stat1 construct in the presence of a fixed amount of RARα (50 ng) were cotransfected with pIRE-TK-CAT and pnlsLACZ. Cells were incubated in the absence (□) or in the presence (▪) of an optimal concentration of ATRA (10−6 mol/L). The fold-increase relative to control conditions is indicated in parenthesis. (C) PSG5-Stat1 (0.5 μg), RARα (50 ng) were cotransfected with pIRE-TK-CAT and pnlsLACZ. Cells were incubated with increasing concentrations of ATRA, as indicated. (D) 50 ng each of RARα or PML-RAR were cotransfected with pIRE-TK CAT and pnlsLACZ in the absence or in the presence of PSG5-Stat1 (0.5 μg), as indicated. Cells were incubated with medium alone (□) or medium containing 10−6 mol/L ATRA (▪). All the results are the mean ± SD of three replicate dishes and are expressed as relative CAT activity, which is the ratio of CAT/β-galactosidase activity. All the data shown are representative of at least three independent experiments.
In the absence of Stat1α cDNA, ATRA treatment of pIRE-TK-CAT transfected cells does not cause an alteration in the basal level of expression of the reporter gene regardless of the fact that RARα cDNA (50 ng) is included in the transfection mixture, as shown in Fig 6B. As illustrated by the same figure, following cotransfection of RARα and 0.1 or 0.5 μg of Stat1α cDNA, ATRA enhances the expression of pIRE-TK-CAT relative to what is observed in the absence of the retinoid. The effect of ATRA is lost when 5.0 μg of Stat1α are cotransfected, at least partly because of an increase in the retinoid-independent activation of the reporter gene. Thus, ATRA-dependent activation of the pIRE-TK-CAT reporter gene is dependent on the presence of optimal concentrations of Stat1α and RARα proteins. As shown in Fig 6C, following cotransfection of optimal amounts of Stat1α and RARα cDNAs, the pIRE-TK-CAT reporter gene is activated by ATRA in a dose-dependent way. The dose-response curve is steep and maximal activation is observed at 10−8 mol/L and maintained up to 10−6 mol/L.
The experiment shown in Fig 6D was performed to assess whether RARα and PML-RAR have a differential effect on Stat1 activation. The experiment was conducted by transfecting equal amounts (50 ng) of plasmids expressing each of the two retinoic acid receptor cDNAs. In these conditions the level of RARα and PML-RAR expressed is similar, as assessed by Western blotting analysis (data not shown). In addition, the quantity of receptor expressed is saturating, at least for retinoic acid-responsive element containing reporter genes.29-32 Our data show that activation of the pIRE-TK-CAT reporter gene is observed in the presence of RARα but not in the presence of PML-RAR. In the absence of Stat1α cDNA expression, neither unliganded nor liganded RARα or PML-RAR causes any effect on pIRE-CAT reporter gene activity. When optimal concentrations of Stat1α cDNA are cotransfected, ATRA significantly increases pIRE-CAT activity in the presence of RARα, whereas no significant effects on the reporter gene are observed in the presence of PML-RAR.
ATRA upregulates IRF1 and OAS gene expression in NB4 cells.To evaluate whether Stat1α and Stat2 induction and/or activation has any consequence on the expression of IFN-regulated genes in NB4 cells, the levels of IRF1 and OAS mRNAs were studied. IRF1 and OAS genes are regulated by both type I and type II IFNs in various cellular contexts.26,34 In addition, IRF1 is a tumor suppressor gene.35 Figure 7 illustrates that treatment of NB4 cells with ATRA (10−6 mol/L) for various lengths of time causes a rapid and progressive accumulation of a major 2.1-kb IRF1 transcript relative to what is observed in basal conditions. Substantial amounts of this mRNA species are already observed following 2 hours of treatment with the retinoid. In the absence of ATRA, NB4 cells do not express detectable amounts of OAS mRNA species. A minimum of 72-hours incubation with the retinoid is necessary to determine the appearance of two major OAS mRNA species (1.6 and 1.8 kb) and several other longer transcripts of much lower abundance. However, induction of OAS mRNAs is much more evident at 96 hours.
Effects of ATRA on the levels of IRF1 and OAS mRNAs in NB4 cells. NB4 cells (4 × 105/mL) were treated for the indicated lengths of time with 10−6 mol/L ATRA. Total RNA was isolated and subjected (20 μg/lane) to Northern blot analysis. Filters were sequentially hybridized with cDNA probes coding for IRF1 and β-actin or OAS and β-actin. A Northern blot representative of two independent experiments is shown.
Effects of ATRA on the levels of IRF1 and OAS mRNAs in NB4 cells. NB4 cells (4 × 105/mL) were treated for the indicated lengths of time with 10−6 mol/L ATRA. Total RNA was isolated and subjected (20 μg/lane) to Northern blot analysis. Filters were sequentially hybridized with cDNA probes coding for IRF1 and β-actin or OAS and β-actin. A Northern blot representative of two independent experiments is shown.
IFNα has been recently reported to upregulate the expression of OAS and other IFN-responsive genes in NB4 cells.36 As a first step in assessing whether ATRA and IFNα regulate IRF1 and OAS genes through similar or different pathways, we studied the combined effects of the retinoid and the cytokine. As shown in Fig 8A, treatment of NB4 cells for 4 days with ATRA (10−6 mol/L) and IFNα (1,000 U/mL) causes an almost equivalent increase in the levels of IRF1 mRNA relative to baseline. Contemporaneous treatment with the retinoid and the cytokine does not cause an enhancement in the accumulation of the IRF1 transcript. Figure 8B shows that ATRA is a more potent stimulus than IFNα in elevating OAS mRNA and that the combination of ATRA and IFNα is no more effective than the retinoid alone. Taken together these data suggest that the retinoid and the cytokine act through the same or similar intracellular pathways.
Effects of ATRA, IFNα, and the combination of the two compounds on the levels of IRF1 and OAS mRNAs in NB4 cells. NB4 cells (4 × 105/mL) were treated for 4 days with medium alone (Medium) and medium containing ATRA (10−6 mol/L), IFNα (1,000 U/mL), or the combination of the two compounds. Total RNA was isolated and subjected (20 μg/lane) to Northern blot analysis. Filters were sequentially hybridized with cDNA probes coding for IRF1 and β-actin or OAS and β-actin. A Northern blot representative of three independent experiments is shown.
Effects of ATRA, IFNα, and the combination of the two compounds on the levels of IRF1 and OAS mRNAs in NB4 cells. NB4 cells (4 × 105/mL) were treated for 4 days with medium alone (Medium) and medium containing ATRA (10−6 mol/L), IFNα (1,000 U/mL), or the combination of the two compounds. Total RNA was isolated and subjected (20 μg/lane) to Northern blot analysis. Filters were sequentially hybridized with cDNA probes coding for IRF1 and β-actin or OAS and β-actin. A Northern blot representative of three independent experiments is shown.
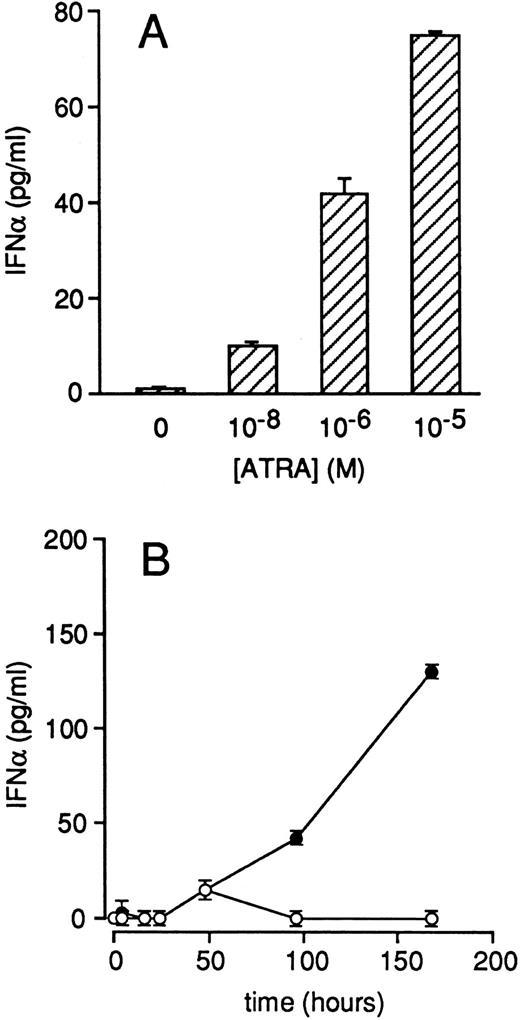
ATRA increases IFNα secretion by NB4 cells.IRF1 is not only an IFN-regulated gene but also a transacting factor that induces the transcriptional activity of the IFNα and IFNβ genes.37 Therefore, we evaluated whether ATRA treatment is capable of augmenting the amounts of IFNα secreted in the growth medium of NB4 cells. As illustrated in Fig 9A, NB4 cells grown in the presence of medium alone secrete amounts of IFNα, which are just above the limit of detection of the immunoassay employed. A significant and dose-dependent increase in IFNα secretion is observed after treatment of NB4 cells with ATRA for 4 days. Figure 9B shows that ATRA-dependent induction of IFNα secretion is a late event requiring at least 4 days to be significant. However, from the fourth day on, the accumulation of the cytokine in the medium is almost linear. In the same experimental conditions, we did not observe significant synthesis of IFNγ by NB4 cells.
Effects of ATRA on the secretion of IFNα in the growth medium of NB4 cells. NB4 cells (4 × 105/mL) were treated for 4 days with medium containing the indicated concentrations of ATRA (A) or with medium alone (○) and medium containing 10−6 mol/L ATRA (•) for the indicated amount of time (B). The concentration of IFNα released in the medium was measured with a specific ELISA. Each experimental value is the mean ± SD of three separate culture dishes.
Effects of ATRA on the secretion of IFNα in the growth medium of NB4 cells. NB4 cells (4 × 105/mL) were treated for 4 days with medium containing the indicated concentrations of ATRA (A) or with medium alone (○) and medium containing 10−6 mol/L ATRA (•) for the indicated amount of time (B). The concentration of IFNα released in the medium was measured with a specific ELISA. Each experimental value is the mean ± SD of three separate culture dishes.
DISCUSSION
In this report, we provide evidence that ATRA induces the expression of Stat1α, Stat2, and to a lesser extent of p48. The retinoid does not cause any change in the levels of Jak1, Jak2, and Tyk2 tyrosine kinases. These effects have been documented in NB4, an ATRA-sensitive cell line that recapitulates most of the characteristics of APL blasts, including granulocytic maturation upon challenge with the retinoid.18 Induction of Stat1α is also observed in a subgroup of freshly isolated APL cells, as well as in HL-60 and U937, two ATRA responsive myeloid cell lines which do not present the APL-specific t(15:17) balanced chromosomal translocation. Thus, our results indicate that ATRA-sensitivity is necessary and sufficient to permit Stat1α induction by the retinoid. In addition, they show that the phenomenon is not necessarily connected with the APL-specific expression of PML-RAR, the product of the t(15:17) chromosomal rearrangement. Interestingly, the subgroup of APL patients that do not respond to ATRA with a significant elevation of Stat1α protein responds to G-CSF, another granulocyte cyto-differentiating agent. The modulation of Stat1α by G-CSF is enhanced by cotreatment with ATRA. The reasons for this behavior are as yet unknown. Nevertheless, it is relevant to remind that G-CSF has been shown to potentiate granulocytic differentiation triggered by ATRA on APL cells23 and this effect is more evident in some patients than in others (M.G. and E.G., unpublished observations, May 1996). Treatment of NB4 cells for 4 days with optimal concentrations of other granulocytic cyto-differentiating agents like dimethylsulfoxide (1.3% vol/vol) and sodium butyrate (0.4 mmol/L) does not alter the level of expression of Stat1α protein (I.F., M.T., and E.G., unpublished observations, July 1996).8 This suggests that Stat1α induction is not necessarily associated with granulocytic maturation, although the phenomenon may be involved in the differentiation program triggered by ATRA.
Although we cannot rule out translational effects, induction of Stat1α protein is primarily the consequence of an increase in the levels of the respective transcript, as shown by the Northern blot experiments presented. The process is likely to be transcriptional in nature, as suggested by the rapid kinetics of mRNA accumulation and by its sensitivity to actinomycin D inhibition (data not shown). In addition, the process does not require de novo protein synthesis, as indicated by insensitivity to cycloheximide (data not shown). Thus, the Stat1 gene seems to be a direct target for the activity of ATRA and it will be important to establish whether its regulatory regions contain a canonical consensus sequence for the binding of RARs.
In NB4 cells, following treatment with ATRA, induction of Stat1α is preceeded by a rapid augmentation in the tyrosine phosphorylation of the protein. Although NB4 treatment with the retinoid has been reported to cause increased tyrosine kinase activity,38 the specific tyrosine kinases involved in the ATRA-dependent phosphorylation of Stat1α are as yet unidentified. Moreover, it is yet to be established whether tyrosine phosphorylation is dependent on the activation of nuclear retinoic acid receptors or it is a receptor-independent phenomenon. As to the possible tyrosine kinases involved, Jak1, Jak2, and Tyk2, which are believed to directly phosphorylate Stat proteins following activation of the IFN receptors by their respective ligands,11 do not seem to be targets for ATRA. In fact, following treatment with ATRA (10−6 mol/L) for 5, 15, 30, and 60 minutes, NB4 cells do not show any evidence of retinoid-dependent Jak1, Jak2, or Tyk2 tyrosine phosphorylation (I.F., M.T., and E.G., unpublished observations, July 1996).8 As to retinoic acid receptor(s) dependence, a mechanism involving both a receptor-dependent and a receptor-independent component is possible. A receptor-independent component may be active during the early phases of the process of Stat1α activation. In fact, increased tyrosine phosphorylation of Stat1α is already observed at 15 minutes, which is probably too short a period of time to permit retinoic acid receptor-regulated gene expression. This contention is further supported by the fact that increased phosphorylation of Stat1α is also observed in HL-60 cells and this effect is maximal at 5 minutes. By contrast, in NB4 cells, a receptor-dependent component may be involved in maintaining increased tyrosine phosphorylation of Stat1α, which is long-lasting and still observed at 24 hours. This idea is consistent with the data obtained in COS-7 cells (see Fig 6), where ATRA-dependent Stat1α activation of an IFN-responsive reporter gene requires fortification of the system with RARα.
In NB4 cells, the consequence of Stat1α induction and increased tyrosine phosphorylation of the protein by ATRA is augmented formation of Stat1-containing nuclear complexes capable of binding to ISRE and pIRE sequences. It is worthwhile noticing that ISRE binding complexes, which consist of Stat1/Stat2 heterodimers, are rapidly induced by ATRA despite the fact that the retinoid augments the basal tyrosine phosphorylation level of Stat1α but not that of Stat2. This suggests that the activation state of Stat1α is limiting during the early phases of the assembly of ISRE-binding complexes following treatment of NB4 cells with ATRA. ATRA-dependent induction of ISRE and pIRE binding complexes can lead to the transcription of genes that are normally under the control of IFNs, as supported by the transient transfection experiments performed in COS-7 fibroblasts. In this cell system, we observed that Stat1α-dependent activation of a reporter gene containing functional Stat1α binding sites by ATRA requires the presence of RARα. Interestingly, in the same experimental context, RARα cannot be effectively substituted for by PML-RAR. Although we cannot exclude that PML-RAR-dependent Stat1/ATRA interaction may be seen in different experimental conditions and although caution should be exercised in extrapolating this finding to the APL situation, our data suggest that the residual amount of RARα may play a major role in the ATRA-dependent transcriptional activation of Stat1 in NB4 cells. Because the two retinoic acid receptors differ at their amino-terminus, it is possible that the functional domains important for Stat1α activation reside in the portion of RARα which is absent in PML-RAR.
The potential for ATRA to induce the expression of IFN-responsive genes by a mechanism that bypasses the membrane receptor of the cytokine is further suggested by upregulation of IRF1 and OAS gene expression following treatment of NB4 cells with the retinoid. IRF1 and OAS are two IFN-stimulated genes that contain functional regulatory sequences capable of binding Stat1α in the form of a homodimer and as an ISGF3 complex, respectively.20,39 The effect of ATRA on these two genes is transcriptional in nature being blocked by RNA polymerase II inhibitors (unpublished results, May 1996).8 Since neither the IRF1, nor the OAS gene are known to contain ATRA-responsive regulatory elements, it is unlikely that ATRA-dependent upregulation is directly mediated by the classical pathway involving nuclear retinoic acid receptors. It is more likely that Stat1α activation and/or induction caused by ATRA are instrumental for the increase in the transcriptional rate of the IRF1 and OAS genes. This contention is further supported by the fact that upregulation of IRF1 and OAS transcripts by ATRA and IFNα seems to proceed through common biochemical pathways, as suggested by the lack of evident additive or synergistic effects between the retinoid and the cytokine. Although IRF1 mRNA induction is a very early event, appearance of the OAS transcript is a rather late phenomenon. Thus, only a relatively fast mechanism like tyrosine phosphorylation and activation of Stat1α may explain the ATRA-dependent upregulation of IRF1 gene expression. By contrast, both induction and activation of Stat1α by ATRA may be implicated in the upregulation of OAS mRNAs. It is interesting to notice that ATRA causes an induction not only of two IFN-responsive genes but also increased synthesis and secretion of IFNα itself. This raises the question as to whether Stat1α activation and induction, as well as upregulation of IRF1 and OAS gene expression are secondary to an IFNα autocrine effect. Given the slow kinetics of OAS mRNA accumulation, this is the only gene for which ATRA-induced IFN secretion may be important.
In conclusion, the data reported in this paper indicate that ATRA is capable of modulating the levels and the state of activation of some of the components of the IFN intracellular signaling pathways in APL and other myeloid leukemic cells. In addition, they indicate that ATRA can bypass IFN/IFN-receptor interactions and induce the expression of IFN-regulated genes. Our data add a further level of complexity to the cross-talk between retinoids and IFN-dependent signaling pathways already observed in various cellular contexts.16,17,40 Furthermore, since Stat1α phosphorylation and activation is coupled to receptor of cytokines and growth factors other than IFNs,11 it is possible that ATRA mimicks or modulates the biological activity of these compounds as well. Finally our results suggest that Stat1α induction or activation may be involved in the cyto-differentiating or antiproliferative effect triggered by ATRA in APL cells. With respect to the last point, Stat1 has been recently reported to induce p21,41 an inhibitor of cyclin-dependent kinases, which are proteins allowing progression along the cell cycle. Hence, the protein may represent a target of therapeutic intervention not only in the cyto-differentiating but also in the antiproliferative treatment of APL and other myeloid types of leukemia. Studies are in progress to elucidate all these points.
ACKNOWLEDGMENT
We thank Dr M. Lanotte (Unite' INSERM 301, “Genetique cellulaire et moleculaire des leucemies,” Hopital St Louis, Paris, France) for supplying us with the NB4 acute promyelocytic cell line. We are grateful to Dr P.G. Pelicci (Policlinico Monteluce, Universita' di Perugia, Italy ) for the kind gift of the plasmids pSG5-RARα and pSG5-PML-RAR. We thank Dr A. Weisz (University of Naples, Italy) for providing us with the reporter construct pnlsLACZ. We are also grateful to Prof S. Garattini and Dr M. Salmona for the critical reading of the manuscript and to Dr A. Sica for the helpful discussion on many aspects of the experiments contained in this report. M.L.C is a recipient of a fellowship from “La via di Natale.”
Supported in part by Grants from the Consiglio Nazionale delle Ricerche (CNR), Progetto Finalizzato “Ingegneria Genetica” and from the Associazione per la Ricerca contro il Cancro (AIRC).
Address reprint requests to Enrico Garattini, MD, Molecular Biology Unit, Centro Daniela e Catullo Borgomaainerio, Istituto di Ricerche Farmacologiche “Mario Negri,” via Eritrea 62, 20157 Milan, Italy.

![Fig. 1. Effects of ATRA and IFNs on the expression of Stat, p48, Jak 1, Jak 2, and Tyk 2 proteins in NB4 and HeLa cells. NB4 (4 ×105/mL) or HeLa (a confluent 25 cm2 flask) cells were treated for 4 days with medium alone (Medium) and medium containing ATRA (10−6 mol/L), IFNα (1,000 U/mL), and IFNγ (1,000 U/mL) as indicated. Cells were harvested, lysed, and subjected to Western blot analysis, using antibodies specific for Stat 1α, Stat 2, Stat 3, and Stat 4 (A), p48 (B), Jak1, Jak 2, and Tyk 2 (C). For Stat 3 and Jak 1, 100 μg of protein/lane were used, whereas for all the other samples, 10 mg of protein/lane were used. In (B), the apparent molecular weight of standard proteins (carbonic anhydrase [28 kD], ovalbumin [43 kD], bovine serum albumin [69 kD], and phosphorylase B [105 kD]) is indicated on the right. The apparent molecular weight of Stat proteins, Jak 1, Jak 2, and Tyk 2 is indicated on the right of (A) and (C). Western blots representative of at least two independent experiments are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/3/10.1182_blood.v89.3.1001/8/m_bl_0032f1.jpeg?Expires=1763524633&Signature=x2bSoDXhFXcbasXDkkXMQ6Nrl~HRnGENtDop8DVwLAvOu6Ah7BpXtbtIdYohbDQGcX-2VGBeOYr6Wt9qnJxB4YPppAMIaZ6ea2JJLbQxfSQHEFmmbKC0FbNsrEcg3aN~FthnrCkHNkrouBTLbSfpSmVQtf~NUm7g6EshbqjLs3zZVt-aEYWTxlKIwrj8tDXD8fOtfA22iMEwgmVPGYWVdDz690jCpH7N4qq7IjHs9ACwaG6tS1-VS386Kf1Ea9X5KSxoC-qAq8YksW0JBjemFicZ7YzTJUw8sKUoBg59jApemeDyOXqwi2gunXcXuaqga1Ft-MYD-kgeVkIGcvDp8w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. Dose- and time-dependent induction of Stat 1α protein and Stat1 mRNA by ATRA in NB4 cells. (A) NB4 cells (4 × 105/mL) were treated for 4 days with medium alone (medium) and medium containing the indicated concentrations of ATRA. Cells were harvested, lysed and the cell extracts (10 μg of protein/lane) subjected to Western blot analysis, using antibodies specific for Stat 1α. (carbonic anhydrase [28 kD], ovalbumin [43 kD], bovine serum albumin [69 kD], phosphorylase B [105 kD], and myosin heavy chain [215 kD]). A Western blot representative of two independent experiments is shown. (B) NB4 cells (4 × 105/mL) were treated for the indicated lengths of time with 10−6 mol/L ATRA. Western blot analysis was conducted as described in (A). The apparent molecular weight of standard proteins (carbonic anhydrase [28 kD], ovalbumin [43 kD], bovine serum albumin [69 kD], phosphorylase B [105 kD], and myosin heavy chain [215 kD]) is indicated on the right. A Western blot representative of three independent experiments is shown. (C) NB4 cells (4 × 105/mL) were treated for the indicated lengths of time with 10−6 mol/L ATRA. Total RNA was isolated and subjected (20 μg/lane) to Northern blot analysis. Filters were sequentially hybridized with cDNA probes coding for Stat 1α and β-actin. A Northern blot representative of two independent experiments is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/3/10.1182_blood.v89.3.1001/8/m_bl_0032f2.jpeg?Expires=1763524633&Signature=YfIztgXW7Z0HHsHA-20r3KL~0JPEWoBZUiA0c~-jM4YL~zQqRQg0kZwjBoGjf6KIoNROChRqeNalpTmC7CwTPOwVK7Exq1iMT3zchnzu0d1pQSuNTpPFNXaSuQQzXe7nO5aTZ7uOr5iksdDDGkLc5FQ9tH5fiwCh4LdV1JtRl7ZRor8ccsoZ~GPA-nqpdbWUf9NkeRzS2Mu7QdwtLyv~v02y79NYxiPP7a0lUD4Xawr8G0FrG7mfZajjBtGb3RjFz4tic0Eyjwa00hAxqjJzK0YLJLllrhiTYQ2MwijuiKyljM2ClI7HCqJinuLbiFy~TcRKR1NTQLCK~3YQstoBrg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal