Abstract
Acute promyelocytic leukemia (APML) is characterized by abnormal myeloid development, resulting an accumulation of leukemic promyelocytes that are often highly sensitive to retinoic acid. A balanced t(15; 17) (q22; q21) reciprocal chromosomal translocation is found in approximately 90% of APML patients; this translocation fuses the PML gene on chromosome 15 to the retinoic acid receptor α (RARα) gene on chromosome 17, creating two novel fusion genes, PML-RARα and RARα-PML. The PML-RARα fusion gene product, which is expressed in virtually all patients with t(15; 17), is thought to play a direct role in the pathogenesis of APML. To determine whether PML-RARα is sufficient to cause APML in an animal model, we used the promyelocyte-specific targeting sequences of the human cathepsin G (hCG) gene to direct the expression of a PML-RARα cDNA to the early myeloid cells of transgenic mice. Mice expressing the hCG–PML-RARα transgene were found to have altered myeloid development that was characterized by increased percentages of immature and mature myeloid cells in the peripheral blood, bone marrow, and spleen. In addition, approximately 30% of transgene-expressing mice eventually developed acute myeloid leukemia after a long latent period. The splenic promyelocytes of mice with both the nonleukemic and leukemic phenotypes responded to all-trans retinoic acid (ATRA) treatment, which caused apoptosis of myeloid precursors. Although low-level expression of the hCG–PML-RARα transgene is not sufficient to directly cause acute myeloid leukemia in mice, its expression alters myeloid development, resulting in an accumulation of myeloid precursors that may be susceptible to cooperative transforming events.
ASPECIFIC t(15; 17) chromosomal translocation is detected in most cases of acute promyelocytic leukemia (APML) and is often the only cytogenetic abnormality observed.1,2 The breakpoints of this translocation have been localized to the PML gene on chromosome 15 and the RARα gene on chromosome 17.3-8 Transcripts corresponding to the reciprocal fusion genes generated by this translocation (PML-RARα and RARα-PML) have been detected in the leukemic blasts of APML patients.4,5,8-10 Expression of PML-RARα has been detected in virtually all APML patients with the t(15; 17) translocation; 70% to 80% of these patients also express RARα-PML.4,5,8-10 In addition, expression of PML-RARα has been shown to produce the phenotypic features of APML blasts in several myeloid cell lines.11 12 Thus, it has been suggested that PML-RARα may be directly involved in the pathogenesis of APML.
The RARα gene encodes a hormone-inducible nuclear receptor that has been shown to be involved in myeloid development.13-16 The PML-RARα fusion gene product retains the critical functional domains of the normal RARα protein, including the domains for DNA binding, dimerization, and retinoic acid binding.4-6,8 Therefore, it has been suggested that PML-RARα may promote leukemogenesis by interfering with normal RARα signaling pathways. In transient transfection assays, PML-RARα has been shown to interfere with the activation of retinoic acid-responsive reporter genes by RARα.5,17 This effect is presumably mediated at the level of DNA binding. PML-RARα homodimers or PML-RARα/RXR heterodimers, both of which form in vitro, may directly compete with RARα/RXR heterodimers for binding to retinoic acid response elements (RAREs); alternatively, PML-RARα may act indirectly by sequestering RXRs, thus preventing RARα/RXR heterodimer formation.18 The rearrangement of the RARα gene associated with this disease is intriguing considering that APML blasts are sensitive to all-trans retinoic acid (ATRA) in vitro and in vivo.19-23 In addition, ATRA is capable of inducing complete remissions in most APML patients with the t(15; 17).19,20,22,23 Furthermore, the PML-RARα fusion protein itself is likely to be responsible for ATRA sensitivity.11
Alternatively, PML-RARα may promote leukemogenesis by interfering with normal PML function. Although the exact cellular functions of this protein are not yet understood, PML has been characterized as a tumor suppressor; disruption of this tumor suppressor by the t(15,17) translocation may be required to produce APML.24,25 PML appears to be expressed in a variety of types,26 but expression of PML-RARα under the control of PML regulatory elements is associated only with myeloid transformation. Analysis of the predicted amino acid sequence of PML showed three N-terminal cysteine/histidine-rich regions, the first of which has high homology to the RING domain characteristic of a novel family of nuclear proteins, and a putative coiled-coil domain in the C-terminus that may mediate dimerization.3-8 In APML blasts, the normal subcellular localization of PML (in discrete nuclear structures known as PML oncogenic domains [PODs]) is disrupted by the expression of PML-RARα.27-31 In fact, a direct interaction between the fusion protein and PML has been suggested to be the cause of PML delocalization from PODs.28 Remarkably, ATRA treatment of APML blasts causes PML to be restored to its normal POD structures27-29,31; however, it is not yet clear whether disruption of PML localization from PODs is directly important for the pathogenesis of APML.
Because APML is associated with overabundant promyelocytes, a progenitor cell with a committed phenotype may be the target for transformation.32 The promyelocyte stage of development is characterized by the synthesis of the primary (azurophil) granules; the enzymes that are packaged within these granules are synthesized during this stage of development. We have shown that the gene encoding one of these enzymes, cathepsin G (CG), is expressed exclusively in promyelocytes.33,34 We further showed that all of the sequences required for the early myeloid-specific expression of the human CG (hCG) gene in transgenic mice are present within a 6-kb genomic fragment.35 Using these sequences, we wanted to determine whether targeted expression of PML-RARα in the early myeloid cells of transgenic mice would recapitulate the APML disease phenotype. In this study, we have determined that mice expressing PML-RARα have altered myeloid development; however, low levels of PML-RARα expression are not sufficient to directly cause myeloid leukemia. Acute myeloid leukemia does develop after a long latent period in many mice, suggesting that a second hit is required for the development of APML.
MATERIALS AND METHODS
Construction of the transgene.The neo cassette was removed from the −76 hCG-neo plasmid36 by digestion with HindIII. A synthetic polylinker (60 bp in length) designed with HindIII ends was then subcloned downstream from the CG promoter. The 3.5-kb hCG gene was generated by polymerase chain reaction (PCR) with 5′ Sph I and 3′ BamHI ends for directional cloning into the polylinker. Approximately 1 kb of sequence from each end of the PCR-generated hCG gene was verified using the Sequenase kit (US Biochemical, Cleveland, OH). The 2.4-kb 5′ flank of the hCG gene was subcloned into the EcoRI site at −76 of the hCG promoter. The PML-RARα cDNA was then subcloned as a blunt-ended fragment into the Not I site of the polylinker. The completed 9-kb transgene was released from pUC 9 sequences by codigestion with Sal I and BamHI. Transgenic mice were then generated using this purified fragment as previously described.35
RNA analysis by reverse transcriptase-PCR (RT-PCR).Total cellular RNA was purified from bone marrow cells isolated from the femurs of mice as described.37 One microgram of RNA was then analyzed by RT-PCR (Perkin-Elmer, Norwalk, CT) according to the manufacturer's instructions. One-fourth of the total reaction volume was then subjected to agarose gel electrophoresis. The fractionated PCR products were transferred to Hybond (Amersham, Arlington Heights, IL), which was then hybridized with a random-primer–labeled probe, washed, and exposed to x-ray film. Exposure time for the blot hybridized with the hCG exon 2 probe was overnight at 23°C. The blot hybridized with the murine CG (mCG) exon 4 probe was exposed for 5 minutes at 23°C.
RNA analysis by S1 protection.Portions of spleens were homogenized and total cellular RNA was prepared as previously described.37 End-labeled probes for mCG and mβ2-microglobulin were prepared and used in S1 nuclease protection assays as described.34 The murine β-actin probe was labeled at a Bgl II site in the predicted exon 2; correctly spliced mRNA protects a 135-nt fragment. The hCG–PML-RARα probe was labeled at an Avr II site in the PML portion of the transgene. Correctly spliced human PML mRNA protects a probe fragment of 275 nt from S1 digestion, whereas correctly initiated hCG–PML-RARα mRNA protects a probe fragment of 320 nt.
Assays for hematopoietic colony formation.Single-cell suspensions of isolated spleens were prepared by homogenization. Nucleated cells (1 × 105) were dispersed in Methocult 3430 complete media (Stem Cell Technologies, Inc, Vancouver, British Columbia, Canada) and plated in quadruplicate. Total colony numbers were quantified after 7 days of growth at 37°C in a humidified incubator with 5% CO2 .
Morphologic and histochemical analyses.Peripheral blood smears or bone marrow cells prepared as previously described35 were stained with modified Wright's Giemsa (Sigma, St Louis, MO). Six-micron–thick sections of a frozen spleen from a nontransgenic or a transgenic mouse were histochemically stained with the leukocyte peroxidase (myeloperoxidase) kit according to manufacturer's instructions (Sigma).
Intact spleens and livers from either nontransgenic or transgenic mice were isolated, fixed in 10% buffered formalin (Baxter Diagnostics, Deerfield, IL), and subsequently embedded in paraffin. Six-micron–thick sections of these organs were then deparaffinized and stained with hematoxylin and eosin.
Kaplan-Meyer analysis.Total numbers of nontransgenic or transgenic mice from founder lines 83, 135, and 137 that had postmortem pathologic evidence of myeloid leukemia (defined as massive splenomegaly, leukocytosis, organ infiltration with immature myeloid cells, and clinical illness) were recorded. A Kaplin-Meyer plot was generated using Sigma Plot 3.0 for Windows software (Jandel Scientific, San Rafael, CA).
Tumor formation in SCID mice.A total of 4 or 5 × 107 splenocytes isolated from transgenic mice with the leukemic phenotype (or from nontransgenic, wild-type littermates) were injected into C3H SCID mice intraperitoneally. These mice were then closely monitored for signs of illness, which occurred at 5 to 6 weeks postinjection in recipients of leukemic splenocytes. At this time, necropsies were performed. Abnormal tissues were processed for histopathologic analysis as described above. Recipients of wild-type splenocytes, although showing no signs of illness, were killed and analyzed in parallel.
ATRA treatment of splenocytes.Total splenocytes were cultured in RPMI 1640 + 20% fetal calf serum (FCS) + 10 μ/mL penicillin G and 10 μg/mL streptomycin sulfate with or without 1 μmol/L ATRA (Sigma) for 48 hours. At 0, 24, and 48 hours, approximately 2 × 107 cells were harvested. Total cellular RNA was prepared as described37; end-labeled probes were prepared and S1 nuclease protection assays were performed as previously described.34
RESULTS
Production of transgenic mice expressing PML-RARα in early myeloid cells.We constructed a transgene containing a PML-RARα cDNA under the control of sequences that regulate the promyelocyte-specific expression of the hCG gene. As mentioned above, these sequences are capable of directing the expression of hCG to the early myeloid cells of transgenic mice.35 The PML-RARα cDNA was subcloned into the 5′ untranslated region of the 6-kb hCG genomic fragment (Fig 1A). Expression of the PML-RARα cDNA cassette used in this construct was previously shown to produce phenotypic features of APML blasts when expressed in U937 cells.11 During the construction of this transgene, the initiator ATG of the hCG gene was mutated to TTG to prevent initiation of hCG protein translation. Eight stable founder lines of mice were generated, containing 1 to 15 copies of the hCG–PML-RARα transgene.
(A) Diagram of the hCG–PML-RARα transgene. Transcription begins at the arrow, the transcriptional start site of the hCG gene. The hCG sequences extend upstream approximately 2.4 kb from the start site. The solid black boxes represent the five exons of the hCG gene; the polyadenylation signal is provided by the hCG gene. Sizes of DNA fragments used in constructing the transgene are denoted below the diagram. (B) RT-PCR analysis of bone marrow RNA. The diagram illustrates the portion of the transgene-specific mRNA that is amplified. The sequences of the forward and reverse primers are also shown. PCR products that hybridize to an hCG exon 2-specific probe shown in the upper autoradiogram indicate the founder lines that express the transgene. In the lower autoradiogram, RNA quality is controlled using primers specific for endogenous mCG exon 4 and a probe specific for detecting these sequences. The RNA source and transgene status is indicated above each lane (founder line/transgene copy number: 67/5, 71/1, 72/15, 83/2, 126/1, 135/4, 137/1, 148/1).
(A) Diagram of the hCG–PML-RARα transgene. Transcription begins at the arrow, the transcriptional start site of the hCG gene. The hCG sequences extend upstream approximately 2.4 kb from the start site. The solid black boxes represent the five exons of the hCG gene; the polyadenylation signal is provided by the hCG gene. Sizes of DNA fragments used in constructing the transgene are denoted below the diagram. (B) RT-PCR analysis of bone marrow RNA. The diagram illustrates the portion of the transgene-specific mRNA that is amplified. The sequences of the forward and reverse primers are also shown. PCR products that hybridize to an hCG exon 2-specific probe shown in the upper autoradiogram indicate the founder lines that express the transgene. In the lower autoradiogram, RNA quality is controlled using primers specific for endogenous mCG exon 4 and a probe specific for detecting these sequences. The RNA source and transgene status is indicated above each lane (founder line/transgene copy number: 67/5, 71/1, 72/15, 83/2, 126/1, 135/4, 137/1, 148/1).
To identify the founder lines expressing the hCG–PML-RARα transgene, total cellular RNA purified from the bone marrow cells of F1 progeny from each founder line was analyzed by RT-PCR followed by Southern blotting (Fig 1B). The primers used in the PCR amplification were designed based on the sequences at the exon junctions of the correctly processed transgene mRNA to eliminate the amplification of unspliced RNA or contaminating DNA in the reaction mix (see diagram, Fig 1B). The blot was hybridized with a probe specific for the amplified product, which included sequences from correctly spliced hCG exon 2 of the chimeric transgene mRNA; we expected this portion of the transgene to be transcribed and spliced. RNA isolated from the human U937 myeloid leukemic cell line, which expresses high levels of CG mRNA,33 provides a positive control for this assay (Fig 1B, lane 1). No signal is detected using bone marrow RNA isolated from a nontransgenic animal (Fig 1B, lane 7), confirming that this assay is specific for transgene expression. Measurable levels of transgene expression are detected in six of the eight founder lines (Fig 1B). The hybridizing signal is not due to contaminating DNA, because lines 67 and 126 (Fig 1B, lanes 3 and 9, respectively) are transgene-positive but do not produce hybridizing RT-PCR products. The results from this assay are qualitative and identify transgene-expressing founder lines; however, detection is in the linear range, because most of the hybridizing signals vary in intensity. Transgene-specific transcripts were at the limits of detection by S1 nuclease protection analysis (with a sensitivity of ∼5 copies of mRNA/cell), indicating that expression levels are very low (data not shown). Mice generated from these founder lines had normal growth, development, and fertility; however, low-level expression of this transgene was sufficient to alter myeloid development in these mice.
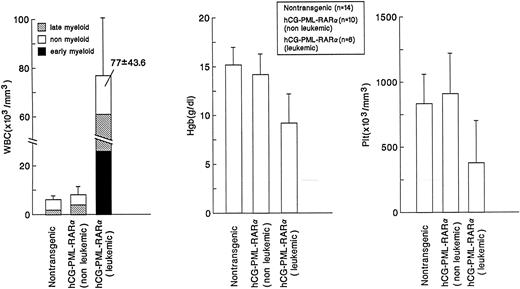
Altered myeloid development in transgene-expressing mice.Virtually all mice that expressed the transgene were found to have an expansion of myeloid cells in their bone marrows and spleens; this phenotype was not dependent on age, sex, or site of transgene integration (ie, the phenotype was observed in 4 independent founder lines). These nonleukemic mice had nearly normal peripheral blood counts, with the exception of a small, but statistically significant, increase in the percentage of morphologically mature myeloid cells (Fig 2). The bone marrow of these mice was packed with myeloid cells at all stages of maturation (Fig 3 and Fig 4A v B); this defect probably caused the extramedullary hematopoiesis found in the interfollicular zones of the spleens (red pulp) of these mice. The expanded red pulp of transgenic spleens contained a substantial increase in the number of cells staining for the myeloid-specific marker myeloperoxidase (Fig 4C v D). Although the red pulp and size of these spleens was expanded, their follicular architecture was preserved (Fig 4D). The presence of increased numbers of hematopoietic progenitors within the spleens of these mice was confirmed using a methylcellulose colony-forming assay: transgenic spleens yielded 86 ± 111 total colonies/105 total spleen cells versus 7 ± 6 colonies/105 total spleen cells in their wild-type littermates (n = 4). The distribution of myeloid and erythroid colonies and colony size were not different between the wild-type and transgenic mice.
Comparison of peripheral blood counts. Values representing the mean ± SD (indicated by error bars) of total white blood cell counts, hemoglobin content, and total platelet counts are shown for nontransgenic or hCG–PML-RARα transgenic mice with nonleukemic or leukemic phenotypes. Statistically significant differences were observed for the percentage of mature myeloid cells in nontransgenic mice versus hCG–PML-RARα (nonleukemic and leukemic) mice (P < .001), the decrease in hemoglobin for hCG–PML-RARα (leukemic) mice (P < .001), and the decrease in platelet counts for hCG–PML-RARα (leukemic) mice (P < .01).
Comparison of peripheral blood counts. Values representing the mean ± SD (indicated by error bars) of total white blood cell counts, hemoglobin content, and total platelet counts are shown for nontransgenic or hCG–PML-RARα transgenic mice with nonleukemic or leukemic phenotypes. Statistically significant differences were observed for the percentage of mature myeloid cells in nontransgenic mice versus hCG–PML-RARα (nonleukemic and leukemic) mice (P < .001), the decrease in hemoglobin for hCG–PML-RARα (leukemic) mice (P < .001), and the decrease in platelet counts for hCG–PML-RARα (leukemic) mice (P < .01).
Comparison of bone marrow differentials. Differentials of 200 cells were performed by two independent observers. Values represent the mean ± SD (indicated by the error bars). hCG–PML-RARα mice have significant (P < .01) increases in early stage myeloid cells (promyelocytes and myelocytes).
Comparison of bone marrow differentials. Differentials of 200 cells were performed by two independent observers. Values represent the mean ± SD (indicated by the error bars). hCG–PML-RARα mice have significant (P < .01) increases in early stage myeloid cells (promyelocytes and myelocytes).
Phenotype of nonleukemic hCG–PML-RARα transgenic mice. (A) Photograph of nontransgenic bone marrow cells stained with Wright's Giemsa. (B) Photograph of bone marrow cells from a nonleukemic, transgene-expressing mouse from founder line 83, stained with Wright's Giemsa. Note that the marrow of this mouse contains predominantly myeloid cells at all stages of differentiation. (C and D) Photographs of spleen sections from (C) a nontransgenic mouse or (D) a nonleukemic, transgene-expressing mouse after histochemical staining for myeloperoxidase activity. A marked increase in cells staining for MPO activity (black stain) is observed in the transgenic spleen.
Phenotype of nonleukemic hCG–PML-RARα transgenic mice. (A) Photograph of nontransgenic bone marrow cells stained with Wright's Giemsa. (B) Photograph of bone marrow cells from a nonleukemic, transgene-expressing mouse from founder line 83, stained with Wright's Giemsa. Note that the marrow of this mouse contains predominantly myeloid cells at all stages of differentiation. (C and D) Photographs of spleen sections from (C) a nontransgenic mouse or (D) a nonleukemic, transgene-expressing mouse after histochemical staining for myeloperoxidase activity. A marked increase in cells staining for MPO activity (black stain) is observed in the transgenic spleen.
We next wanted to determine whether there was an increase in the fraction of promyelocytes within transgenic spleens, using levels of endogenous mCG mRNA as a specific marker. Because expression of CG is restricted to promyelocytes, increases in the levels of mCG mRNA in the spleens of these mice should reflect an increase in the fraction of promyelocytes. RNA isolated from the spleens of both nontransgenic and transgenic, nonleukemic animals from each of the eight founder lines was analyzed using S1 nuclease protection assays (Fig 5). The murine β-actin probe assures that the quality and content of the RNAs are equivalent (Fig 5). PML transcripts from human K562 cell RNA are protected by the PML portion of the transgene-specific probe, showing that this probe can detect basal levels of human PML expression in hematopoietic cells (Fig 5, lane 2). hCG–PML-RARα transgene-specific transcripts are not detectable in any of the spleens using this assay, again reflecting the very low levels of transgene expression. Because there are so few promyelocytes in normal mouse spleens, endogenous mCG transcripts are virtually undetectable (Fig 5, lane 7). However, increased levels of endogenous mCG mRNA are observed in founder lines 83, 72, 148, 137, and 135 (Fig 5, lanes 4, 6, 8, 10, and 11, respectively), suggesting that the spleens of transgene-expressing mice contain an increased percentage of promyelocytes.
S1 nuclease protection analysis of spleen RNAs is isolated from hCG–PML-RARα transgenic mice. Molecular size markers are indicated on the left. The expected positions of probe fragments protected by hCG–PML-RARα, mCG, and mβ actin mRNAs are indicated by the arrows on the right. Founder line and transgene status for each RNA sample are indicated at the bottom. Increases in endogenous mCG mRNA levels are detected in founder lines 83, 72, 148, 137, and 135 (lanes 4, 6, 8, 10, and 11, respectively).
S1 nuclease protection analysis of spleen RNAs is isolated from hCG–PML-RARα transgenic mice. Molecular size markers are indicated on the left. The expected positions of probe fragments protected by hCG–PML-RARα, mCG, and mβ actin mRNAs are indicated by the arrows on the right. Founder line and transgene status for each RNA sample are indicated at the bottom. Increases in endogenous mCG mRNA levels are detected in founder lines 83, 72, 148, 137, and 135 (lanes 4, 6, 8, 10, and 11, respectively).
Development of acute leukemia after a long latent period.Several transgenic mice eventually developed a severe leukemic phenotype. As a result, three founder lines (83, 135, and 137) were expanded for a tumor watch. A Kaplan-Meyer plot depicts the frequency of leukemia-free survival of mice from these three transgene-expressing lines (Fig 6). Approximately 30% of the transgenic mice from these founder lines have developed the leukemic phenotype, but many mice greater than 6 months of age are still at risk (Fig 6). These mice generally presented with severe leukocytosis usually associated with anemia and/or thrombocytopenia (Fig 2). Massive splenomegaly was consistently observed in these animals (Fig 7A). In addition, the peripheral blood (Fig 2) and the bone marrow (Figs 3 and 7B) of these mice contained high percentages of immature myeloid cells with large numbers of azurophil granules, suggesting a massive expansion of the promyelocyte pool. However, myeloid cells with more differentiated phenotypes were noted in both the peripheral blood and the bone marrow (Figs 2 and 3). When compared with nontransgenic spleens (Fig 5C), the splenic architecture of the leukemic mice was completely disrupted by massive expansion of immature myeloid cells (Fig 5D). Infiltration of myeloid precursors into other organs was also commonly noted in these mice. Histopathologic analysis of an enlarged liver isolated from a representative affected mouse showed an accumulation of myeloid precursors (Fig 5E v F ). None of the mice presented with a bleeding diathesis and disseminated intravascular coagulation (DIC) was not apparent at autopsy in any of the mice with the leukemic phenotype. However, coagulation parameters were not measured in any of the mice, and mice were generally killed at the first signs of clinical illness.
Kaplan-Meyer plot displaying the probability of leukemia-free survival in mice derived from founder lines 83, 135, and 137.
Kaplan-Meyer plot displaying the probability of leukemia-free survival in mice derived from founder lines 83, 135, and 137.
Phenotype of leukemic, hCG–PML-RARα transgenic mice. (A) Spleens from a nontransgenic littermate (left) and an hCG–PML-RARα transgenic mouse (no. 135-448) with leukemia (right). (B) Bone marrow cells from a leukemic mouse (no. 137-1106) stained with modified Wright's Giemsa. This marrow from the leukemic mouse consists predominantly of very immature myeloid cells that contain abundant azurophil granules (black arrowheads). (C and D) Spleen sections from a nontransgenic mouse (C) or a transgenic mouse (no. 135-448) (D) with leukemia, stained with hematoxylin and eosin. Note the effacement of follicular structures by the leukemic infiltrate. (E and F ) Liver sections from a nontransgenic mouse (E) or mouse no. 135-448 (F ) stained with hematoxylin and eosin. Note the infiltration of the liver with immature myeloid cells (white arrowheads).
Phenotype of leukemic, hCG–PML-RARα transgenic mice. (A) Spleens from a nontransgenic littermate (left) and an hCG–PML-RARα transgenic mouse (no. 135-448) with leukemia (right). (B) Bone marrow cells from a leukemic mouse (no. 137-1106) stained with modified Wright's Giemsa. This marrow from the leukemic mouse consists predominantly of very immature myeloid cells that contain abundant azurophil granules (black arrowheads). (C and D) Spleen sections from a nontransgenic mouse (C) or a transgenic mouse (no. 135-448) (D) with leukemia, stained with hematoxylin and eosin. Note the effacement of follicular structures by the leukemic infiltrate. (E and F ) Liver sections from a nontransgenic mouse (E) or mouse no. 135-448 (F ) stained with hematoxylin and eosin. Note the infiltration of the liver with immature myeloid cells (white arrowheads).
Expression of the hCG–PML-RARα transgene was routinely detected in leukemic mice. Using the RT-PCR strategy described in Fig 1B, leukemic mouse no. 137-1106 was found to express the transgene in the bone marrow and spleen (data not shown). The bone marrow and splenocytes isolated from leukemic mouse no. 137-399 had levels of transgene expression detectable by S1 nuclease protection; in addition to causing a reduction in the levels of endogenous mCG mRNA, ATRA treatment of no. 137-399 splenocytes induced a simultaneous reduction in transgene expression (data not shown). Transgene expression was also detected in the peripheral blood of leukemic mouse no. 135-448 by S1 nuclease protection (data not shown). Thus, leukemic cells continue to express the transgene.
We next wanted to determine the leukemic potential of the immature myeloid cells in the CG–PML-RARα transgenic mice. Four wild-type, nontransgenic C3H SCID mice were injected intraperitoneally with 5 × 107 splenocytes prepared from a transgenic animal with the leukemic phenotype. Two SCID mice were injected with an equal number of splenocytes isolated from a nontransgenic littermate. After 5 weeks, one of the mice injected with splenocytes from the leukemic animal died and the remaining three mice were visibly ill. Autopsies of these mice showed local intraperitoneal infiltration of leukemic myeloid cells and leukemic infiltrates in the mesenteric lymph nodes, livers, and spleens of these animals (data not shown). Promyelocytes were also observed in the bone marrow and peripheral blood of these SCID mice (data not shown). Mice injected with wild-type splenocytes did not develop any evidence of leukemia. In an independent experiment, splenocytes from a second leukemic mouse generated myeloid neoplasms in five of five evaluable SCID mice within 6 weeks of injection. The ability of these spleen preparations to cause tumors in secondary recipients strongly suggests that at least some of the spleen cells are transformed.
Immature myeloid cells derived from the spleens of hCG–PML-RARα mice are sensitive to ATRA. (A and B) S1 nuclease protection assays using total RNA purified from total splenocytes cultured in the presence or absence of 1 μmol/L ATRA as indicated. Positions of probe bands protected by correctly spliced endogenous mCG or β2 microglobulin mRNAs are shown. Levels of mCG transcripts from nonleukemic (A) or leukemic (B) mice both declined substantially within 24 hours of ATRA treatment. Splenocytes from the nontransgenic littermates of these mice did not have any detectable mCG mRNA at any point during the course of the experiments, as expected. (C and D) Splenocytes from a leukemic mouse (FL no. 135) expressing the hCG–PML-RARα transgene incubated in the absence (C) or presence (D) of 1 μmol/L ATRA for 48 hours. Large clusters of immature myeloid cells (large arrows in [C]) involute in the presence of ATRA (D, large arrow). A macrophage laden with possible nuclear fragments is invading the apoptotic cell cluster (black arrow head). Several cells with fragmented, apoptotic nuclei are seen in the ATRA-treated culture (small arrows).
Immature myeloid cells derived from the spleens of hCG–PML-RARα mice are sensitive to ATRA. (A and B) S1 nuclease protection assays using total RNA purified from total splenocytes cultured in the presence or absence of 1 μmol/L ATRA as indicated. Positions of probe bands protected by correctly spliced endogenous mCG or β2 microglobulin mRNAs are shown. Levels of mCG transcripts from nonleukemic (A) or leukemic (B) mice both declined substantially within 24 hours of ATRA treatment. Splenocytes from the nontransgenic littermates of these mice did not have any detectable mCG mRNA at any point during the course of the experiments, as expected. (C and D) Splenocytes from a leukemic mouse (FL no. 135) expressing the hCG–PML-RARα transgene incubated in the absence (C) or presence (D) of 1 μmol/L ATRA for 48 hours. Large clusters of immature myeloid cells (large arrows in [C]) involute in the presence of ATRA (D, large arrow). A macrophage laden with possible nuclear fragments is invading the apoptotic cell cluster (black arrow head). Several cells with fragmented, apoptotic nuclei are seen in the ATRA-treated culture (small arrows).
ATRA sensitivity of transgenic promyelocytes.Because most APML patients enter complete remission when treated with ATRA,19,20,22 23 we assessed the ATRA responsiveness of myeloid precursors from both nonleukemic and leukemic mice. Splenocytes were cultured in the absence or presence of 1 μmol/L ATRA for a period of 48 hours. RNA was purified from these cells after 0, 1, or 2 days and analyzed using S1 nuclease protection assays (Fig 8A and B). A probe specific for murine β2 microglobulin controlled for RNA quality and content in these experiments (Fig 8A and B). The levels of mCG transcripts are initially very high in both nonleukemic (Fig 8A, lane 3) and leukemic splenocytes (Fig 8B, lane 1), reflecting an increase in the percentage of splenic promyelocytes. In response to ATRA, mCG transcripts declined after 1 day and were virtually undetectable after 2 days (Fig 8A and B). Thus, the promyelocytes present in the spleens of these transgenic mice are sensitive to ATRA.
The decline of mCG transcripts induced by ATRA could be explained by at least two mechanisms: first, ATRA may be inducing terminal differentiation of the myeloid precursors,13-16 resulting in a loss of mCG expression as cells mature33,34; alternatively, ATRA may induce apoptosis of promyelocytes, causing a reduction in the fraction of these cells that normally express mCG. Analysis of untreated splenocytes from a leukemic mouse after 2 days showed large clusters of self-adherent myeloid cells with an undifferentiated morphology (Fig 8C, large arrows). ATRA treatment caused virtually all of these clusters to undergo apoptosis and involute (Fig 8D, large arrow); macrophages invading these clusters contained numerous inclusions that probably represent nuclear fragments. These findings were reproduced in four independent experiments. Levels of c-fgr transcripts, which increase during terminal myeloid differentiation,38 did not change with ATRA treatment (data not shown). Similarly, flow cytometric analysis of cultured splenocytes did not detect a significant change in the fraction of cells expressing Gr-1, a murine myeloid antigen that is also induced upon maturation (data not shown).
DISCUSSION
In this report, we describe the production of transgenic mice containing a chimeric PML-RARα fusion cDNA under the control of sequences that target the expression of this cDNA to promyelocytes. Although levels of transgene expression were low, these mice developed an accumulation of myeloid cells in the marrow, which presumably led to the development of extramedullary hematopoiesis in the spleen. This altered myeloid development increased the susceptibility of these animals to the development of acute myeloid leukemia after a long latent period. This leukemic phenotype could be transferred to SCID mice, suggesting that cells from these transgenic mice are transformed. In vitro, splenic promyelocytes expressing the transgene virtually disappeared after 2 days of incubation with ATRA; these promyelocytes are probably eliminated due to ATRA-induced apoptosis. Our results suggest that the PML-RARα fusion protein plays a direct role in altering the process of myeloid development.
The leukemic phenotype of these transgenic mice has similarities and differences when compared with human patients with APML. Thrombocytopenia and anemia, which are characteristic of APML in humans, were consistently observed in these leukemic mice. Both human APML patients and leukemic mice display low percentages of bone marrow blasts and high percentages of bone marrow promyelocytes. Leukemic cells of human APML patients are sensitive to ATRA both in vitro and in vivo. Likewise, promyelocytes from the spleens of our transgenic mice are sensitive to ATRA in culture. The in vivo ATRA sensitivity of our mice is currently being evaluated. However, the severe leukocytosis and organ infiltration observed in our leukemic mice is not a common characteristic of human APML patients and neither is the full range of myeloid maturation in the bone marrow. These differences suggest that targeted expression of PML-RARα to the early myeloid cells of transgenic mice is not sufficient to fully recapitulate the human APML phenotype.
Two other groups have recently attempted to produce animal models of APML. Altabef et al39 reported that retroviral transfer of PML-RARα can transform hematopoietic progenitors and induce acute leukemias in chickens. The vector used in this system produced a fusion protein, with a portion of the retroviral gag protein fused to the N-terminus of PML-RARα. Bone marrow cells infected in culture exhibited a nuclear staining pattern of the gag–PML-RARα protein similar to that of PML-RARα in APML blasts. When chick embryos were infected with the PML-RARα–containing retrovirus, they developed acute leukemia within 2 weeks of hatching. Early et al40 generated transgenic mice with a PML-RARα cDNA driven by the CD11b promoter. The peripheral blood total leukocyte counts and neutrophil counts of these transgenic mice were indistinguishable from control mice. However, peripheral blood leukocytes from these transgenic mice produced fewer colonies in vitro than control mice, most notably in response to granulocyte-macrophage colony-stimulating factor (GM-CSF ). This defect was extended in vivo by showing that only 48% of transgenic mice recovered from a single sublethal dose of radiation. Lack of survival was associated with leukopenia in the transgenic mice; in addition, ATRA treatment of these irradiated transgenic mice enhanced granulocytic recovery. The results of these studies also show that expression of PML-RARα can alter hematopoiesis.
However, neither of these models recapitulate the APML phenotype observed in humans. The CD11b–PML-RARα transgenic mice were phenotypically normal and did not develop leukemia, even after 2 years of observation.40 In the chicken model, the cells transformed in vitro or in vivo resembled multipotential hematopoietic progenitors, rather than the committed precursors characteristic of the human disease.39 In addition, progenitors were transformed only by gag–PML-RARα fusion proteins that had acquired mutations in two domains of PML; no transformed cells were isolated that contained the unaltered form of PML-RARα.39 These mutations have not yet been detected in patients with APML.39 Furthermore, the transformed chicken progenitors were not sensitive to ATRA, which could be due to the mutations acquired in gag–PML-RARα protein or to differences inherent in the avian system.39
The target cell for transformation in APML is likely to be a committed myeloid progenitor.32 Thus, expression of PML-RARα at an early stage of myeloid development may be crucial for leukemogenesis. The CD11b promoter is activated during the late stages of myeloid development and is capable of directing the expression of reporter genes to other hematopoietic lineages in transgenic mice41,42; this may account for the panleukopenia observed in the irradiated CD11b–PML-RARα transgenic mice. Importantly, expression of PML-RARα in late-stage myeloid cells does not produce leukemia in these mice. Retroviral long terminal repeats (LTRs) were used to drive expression of gag–PML-RARα in transduced chicken bone marrow cells.39 Although uncommitted progenitors are transformed by the mutated gag–PML-RARα proteins, their morphology suggests that they may not be the avian equivalents of the target cells in the human disease. In our model, targeting expression of PML-RARα to promyelocytes via CG regulatory sequences produced mice that had a leukemic phenotype that was similar in many respects to the human disease. Comparison of these models suggests that the stage during myeloid development at which PML-RARα is expressed is critical for producing an APML-like phenotype.
The time period required for the development of acute leukemia among hCG–PML-RARα transgenic mice was highly variable, but disease progression was rapid. The leukemic phenotype could be transferred to SCID recipients, which consistently developed clinical illness and myeloid leukemia in a period of 5 to 6 weeks. These recipients may therefore prove useful for testing potential therapeutic agents. A similar SCID ascites model has recently been developed using the human NB4 cell line,43 which was derived from a patient with APML.
The long latency required for hCG–PML-RARα transgenic mice to develop the leukemic phenotype may reflect the low levels of transgene expression observed in our founder lines. It is possible that increasing expression levels of the transgene may decrease the latency for development of leukemia. We are currently addressing this issue by creating mice homozygous for the hCG–PML-RARα transgene, which doubles the transgene dosage. In addition, we are using knock-in technology to target a single copy of the PML-RARα cDNA to the mCG locus; this approach may yield higher expression levels, because the PML-RARα cDNA will be under control of the endogenous mCG promoter and because integration site effects on transgene expression will be eliminated. Nevertheless, our results prove that even low levels of hCG–PML-RARα transgene expression can have a profound effect on myeloid development.
The long latency associated with the development of leukemia in hCG–PML-RARα transgenic mice suggests that PML-RARα is not sufficient to directly cause acute myeloid leukemia in transgenic mice. Long latencies are also characteristic of many other transgenic mouse models involving the cell-type–specific expression of an oncogene; eg, mice expressing SV40 large T-antigen under the control of the myeloid-specific gp91-phox promoter develop histiocytic malignancies only after 6 to 12 months.44 Similarly, lck-Ttg-1 transgenic mice develop thymic tumors after a latent period of 8 to 12 months.45 Long latencies suggest that secondary genetic events may be required for tumor formation. In any case, the development of the leukemic phenotype in hCG–PML-RARα transgenic mice is entirely transgene dependent.
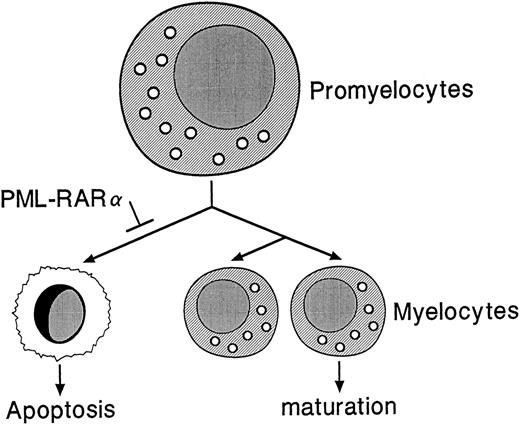
PML-RARα may indirectly contribute to leukemogenesis by causing an accumulation of promyelocytes in the bone marrows and spleens of hCG–PML-RARα transgenic mice, thus increasing the number of myeloid precursors susceptible to secondary mutations. PML-RARα may influence a normal decision that promyelocytes make, ie, a decision to die versus divide and differentiate (Fig 9). PML-RARα may promote the survival of promyelocytes by somehow inhibiting the death pathway. Interestingly, PML-RARα has been shown to confer resistance to apoptosis in U937 clones under conditions of serum deprivation and also to reduce the frequency of commitment to apoptosis upon growth factor deprivation of GM-CSF–dependent TF-1 cells11,46; this model is therefore consistent with the results obtained from our hCG–PML-RARα mice.
Proposed model for the role of PML-RARα in altered myeloid development. PML-RARα expression in promyelocytes may block a normal apoptotic pathway, allowing some promyelocytes to survive and accumulate second hits that could lead to the development of acute leukemia.
Proposed model for the role of PML-RARα in altered myeloid development. PML-RARα expression in promyelocytes may block a normal apoptotic pathway, allowing some promyelocytes to survive and accumulate second hits that could lead to the development of acute leukemia.
The long latency observed in this model may be due to the requirement for a second genetic hit. The t(15,17) translocation results in disruption of single alleles of PML and RARα, which may contribute to the APML phenotype in patients; notably, our hCG–PML-RARα mice have two normal alleles of both PML and RARα. Although RARα has been shown to be involved in ATRA-induced myeloid differentiation, mice deficient in RARα have no apparent defect in myeloid development.47,48 This does not preclude the possibility that PML-RARα may promote leukemogenesis in conjunction with the disruption of one allele of PML, one allele of RARα, or both. In fact, a model whereby the disruption of an allele of PML by the t(15,17) translocation cooperates with expression of PML-RARα to produce APML has been suggested.24 Paggi et al49 have shown that mutations in Rb are more frequently associated with APML than any other form of myeloid leukemia; the Rb status of these leukemic mice has not yet been examined. Finally, it is not known whether the expression of the reciprocal RARα-PML fusion gene (found in 70% to 80% of APML patients) may play a role in the pathogenesis of APML.9,10 50 We have generated transgenic mice expressing a RARα-PML cDNA to address this possibility, and we will produce mice coexpressing RARα-PML and PML-RARα. Clearly, the identification of secondary mutations that promote leukemogenesis in the hCG–PML-RARα mice will shed new light on the mechanisms of transformation that lead to APML.
ACKNOWLEDGMENT
The authors thank Pam Goda for excellent care of these animals and members of the Ley laboratory for their critical comments. Nancy Reidelberger provided expert assistance with the preparation of this manuscript.
Supported by National Institutes of Health Grant No. CA49712 (to T.J.L.).
Address reprint requests to Timothy J. Ley, MD, Washington University Medical School, Departments of Internal Medicine and Genetics, Division of Bone Marrow Transplantation and Stem Cell Biology, 660 S Euclid, Box 8007, St Louis, MO 63110.








![Fig. 8. Immature myeloid cells derived from the spleens of hCG–PML-RARα mice are sensitive to ATRA. (A and B) S1 nuclease protection assays using total RNA purified from total splenocytes cultured in the presence or absence of 1 μmol/L ATRA as indicated. Positions of probe bands protected by correctly spliced endogenous mCG or β2 microglobulin mRNAs are shown. Levels of mCG transcripts from nonleukemic (A) or leukemic (B) mice both declined substantially within 24 hours of ATRA treatment. Splenocytes from the nontransgenic littermates of these mice did not have any detectable mCG mRNA at any point during the course of the experiments, as expected. (C and D) Splenocytes from a leukemic mouse (FL no. 135) expressing the hCG–PML-RARα transgene incubated in the absence (C) or presence (D) of 1 μmol/L ATRA for 48 hours. Large clusters of immature myeloid cells (large arrows in [C]) involute in the presence of ATRA (D, large arrow). A macrophage laden with possible nuclear fragments is invading the apoptotic cell cluster (black arrow head). Several cells with fragmented, apoptotic nuclei are seen in the ATRA-treated culture (small arrows).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/2/10.1182_blood.v89.2.376/4/m_bl_0048f8c.jpeg?Expires=1763486178&Signature=zQaXEJgoAE1T4nF5jB6I5KDNW9Kn6FhZi4Kb9qLVG-QJLDzq9YUWSp6GHqd17vX~FF1TkKeeQ6DLMN8LQxRyM1AiOrbGvnKphql-vqSMjCP7K4Lc6HESMTY0weDdcarysLq4Y76SpVlf6eEq2cdoXBOs8TC-UsT8xos8tmx7V7vVhZz3VnVLP2HPw-sPkZcY0ABGAQO3n78SVS94bdBJyak45j1XvnuKt49uKEQrDur0-9OlugcUqOPsr0uo1E0UuTMASBTI7Xfee6isW4C4s8wowx9O~S6rRl0Nq7ShLY~kBWEQINV96K1NLuLA2MhY5JsSYFvmbKsiJx8lYIe9xA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal